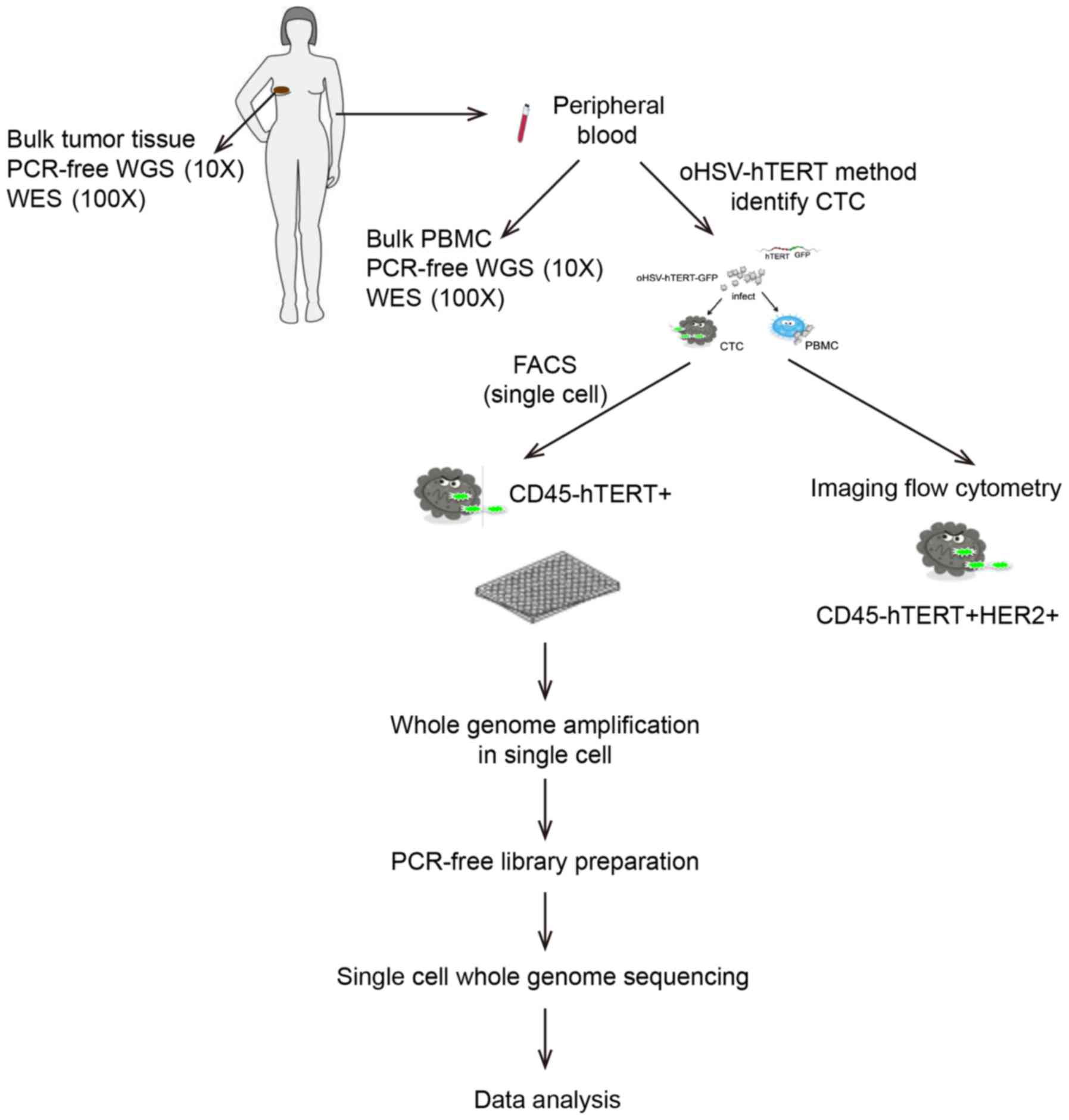
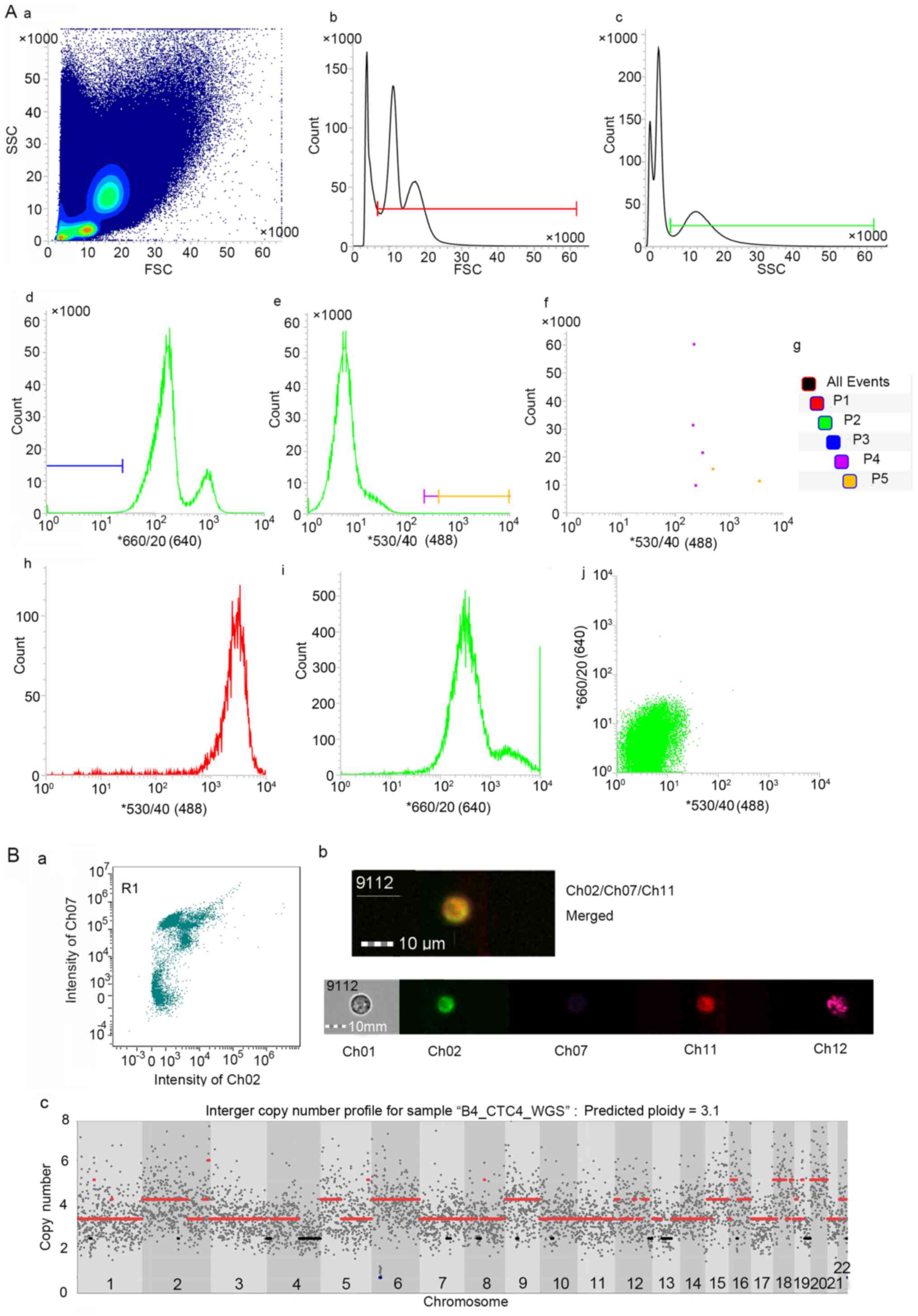
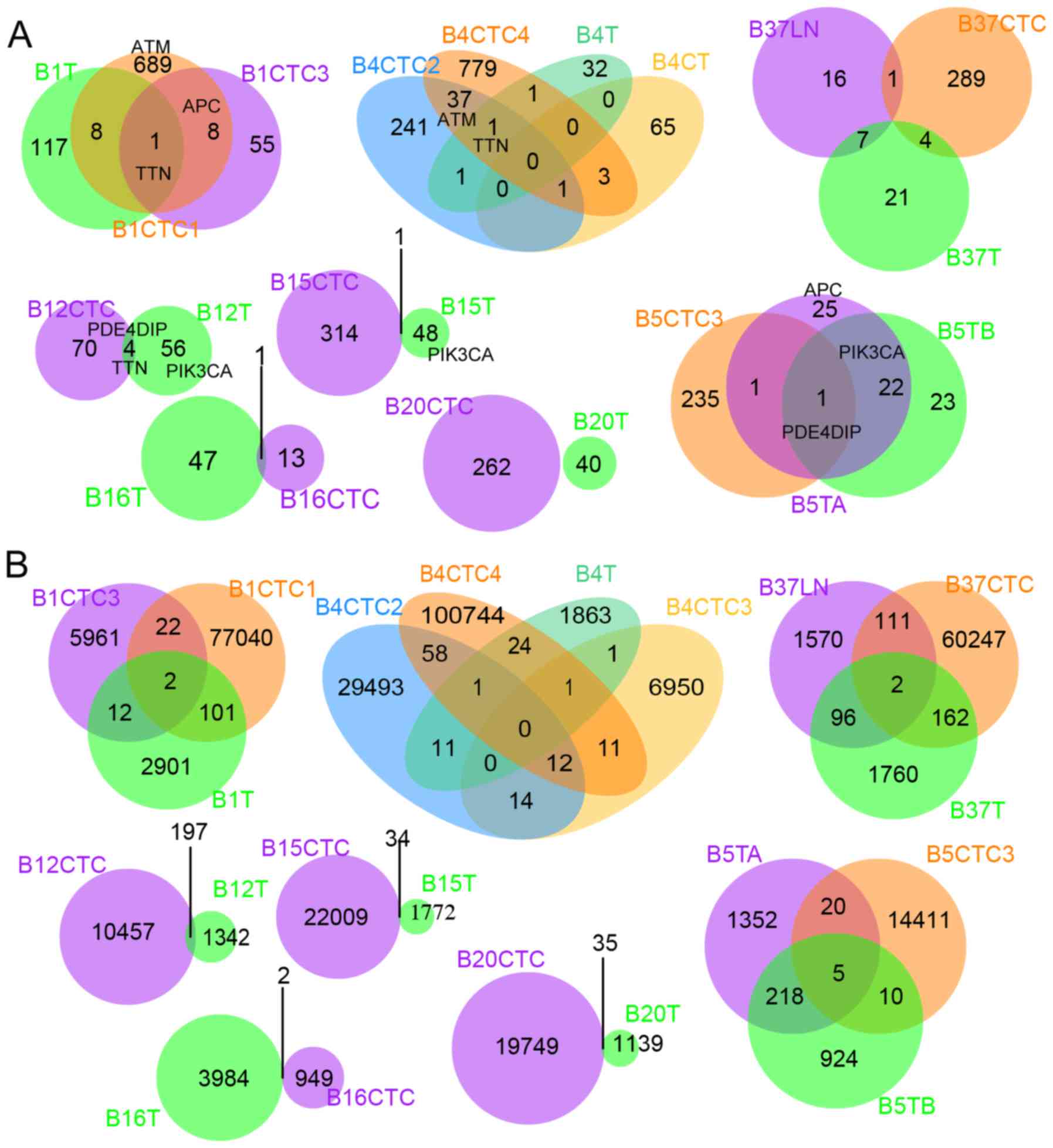
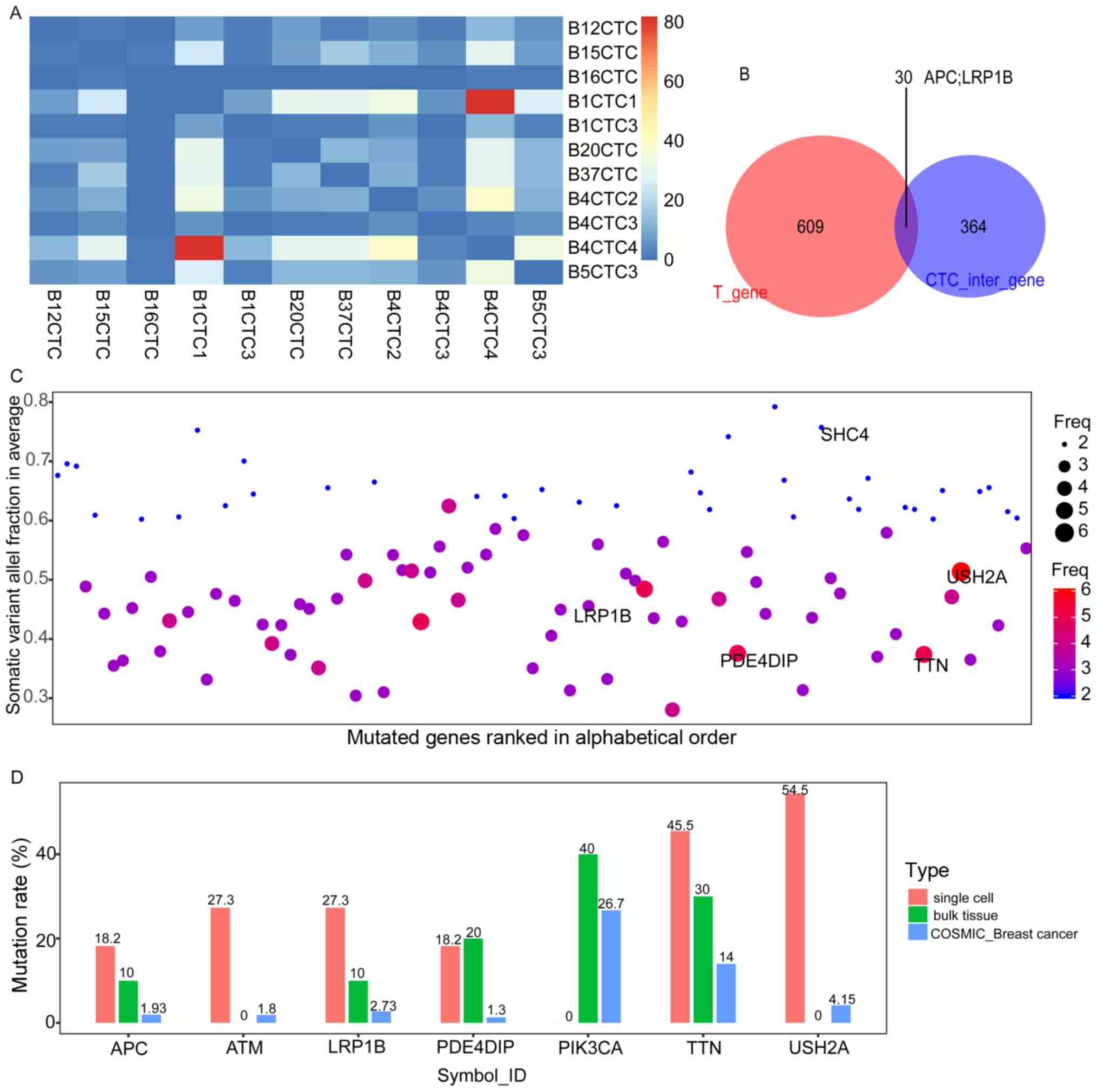
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Giuliano M, Giordano A, Jackson S, De
Giorgi U, Mego M, Cohen EN, Gao H, Anfossi S, Handy BC, Ueno NT, et
al: Circulating tumor cells as early predictors of metastatic
spread in breast cancer patients with limited metastatic
dissemination. Breast Cancer Res. 16:4402014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Magbanua MJ, Carey LA, DeLuca A, Hwang J,
Scott JH, Rimawi MF, Mayer EL, Marcom PK, Liu MC, Esteva FJ, et al:
Circulating tumor cell analysis in metastatic triple-negative
breast cancers. Clin Cancer Res. 21:1098–1105. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Klein CA: Selection and adaptation during
metastatic cancer progression. Nature. 501:365–372. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Deng G, Krishnakumar S, Powell AA, Zhang
H, Mindrinos MN, Telli ML, Davis RW and Jeffrey SS: Single cell
mutational analysis of PIK3CA in circulating tumor cells and
metastases in breast cancer reveals heterogeneity, discordance, and
mutation persistence in cultured disseminated tumor cells from bone
marrow. Bmc Cancer. 14:4562014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu M, Bardia A, Wittner BS, Stott SL, Smas
ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, et al:
Circulating breast tumor cells exhibit dynamic changes in
epithelial and mesenchymal composition. Science. 339:580–584. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Feng T, Wang Y, Seiler M and Hu Z:
Functional characterization of breast cancer using pathway
profiles. Bmc Med Genomics. 7:452014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Polzer B, Medoro G, Pasch S, Fontana F,
Zorzino L, Pestka A, Andergassen U, Meier-Stiegen F, Czyz ZT,
Alberter B, et al: Molecular profiling of single circulating tumor
cells with diagnostic intention. EMBO Mol Med. 6:1371–1386. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang W, Bao L, Yang S, Qian Z, Dong M,
Yin L, Zhao Q, Ge K, Deng Z, Zhang J, et al: Tumor-selective
replication herpes simplex virus-based technology significantly
improves clinical detection and prognostication of viable
circulating tumor cells. Oncotarget. 7:39768–39783. 2016.PubMed/NCBI
|
|
10
|
Zong C, Lu S, Chapman AR and Xie XS:
Genome-wide detection of single-nucleotide and copy-number
variations of a single human cell. Science. 338:1622–1626. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
de Bourcy CF, De Vlaminck I, Kanbar JN,
Wang J, Gawad C and Quake SR: A quantitative comparison of
single-cell whole genome amplification methods. PLoS One.
9:e1055852014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Martin M: Cutadapt removes adapter
sequences from high-throughput sequencing reads. Embnet J.
17:10–12. 2011. View Article : Google Scholar
|
|
13
|
Li H and Durbin R: Fast and accurate short
read alignment with burrows-wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The genome analysis Toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Koboldt DC, Zhang Q, Larson DE, Shen D,
McLellan MD, Lin L, Miller CA, Mardis ER, Ding L and Wilson RK:
VarScan 2: Somatic mutation and copy number alteration discovery in
cancer by exome sequencing. Genome Res. 22:568–576. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Roth A, Ding J, Morin R, Crisan A, Ha G,
Giuliany R, Bashashati A, Hirst M, Turashvili G, Oloumi A, et al:
JointSNVMix: A probabilistic model for accurate detection of
somatic mutations in normal/tumour paired next-generation
sequencing data. Bioinformatics. 28:907–913. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ramos AH, Lichtenstein L, Gupta M,
Lawrence MS, Pugh TJ, Saksena G, Meyerson M and Getz G: Oncotator:
Cancer variant annotation tool. Hum Mutat. 36:E2423–E2429. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Silva GO, He X, Parker JS, Gatza ML, Carey
LA, Hou JP, Moulder SL, Marcom PK, Ma J, Rosen JM and Perou CM:
Cross-species DNA copy number analyses identifies multiple 1q21-q23
subtype-specific driver genes for breast cancer. Breast Cancer Res
Treat. 152:347–356. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gulbahce N, Magbanua MJM, Chin R, Agarwal
MR, Luo X, Liu J, Hayden DM, Mao Q, Ciotlos S, Li Z, et al:
Quantitative whole genome sequencing of circulating tumor cells
enables personalized combination therapy of metastatic cancer.
Cancer Res. 77:4530–4541. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Palmisano GL, Pistillo MP, Capanni P, Pera
C, Nicolò G, Salvi S, Perdelli L, Pasciucco G and Ferrara GB:
Investigation of HLA class I downregulation in breast cancer by
RT-PCR. Hum Immunol. 62:133–139. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kaneko K, Ishigami S, Kijima Y, Funasako
Y, Hirata M, Okumura H, Shinchi H, Koriyama C, Ueno S, Yoshinaka H
and Natsugoe S: Clinical implication of HLA class I expression in
breast cancer. BMC Cancer. 11:4542011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu X, Hou Y, Yin X, Bao L, Tang A, Song L,
Li F, Tsang S, Wu K, Wu H, et al: Single-cell exome sequencing
reveals single-nucleotide mutation characteristics of a kidney
tumor. Cell. 148:886–895. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ni X, Zhuo M, Su Z, Duan J, Gao Y, Wang Z,
Zong C, Bai H, Chapman AR, Zhao J, et al: Reproducible copy number
variation patterns among single circulating tumor cells of lung
cancer patients. Proc Natl Acad Sci USA. 110:21083–21088. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y, Waters J, Leung ML, Unruh A, Roh
W, Shi X, Chen K, Scheet P, Vattathil S, Liang H, et al: Clonal
evolution in breast cancer revealed by single nucleus genome
sequencing. Nature. 512:155–160. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gao Y, Ni X, Guo H, Su Z, Ba Y, Tong Z,
Guo Z, Yao X, Chen X, Yin J, et al: Single-cell sequencing
deciphers a convergent evolution of copy number alterations from
primary to circulating tumour cells. Genome Res. 27:1312–1322.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim N, Hong Y, Kwon D and Yoon S: Somatic
mutaome profile in human cancer tissues. Genomics Inform.
11:239–244. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Forbes SA, Bindal N, Bamford S, Cole C,
Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al:
COSMIC: Mining complete cancer genomes in the catalogue of somatic
mutations in cancer. Nucleic Acids Res. 39:D945–D950. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ross CA: The Trophoblast Model of Cancer.
Nutr Cancer. 67:61–67. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Alix-Panabieres C, Cayrefourcq L, Mazard
T, Maudelonde T, Assenat E and Assou S: Molecular portrait of
metastasis-competent circulating tumor cells in colon cancer
reveals the crucial role of genes regulating energy metabolism and
DNA repair. Clin Chem. 63:700–713. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fnu S, Williamson EA, De Haro LP,
Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff
JA and Hromas R: Methylation of histone H3 lysine 36 enhances DNA
repair by nonhomologous end-joining. Proc Natl Acad Sci USA.
108:540–545. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang J, Wang HT and Li BG: Prognostic
significance of circulating tumor cells in small-cell lung cancer
patients: A meta-analysis. Asian Pac J Cancer Prev. 15:8429–8433.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Goldkorn A, Ely B, Quinn DI, Tangen CM,
Fink LM, Xu T, Twardowski P, Van Veldhuizen PJ, Agarwal N, Carducci
MA, et al: Circulating tumor cell counts are prognostic of overall
survival in SWOG S0421: A phase III trial of docetaxel with or
without atrasentan for metastatic castration-resistant prostate
cancer. J Clin Oncol. 32:1136–1142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Romiti A, Raffa S, Di Rocco R, Roberto M,
Milano A, Zullo A, Leone L, Ranieri D, Mazzetta F, Medda E, et al:
Circulating tumor cells count predicts survival in colorectal
cancer patients. J Gastrointestin Liver Dis. 23:279–284.
2014.PubMed/NCBI
|
|
36
|
Lohr JG, Adalsteinsson VA, Cibulskis K,
Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ,
Shalek AK, Satija R, et al: Whole-exome sequencing of circulating
tumor cells provides a window into metastatic prostate cancer. Nat
Biotechnol. 32:479–484. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Weissenstein U, Schumann A, Reif M, Link
S, Toffol-Schmidt UD and Heusser P: Detection of circulating tumor
cells in blood of metastatic breast cancer patients using a
combination of cytokeratin and EpCAM antibodies. BMC Cancer.
12:2062012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Papadaki MA, Kallergi G, Zafeiriou Z,
Manouras L, Theodoropoulos PA, Mavroudis D, Georgoulias V and
Agelaki S: Co-expression of putative stemness and
epithelial-to-mesenchymal transition markers on single circulating
tumour cells from patients with early and metastatic breast cancer.
BMC Cancer. 14:6512014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kanwar N, Hu P, Bedard P, Clemons M,
McCready D and Done SJ: Identification of genomic signatures in
circulating tumor cells from breast cancer. Int J Cancer.
137:332–344. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Meng S, Tripathy D, Frenkel EP, Shete S,
Naftalis EZ, Huth JF, Beitsch PD, Leitch M, Hoover S, Euhus D, et
al: Circulating tumor cells in patients with breast cancer
dormancy. Clin Cancer Res. 10:8152–8162. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Aktas B, Tewes M, Fehm T, Hauch S, Kimmig
R and Kasimir-Bauer S: Stem cell and epithelial-mesenchymal
transition markers are frequently overexpressed in circulating
tumor cells of metastatic breast cancer patients. Breast Cancer
Res. 11:R462009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bingham C, Fernandez SV, Fittipaldi P,
Dempsey PW, Ruth KJ, Cristofanilli M and Alpaugh Katherine R:
Mutational studies on single circulating tumor cells isolated from
the blood of inflammatory breast cancer patients. Breast Cancer Res
Treat. 163:219–230. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mu Z, Benali-Furet N, Uzan G, Znaty A, Ye
Z, Paolillo C, Wang C, Austin L, Rossi G, Fortina P, et al:
Detection and characterization of circulating tumor associated
cells in metastatic breast cancer. Int J Mol Sci. 17:E16652016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fagiani E, Giardina G, Luzi L, Cesaroni M,
Quarto M, Capra M, Germano G, Bono M, Capillo M, Pelicci P and
Lanfrancone L: RaLP, a new member of the Src homology and collagen
family, regulates cell migration and tumor growth of metastatic
melanomas. Cancer Res. 67:3064–3073. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Turco MY, Furia L, Dietze A, Diaz
Fernandez L, Ronzoni S, Sciullo A, Simeone A, Constam D, Faretta M
and Lanfrancone L: Cellular heterogeneity during embryonic stem
cell differentiation to epiblast stem cells is revealed by the
ShcD/RaLP adaptor protein. Stem Cells. 30:2423–2436. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ting DT, Wittner BS, Ligorio M, Jordan
Vincent N, Shah AM, Miyamoto DT, Aceto N, Bersani F, Brannigan BW,
Xega K, et al: Single-cell RNA sequencing identifies extracellular
matrix gene expression by pancreatic circulating tumor cells. Cell
Rep. 8:1905–1918. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Friedl P and Wolf K: Proteolytic
interstitial cell migration: A five-step process. Cancer Metastasis
Rev. 28:129–135. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
von Nandelstadh P, Gucciardo E, Lohi J, Li
R, Sugiyama N, Carpen O and Lehti K: Actin-associated protein
palladin promotes tumor cell invasion by linking extracellular
matrix degradation to cell cytoskeleton. Mol Biol Cell.
25:2556–2570. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Akalay I, Janji B, Hasmim M, Noman MZ,
André F, De Cremoux P, Bertheau P, Badoual C, Vielh P, Larsen AK,
et al: Epithelial-to-mesenchymal transition and autophagy induction
in breast carcinoma promote escape from T-cell-mediated lysis.
Cancer Res. 73:2418–2427. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Najmeh S, Cools-Lartigue J, Rayes RF,
Gowing S, Vourtzoumis P, Bourdeau F, Giannias B, Berube J, Rousseau
S, Ferri LE and Spicer JD: Neutrophil extracellular traps sequester
circulating tumor cells via β1-integrin mediated interactions. Int
J Cancer. 140:2321–2330. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Demers M, Krause DS, Schatzberg D,
Martinod K, Voorhees JR, Fuchs TA, Scadden DT and Wagner DD: Cancer
predispose neutrophils to release extracellular DNA traps that
contribute to cancer-associated thrombosis. Proc Natl Acad Sci USA.
109:13076–13081. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Goubran HA, Burnouf T, Radosevic M and
El-Ekiaby M: The platelet-cancer loop. Eur J Intern Med.
24:393–400. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hulka BS and Stark AT: Breast cancer:
Cause and prevention. Lancet. 346:883–887. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
May P, Woldt E, Matz RL and Boucher P: The
LDL receptor-related protein (LRP) family: An old family of
proteins with new physiological functions. Ann Med. 39:219–228.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Cowin PA, George J, Fereday S, Loehrer E,
Van Loo P, Cullinane C, Etemadmoghadam D, Ftouni S, Galletta L,
Anglesio MS, et al: LRP1B deletion in high-grade serous ovarian
cancers is associated with acquired chemotherapy resistance to
liposomal doxorubicin. Cancer Res. 72:4060–4073. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Magbanua MJ and Park JW: Isolation of
circulating tumor cells by immunomagnetic enrichment and
fluorescence-activated cell sorting (IE/FACS) for molecular
profiling. Methods. 64:114–118. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Andersen MN, Al-Karradi SN, Kragstrup TW
and Hokland M: Elimination of erroneous results in flow cytometry
caused by antibody binding to Fc receptors on human monocytes and
macrophages. Cytometry A. 89:1001–1009. 2016. View Article : Google Scholar : PubMed/NCBI
|