Introduction
Circulating tumor cells (CTCs) have been identified
in the blood and bone marrow of patients with breast, prostate and
colon cancers (1–3) in as low as 1/100 million or 1 billion
blood cells. Molecular characterization of CTCs may provide a
greater understanding of the disease metastases, identify
aggressive tumors and enable therapeutic selection and monitoring
of the disease for patients undergoing treatment (4,5). A
variety of technologies have been developed to improve detection
and capture of CTCs from peripheral blood, which include
immune-magnetic bead separation using monoclonal antibodies
targeting cell-surface antigens for positive or negative selection,
cell sorting using flow cytometry, filtration-based size
separation, density gradient centrifugation, microfluidic devices
and fast-scan imaging (6–10). For example, CellSearch™ was the
first CTC technology that demonstrated its clinical validity in
predicting progression-free and overall survival of metastatic
cancer patients based on CTC enumeration (3–6).
It is of great interest to go beyond cell
enumeration and further characterize the CTCs by assessing
clinically relevant molecular markers on or within CTCs to gain
insight into the mechanisms of metastasis and best treatment
modalities for patients (1–3,11,12).
For example, significant progress has been made in breast cancer,
including effective hormonal therapy, chemotherapy and targeted
therapies against estrogen receptor (ER) and HER-2. In prostate
cancer, androgen receptor (AR) variant 7 has been implicated in
predicting response to targeted therapies on AR. Established
clinical, pathological features and biomarker status are routinely
used to guide treatment options. It has become critically important
to determine which patients are most likely to benefit from
specific therapies. Detecting such molecular markers using a
minimally-invasive blood test for CTCs has great potential in
clinical practice to guide therapy choice for patients. However,
despite advances in CTC technologies, the low frequency of CTCs in
cancer patients and the extensive background leukocytes have
limited the synergism of biomarkers and CTC technologies (11,12).
We have developed a novel microfluidic device,
Celsee PREP100 that uses a size and deformability-based capturing
mechanism of CTCs (13). The
microfluidic chip has a parallel network of fluidic channels which
contain about 56,000 capture chambers (13,14).
The chip fabrication begins with a silicon master device containing
micro-features that make up a fluidic network (75-µm deep), leading
to individual cell trapping chambers (20×25×30 µm) with a pore size
of 10×8 µm. Each chamber ensures smaller blood cells such as red
blood cells and most of the leukocytes escape while larger cancer
cells get trapped and isolated in the chamber. The manufacturing
process uses standard photo-lithography and deep reactive ion
etching for micro-fabrication. From the master device, a soft
elastomeric negative mold is created by pouring and curing against
the silicon master. The final micro-substrate is created by hot
embossing a plastic plate made of cyclic olefin polymer (COP)
against the elastomeric negative mold. A thin plastic laminate
containing pressure-sensitive adhesive is then laminated against
the COP micro-substrate to create the final microfluidic chip. The
chip is placed on the Celsee PREP100 device for CTC capturing.
Since the device captures cells using a label-free
mechanism, it provides an improved sensitivity in capturing CTCs
and an open platform for investigators to use a variety of
antibodies to identify and characterize CTCs upon capturing
(13,14). In a previous study, we compared CTC
enumeration between the Celsee system and the FDA-cleared
CellSearch system using blood samples from patients with metastatic
prostate cancer. CTC counts were significantly higher using the
Celsee system (14). The captured
CTCs could also be retrieved reproducibly using a back-flow
procedure from the microfluidic chip for further nucleic acid
extraction and molecular analysis. In the present study, we report
the development of a novel protocol for capturing cells using blood
sample retrieval and analysis of the cells using PCR amplicon
sequencing. Using this method, we evaluated the potential of
applying Celsee PREP100 in CTC molecular analysis by analyzing
p.K139fs*3 of TP53 and p.T877A of AR in captured prostate cancer
cell lines PC3 and LNCaP and in captured spiked-in cells in normal
donor blood. The method was also tested successfully in clinical
blood samples from 11 patients with metastatic prostate cancer
successfully. Our results demonstrated the potential of utilizing
CTCs for diagnosis, prognosis and treatment selection of patients
with metastatic malignancy.
Materials and methods
Materials
Prostate cancer cell lines PC3 and LNCaP were
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA) and cultured in Dulbecco's modified Eagle's
medium (DMEM) containing 5% fetal bovine serum (FBS) in a
humidified incubator supplemented with 5% CO2. Upon
confluency, cells were digested with 0.5% trypsin and passaged at a
1:4 ratio. Some cells were resuspended in culture medium, counted
and used for experiments. Blood samples were obtained from healthy
donors. Clinical blood samples from patients with metastatic
prostate cancer were obtained at Henry Ford Health System (Detroit,
MI, USA). The present study was approved by the Institutional
Review Board (IRB) of the Henry Ford Health System and written
consent was obtained from all patients. Following informed consent,
blood samples from patients were acquired in 10 ml BCT Cell-Free
DNA tubes (Streck, La Vista, NE, USA) or EDTA-coated
Vacutainer® tubes (BD Biosciences, Franklin Lakes, NJ,
USA).
Removal of CD45-positive cells from
blood samples and CTC enrichment and staining
To test CTC enrichment efficiency using the Celsee
PREP100 instrument, 250 PC3 and 50 LNCaP cells were spiked into 4
ml of blood from healthy donors. CD45-positive cells were removed
using the RosetteSep Human CD45 Depletion Cocktail (Stemcell
Technologies, Inc., Cambridge, MA, USA) following the
manufacturer's protocol. The upper layer of plasma cells was
collected and added into the inlet funnel of the Celsee PREP100.
Cells were enriched in the microfluidic chip and stained for
cytokeratins, CD45 and nuclei using anti-Pan cytokeratin antibody
(1:100 dilution; cat. no. 914204), anti-CD45 antibody (1:100
dilution; cat. no. 368515; both antibodies were from BioLegend,
Inc., San Diego, CA, USA) and DAPI, using Celsee PREP100 CTC
Immunochemistry kit (Celsee Diagnostics, Plymouth, MI, USA). Cells
enriched on the slides were counted using the Celsee Analyzer
(14). Cytokeratins and
DAPI-positive and CD45-negative cells were counted as CTC cells.
The enrichment efficiency was calculated as the percentage of the
enriched cells of the total spiked-in cells.
Cell retrieval using the Celsee
PREP100 instrument
A different amount of PC3 and LNCaP cells were
spiked into the priming buffer and retrieved in 2 ml
phosphate-buffered saline (PBS) using the Celsee PREP100 instrument
(Celsee Diagnostics) following the protocol provided by the
manufacturer. The cells were then concentrated in 10–50 µl by
centrifugation at 500 × g for 10 min, and counted using a
hemocytometer. The retrieval efficiency was calculated as the
percentage of the retrieved cells of the total spiked-in cells.
Retrieval of spiked-in PC3 and LNCaP
cells in normal donor blood samples
A different amount of PC3 and LNCaP cells were
spiked into 4 ml of blood from healthy donors. After removal of the
CD45-positive cells and enrichment of the spiked-in CTCs as
aforementioned, cells enriched in the microfluidic chip were
retrieved in 2 ml PBS using the Celsee PREP100 instrument (Celsee
Diagnostics) following the manufacturer's protocol. These cells
were then collected by centrifugation at 500 × g for 10 min and
stored at −20°C for future analysis by PCR amplicon sequencing.
Capturing and retrieval of CTCs in
clinical blood samples
An aliquot of 4 ml of blood sample was used to
capture or retrieve CTCs using the Celsee PREP100 instrument
(Celsee Diagnostics) following the protocol provided by the
manufacturer. CTCs were monitored using an inverted fluorescence
microscope. CTC enumeration following antibody labeling was
performed manually. PanCK+/CD45− nucleated
cells were identified as CTCs. Positive and negative controls for
antibody performance and staining were included in each experiment.
After removal of CD45-positive cells and enrichment of the
spiked-in CTCs as aforementioned, cells enriched in the
microfluidic chip were retrieved in 2 ml PBS using the Celsee
PREP100 instrument (Celsee Diagnostics) following the
manufacturer's protocol. These cells were then collected by
centrifugation at 500 × g for 10 min and stored at −20°C for future
analysis by PCR amplicon sequencing.
PCR amplicon sequencing
Mutations p.K139fs*3 of TP53 in PC3 and p.T877A of
AR in LNCaP cells have been previously reported (15,16).
To detect these mutations, nested PCR was employed. Tables I and II lists outer and inner primer sets
designed to amplify each of the mutations. The retrieved cells were
resuspended in 5 µl ddH2O and incubated at 4°C for 10
min, and used as the template for PCR amplification. The PCR assay
was set up in a 20-µl reaction containing 10 µl KAPA HiFi HotStart
Ready Mix (Kapa Biosystems, Inc., Wilmington, MA, USA), 300 nM of
each outer primer and 1 µl dimethyl sulfoxide (DMSO) at the
following cycling conditions: 2 min at 95°C, followed in turn by 3
cycles of 20 sec at 98°C, 20 sec at 64°C and 30 sec at 72°C, 3
cycles of 20 sec at 98°C, 20 sec at 61°C and 30 sec at 72°C, 3
cycles of 20 sec at 98°C, 20 sec at 58°C and 30 sec at 72°C, 35
cycles of 20 sec at 98°C, 20 sec at 57°C and 30 sec at 72°C and 10
min at 72°C. One microliter of amplified PCR products with the
outer primer set was used as the template for the PCR reaction
using the inner primer set under the same conditions. The final PCR
products were examined on 1% agarose gel and subjected to Sanger
sequencing using the inner primers after purification.
 | Table I.Primer sequences used for
amplification of fragments containing mutations of interest. |
Table I.
Primer sequences used for
amplification of fragments containing mutations of interest.
| Mutation | p.K139fs*3 | p.T877A |
|---|
| Forward outer
primer |
CTGAGTGACAGAGCAAGACCCTAT |
AAAATCAGAGGTTGGGGAAGA |
| Reverse outer
primer |
AGTGTTTCTGTCATCCAAATACTCC |
ACAACTTGACACTGGGCCATA |
| Forward inner
primer |
GTTTCTTTGCTGCCGTCTTC |
CACCTCCTTGTCAACCCTGT |
| Reverse inner
primer |
ACACGCAAATTTCCTTCCAC |
TGGGAAGCAAAGTCTGAAGG |
 | Table II.Primer sequences used for
amplification of fragments containing mutations of interest in
clinical blood samples. |
Table II.
Primer sequences used for
amplification of fragments containing mutations of interest in
clinical blood samples.
| Mutation | p.T877A |
|---|
| Forward outer
primer |
AAAATCAGAGGTTGGGGAAGA |
| Reverse outer
primer |
ACAACTTGACACTGGGCCATA |
| Forward inner
primer |
CACCTCCTTGTCAACCCTGT |
| Reverse inner
primer |
TGGGAAGCAAAGTCTGAAGG |
| Forward inner
primer |
AGATTGCGAGAGAGCTGCAT |
| Reverse inner
primer |
TGCCATGGGAGGGTTAGATA |
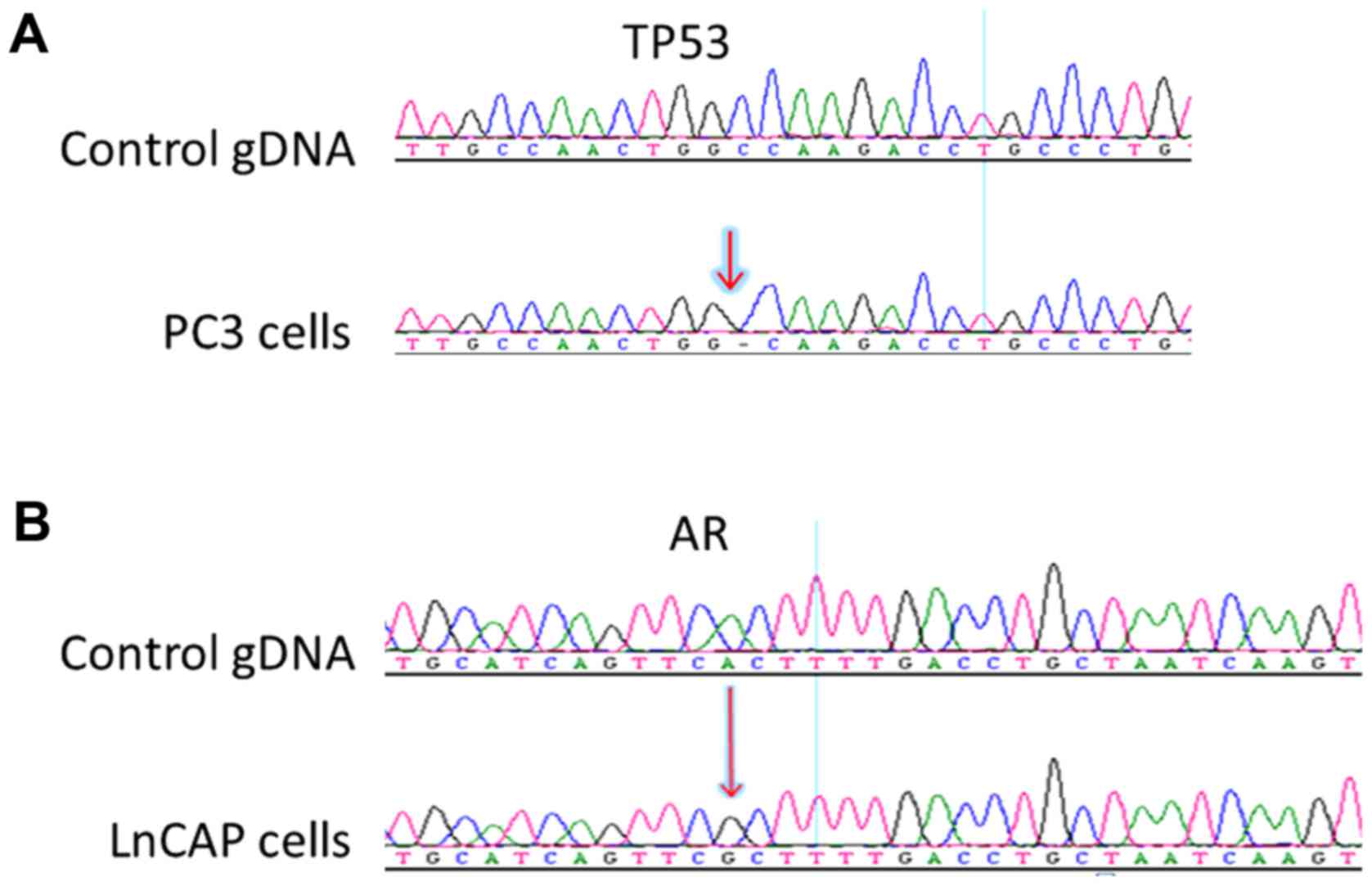
Results
Mutation detection in PC3 and LNCaP
cell lines
PC3 and LNCaP cell lines are established prostate
cancer cells and have been widely used in studies on prostate
cancer. It has been reported that PC3 cells possess a deletion in
the TP53 gene, p.K139fs*3 and LNCaP possess a missense A to G
mutation in the AR gene, p.T887A mutation (15,16).
To detect these mutations, we designed primers (Table I) targeting the mutations, performed
nested PCR and subsequently sequenced the amplicons by Sanger
sequencing. To detect the p.T877A mutation of the AR gene in the
blood samples of patients, a second set of inner primers were used
to improve the sensitivity of the assay (Table II). Fig. 1 reveals the PCR products and
Fig. 2 reveals the Sanger
sequencing results. The p.K139fs*3 and p.T887A mutations were
successfully detected from 28–1,200 PC3 cells, and 10–200 LNCaP
cells, respectively.
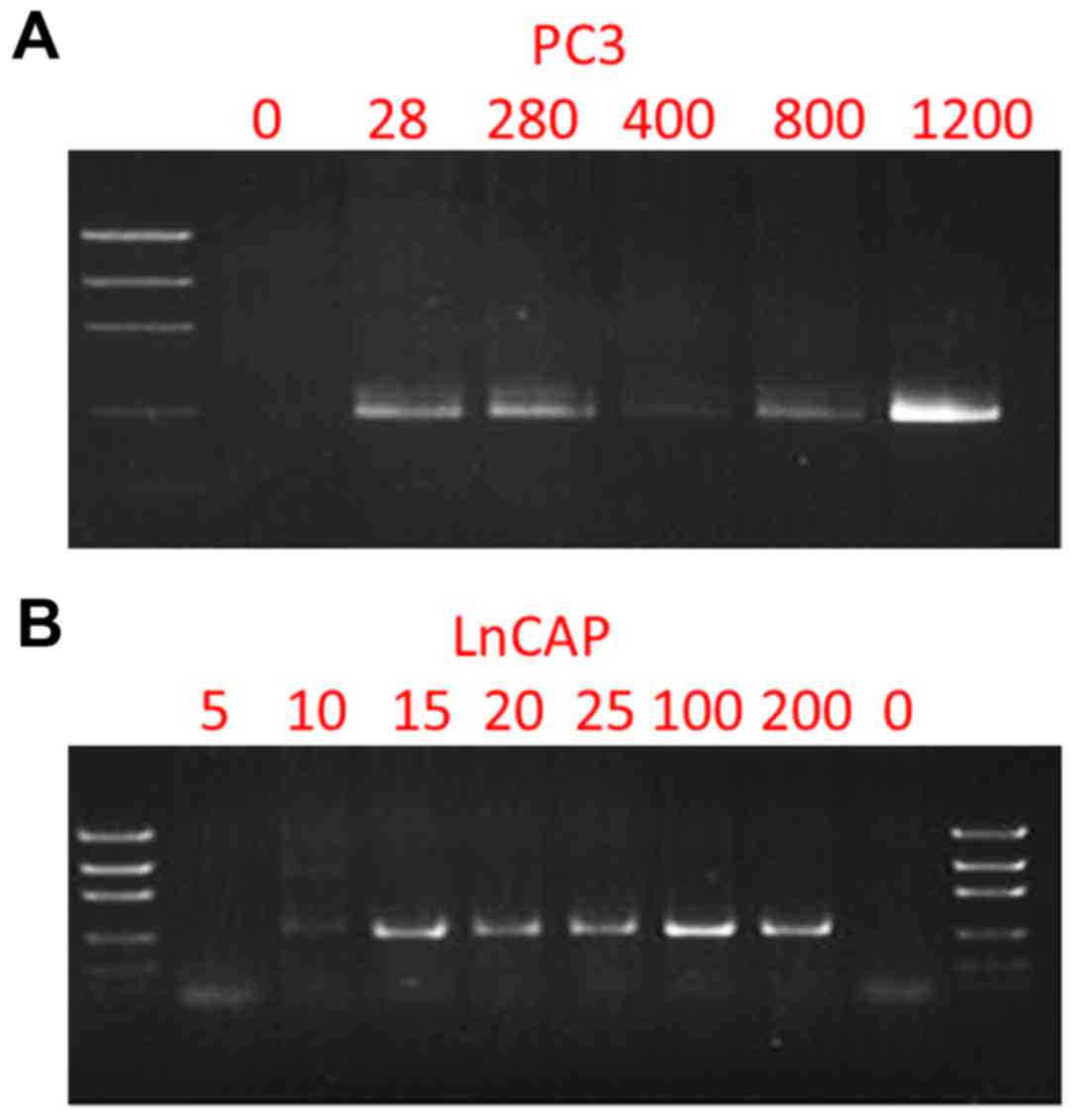 | Figure 1.Nested PCR amplification of the TP53
gene from PC3 cells and the androgen receptor gene from LNCaP
cells. (A) Electrophoresis gel imaging of nested PCR amplicons from
0, 28, 280, 400, 800 and 1,200 PC3 cells. (B) Electrophoresis gel
imaging of nested PCR amplicons from 0, 5, 10, 20, 25, 100 and 200
LNCaP cells. |
Cell retrieval efficiency using Celsee
PREP100
We next tested CTC enrichment efficiency of the
Celsee PREP100 by inputting a total of 110, 220, 330, 440, 550, 880
and 1,100 PC3 cells, and 60, 300, 600, 900, 1,200 and 1,500 LNCaP
cells, respectively. The results are shown in Table III. Overall, the efficiency for
cell retrieval was ~70% with a range from 40 to 121% for PC3 and
LNCaP cells. The observation that some of the cell recovery rates
were >100% was due to the variation of cell counts by
hemocytometer at low cell numbers.
 | Table III.Efficiency of CTC retrieval using
Celsee PREP100. |
Table III.
Efficiency of CTC retrieval using
Celsee PREP100.
| Total input PC3 (no.
of cells) | 110 | 220 | 330 | 440 | 550 | 880 | 1,100 |
| Retrieved PC3 (no. of
cells) | 128 | 150 | 208 | 531 | 360 | 368 | 750 |
| Recovery rate
(%) | 116 | 68 | 63 | 121 | 65 | 42 | 68 |
| Total input LNCaP
(no. of cells) | 60 | 300 | 600 | 900 | 1,200 | 1,200 | 1,500 |
| Retrieved LNCaP (no.
of cells) | 58 | 250 | 618 | 605 | 1,233 | 810 | 600 |
| Recovery rate
(%) | 96 | 83 | 103 | 67 | 103 | 68 | 40 |
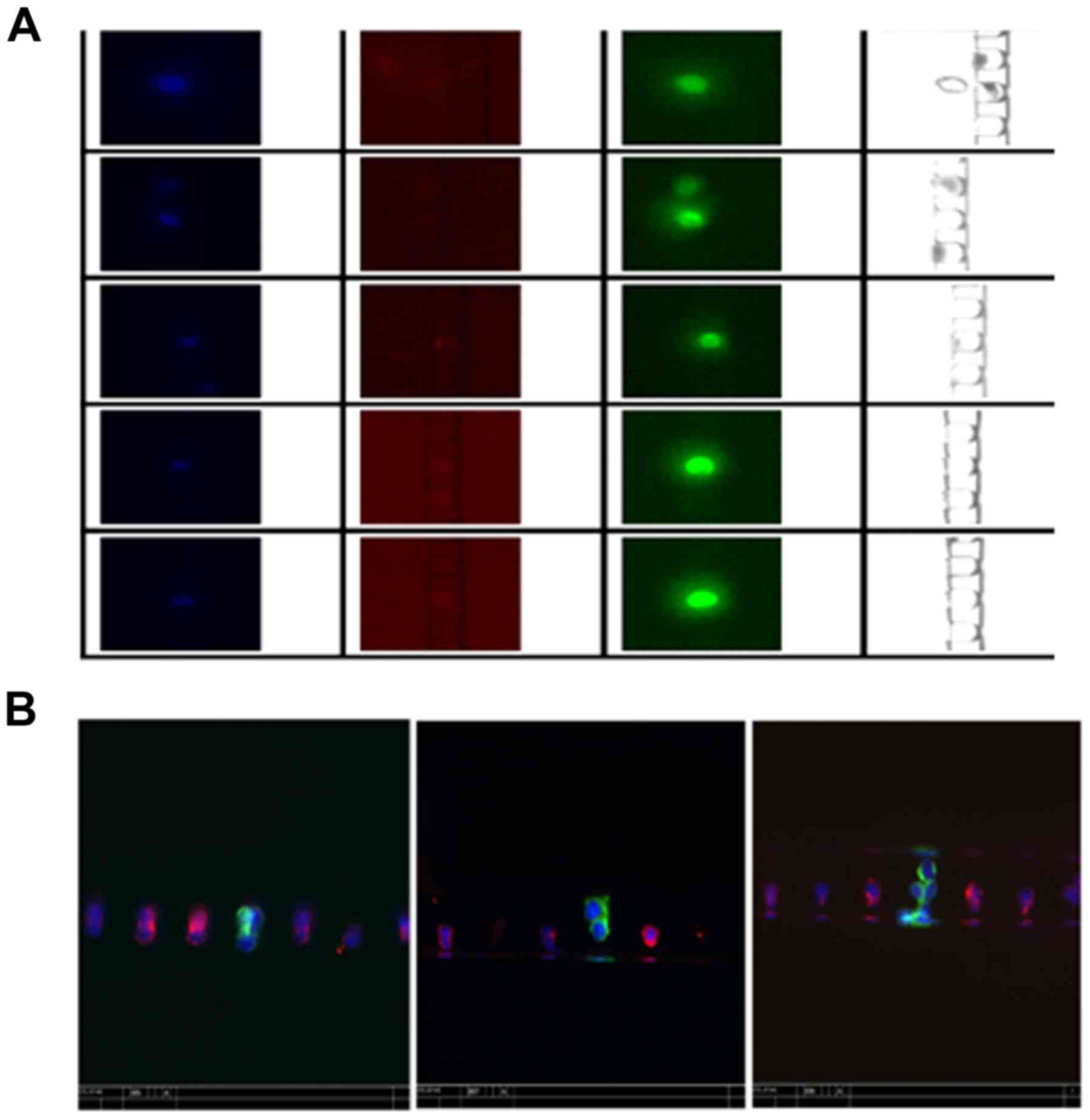
CTC enrichment and retrieval
efficiencies using spiked-in cells in normal donor blood
samples
To test the enrichment and retrieval efficiencies of
CTC in blood samples, we spiked PC3 and LNCaP into 4 ml of whole
blood samples from healthy donors and stained the enriched cells
for cytokeratins, CD45 and nuclei. Fig.
3 reveals typical images of the enriched cells in the
microfluidic chip, where cytokeratins and DAPI-positive (green and
blue) and CD45-negative cells were considered as CTC cells. The
results (Table IV) reveals that
~40% of 250 spiked-in PC3 cells and 74% of 50 spiked-in LNCaP cells
were retrieved after enrichment. Furthermore, the enriched PC3 and
LNCaP cells accounted for ~40 and 25% of the total retrieved cells
and the ratio of captured CTCs to the background cells reached
1:1.5 for PC3 and 1:2.9 for LNCaP cells, suggesting that the
removal of blood cells by the Celsee PREP100 was nearly complete
and the level of remaining leukocytes in the enriched sample was
very low.
 | Table IV.Efficiency of CTC enrichment from
blood samples using Celsee PREP100. |
Table IV.
Efficiency of CTC enrichment from
blood samples using Celsee PREP100.
| Cell types | PC3 | LNCaP |
|---|
| No. of spiked-in
CTC | 250 | 50 |
| No. of total captured
cells | 240 | 146 |
| No. of captured CTC
cells | 96 | 37 |
| No. of background
cells | 144 | 109 |
| CTC recovery rate
(%) | 38 | 74 |
| CTCs vs. background
cells (ratio) | 1:1.50 | 1:2.90 |
CTC enrichment and retrieval using
clinical blood samples
To test the enrichment and retrieval efficiencies of
CTC the blood samples of patients, we stained the enriched cells
for cytokeratins, CD45 and nuclei. Fig.
3 reveals typical images of the enriched cells in the
microfluidic chip, where cytokeratins and DAPI-positive (green and
blue) and CD45-negative cells were considered as CTC cells. For the
patient samples that had CTCs, we processed another 4 ml of the
blood samples and retrieved CTCs for subsequent mutation
analysis.
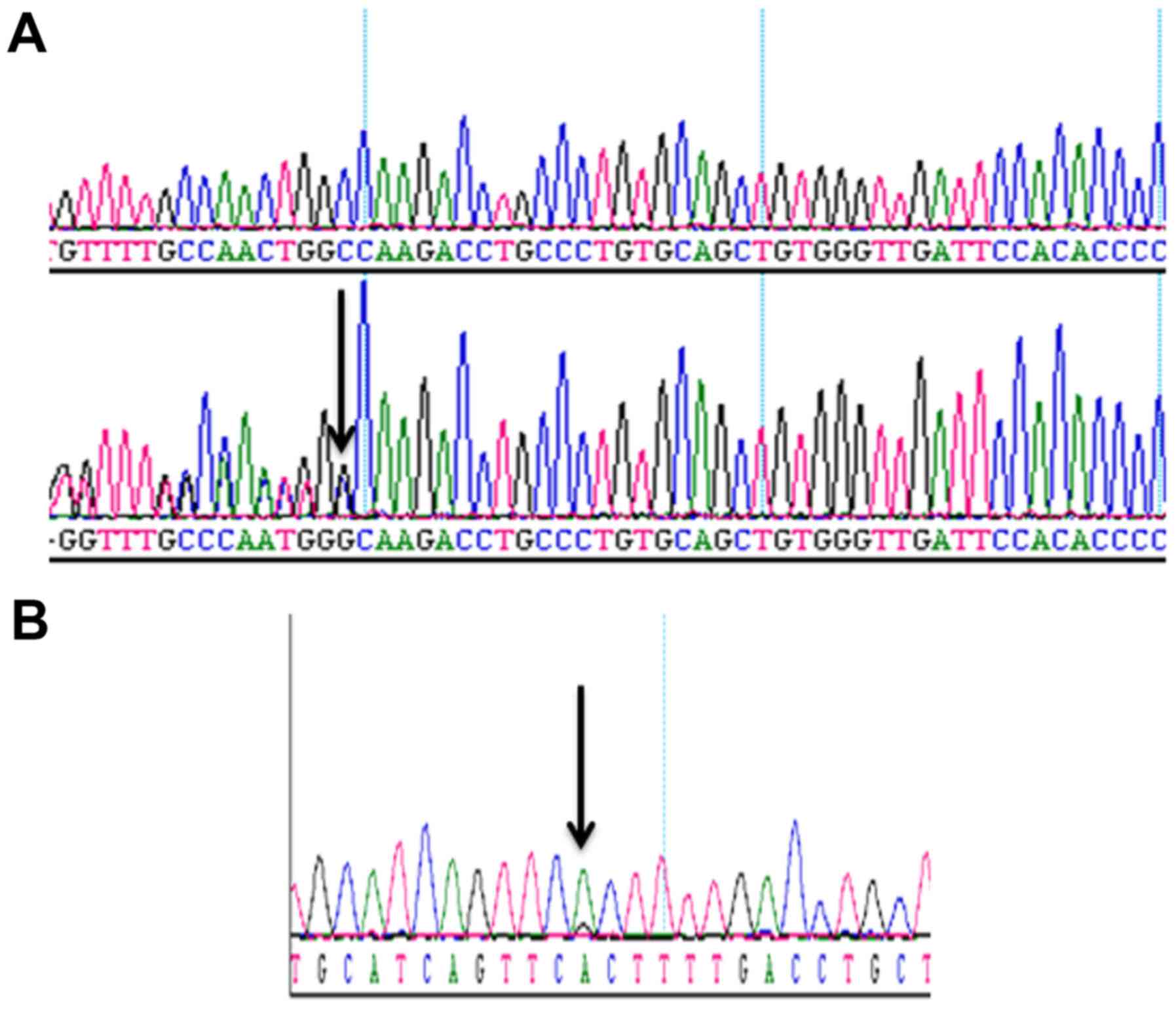
Mutation analysis
In order to test whether or not the mutations
p.K139fs*3 of TP53 in PC3 and p.T877A of AR in LNCaP cells could be
detected in the enriched cells from blood samples, we spiked 50,
100, 300 and 1,000 PC3 cells, and 25, 50, 100 and 250 LNCaP cells
into 4 ml of blood samples, depleted CD45-positive cells, enriched
and retrieved the cancer cells using Celsee PREP100. The cells were
then spun down, resuspended in water and used as templates for PCR
amplification and subsequent Sanger sequencing. Fig. 4 reveals the PCR products. Fig. 5 shows the Sanger sequencing results
of the PCR amplicons, indicating that both mutations could be
successfully detected in the enriched cells. For the CTCs retrieved
from 14 clinical blood samples, we successfully performed the
mutation analysis of p.T877A. The AR mutation (heterozygous
mutation) was identified in the 1 sample (CTC 37) tested (Fig. 6) and all other samples were negative
for this mutation. Sixty-three CTCs were identified in sample CTC
37.
Discussion
Molecular characterization of CTCs has been hindered
by low sensitivity and a high level of background leukocytes of
currently available CTC enrichment technologies. We demonstrated
that CTCs can be readily captured and further characterized with
molecular markers using a simple device, Celsee PREP100. The
protocol we report in the present study enriches and retrieves CTCs
from blood samples based on the fact that tumor cells are larger
and less deformable than normal blood cells. To evaluate the
performance on cell enrichment and retrieval, we prepared prostate
cancer cell lines PC3 and LNCaP and analyzed the captured cells by
PCR amplicon sequencing. We were able to recover an average of 79%
(ranging from 40 to 100%) of 110–1,100 PC3 cells, and 60–1,500
LNCaP cells and detect the p.K139fs*3 deletion of TP53 in PC3 cells
and the p.T877A mutation of AR in LNCaP cells. We also tested these
two types of cells spiked into normal donor blood samples, captured
and retrieved the cells and analyzed the retrieved cells by PCR
amplicon sequencing. We were able to capture ~40% of PC3 cells and
74% of LNCaP cells with the ratio of captured CTCs to the
background leukocytes reaching 1:1.5 for PC3 and 1:2.9 for LNCaP
cells. The p.K139fs*3 deletion and the p.T877A mutation were
detected in the captured spiked-in PC3 and LNCaP cells,
respectively. The method was also tested successfully in clinical
blood samples from patients with metastatic prostate cancer. Our
results demonstrated the potential of CTC molecular
characterization for the diagnosis, prognosis and treatment
selection of patients with metastatic malignancy.
The unique design of the microfluidic chip and
Celsee PREP100 allows separation of CTCs from the background
leukocytes and retrieval of the captured CTCs in a simple fashion.
The variability observed on the recovery rate of cell retrieval is
mainly due to the nature of the manual operation of the protocol.
For example, the pressure and speed of manual pausing and pumping
to retrieve the captured cells could vary from experiment to
experiment and operator to operator. The variable number of
background leukocytes could come from different healthy donors of
the blood samples. To improve the purity of captured cells and the
consistency of the protocol performance, we are developing an
automated pump that can be connected to the Celsee PREP100 to
retrieve captured cells from the microfluidic chip for PCR and
sequencing analyses.
Enrichment of circulating cells can enable a number
of downstream molecular applications. In addition to Sanger
sequencing analysis used in our study, RT-PCR and DNA array assays
on gene expression profiling, as well as NGS analysis on several
DNA and RNA based genomic applications have been explored with
enriched CTCs. Fluorescence in situ hybridization (FISH)
assay on CTCs has also been demonstrated for determining gene
amplification and aberrant copy number changes in cancer cells.
Molecular profiling of CTCs could produce insightful information
towards understanding the heterogeneity and the complexity of
cancer and shed further light on the mechanisms of tumor
metastasis, thus delineating tumor cells that are relevant to
prognosis and therapy choice. With improvements on automation and
standardization, enrichment and characterization of CTCs could
overcome the technical limitations of low sensitivity and high
background leukocytes and become a routine diagnostic tool in
clinical use.
Acknowledgements
The authors thank all the supporting staff in the
hematology/oncology clinic, Henry Ford Health System for their
support in collection of the patient blood samples.
Funding
The present study was supported in part by the pilot
project funding from the Henry Ford Health System Research
Administration.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
RK, CH, NP, CL and YW conceived and designed the
study. RK, JZ, PG, SC and WC performed the experiments. CH
recruited and obtained consent from patients. RK, NP, CL and YW
wrote the manuscript. RK, NP, CL and YW reviewed and edited the
manuscript. All authors read and approved the manuscript and agree
to be accounTable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Institutional Review Board of the Henry Ford Health System.
Consent for publication
Not applicable.
Competing interests
The authors decalre that they have no competing
interests.
References
|
1
|
Pantel K and Alix-Panabières C:
Circulating tumor cells in cancer patients: Challenges and
perspectives. Trends Mol Med. 16:398–406. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Maheswaran S and Haber DA: Circulating
tumor cells: A window into cancer biology and metastasis. Curr Opin
Genet Dev. 20:96–99. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cristofanilli M, Budd GT, Ellis MJ,
Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ,
Terstappen LW, et al: Circulating tumor cells, disease progression,
and survival in metastatic breast cancer. N Engl J Med.
351:781–791. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Punnoose EA, Atwal SK, Spoerke JM, Savage
H, Pandita A, Yeh RF, Pirzkall A, Fine BM, Amler LC, Chen DS and
Lackner MR: Molecular biomarker analyses using circulating tumor
cells. PLoS One. 5:e125712010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Allard WJ, Matera J, Miller MC, Repollet
M, Connelly MC, Rao C, Tibbe AG, Uhr JW and Terstappen LW: Tumor
cells circulate in the peripheral blood of all major carcinomas but
not in healthy subjects or patients with nonmalignant diseases.
Clin Cancer Res. 10:6897–6904. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Riethdorf S, Fritsche H, Müller V, Rau T,
Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Jänicke F, et al:
Detection of circulating tumor cells in peripheral blood of
patients with metastatic breast cancer: A validation study of the
CellSearch system. Clin Cancer Res. 13:920–928. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nagrath S, Sequist LV, Maheswaran S, Bell
DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky
A, et al: Isolation of rare circulating tumour cells in cancer
patients by microchip technology. Nature. 450:1235–1239. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Khoo BL, Warkiani ME, Tan DS, Bhagat AA,
Irwin D, Lau DP, Lim AS, Lim KH, Krisna SS, Lim WT, et al: Clinical
validation of an ultra high-throughput spiral microfluidics for the
detection and enrichment of viable circulating tumor cells. PLoS
One. 9:e994092014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stott SL, Hsu CH, Tsukrov DI, Yu M,
Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK,
et al: Isolation of circulating tumor cells using a
microvortex-generating herringbone-chip. Proc Natl Acad Sci USA.
107:18392–18397. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Saliba AE, Saias L, Psychari E, Minc N,
Simon D, Bidard FC, Mathiot C, Pierga JY, Fraisier V, Salamero J,
et al: Microfluidic sorting and multimodal typing of cancer cells
in self-assembled magnetic arrays. Proc Natl Acad Sci USA.
107:14524–14529. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Powell AA, Talasaz AH, Zhang H, Coram MA,
Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, et
al: Single cell profiling of circulating tumor cells:
Transcriptional heterogeneity and diversity from breast cancer cell
lines. PLoS One. 7:e337882012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sieuwerts AM, Kraan J, Bolt-de Vries J,
van der Spoel P, Mostert B, Martens JW, Gratama JW, Sleijfer S and
Foekens JA: Molecular characterization of circulating tumor cells
in large quantities of contaminating leukocytes by a multiplex
real-time PCR. Breast Cancer Res Treat. 118:455–468. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Riahi R, Gogoi P, Sepehri S, Zhou Y,
Handique K, Godsey J and Wang Y: A novel microchannel-based device
to capture and analyze circulating tumor cells (CTCs) of breast
cancer. Int J Oncol. 44:1870–1878. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gogoi P, Sepehri S, Zhou Y, Gorin MA,
Paolillo C, Capoluongo E, Gleason K, Payne A, Boniface B,
Cristofanilli M, et al: Development of an automated and sensitive
microfluidic device for capturing and characterizing circulating
tumor cells (CTCs) from clinical blood samples. PLoS One.
11:e01474002016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chang H, Jackson DG, Kayne PS,
Ross-Macdonald PB, Ryseck RP and Siemers NO: Exome sequencing
reveals comprehensive genomic alterations across eight cancer cell
lines. PLoS One. 6:e210972011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Veldscholte J, Ris-Stalpers C, Kuiper GG,
Jenster G, Berrevoets C, Claassen E, van Rooij HC, Trapman J,
Brinkmann AO and Mulder E: A mutation in the ligand binding domain
of the androgen receptor of human LNCaP cells affects steroid
binding characteristics and response to anti-androgens. Biochem
Biophy Res Commun. 173:534–540. 1990. View Article : Google Scholar
|