Introduction
It has recently been demonstrated that the
recurrence of cancer in vivo was attributable to the
self-renewal capacity of so called CSCs or cancer initiating cells
(CICs) (1). It has also been
revealed that resistance to conventional therapeutic regimens in
vitro and in vivo was a feature of these cells (2). Thus, it was crucial to assess the
self-renewal of CSC480 cells. A particular set of genes, including
ALDH1A1 and CD44 are known for their role in
maintaining the self-renewal of CSCs.
CD44 is one of the most common surface markers used
to identify CSCs (3). The CD44
glycoprotein is a receptor for a major component of the
extracellular matrix, hyaluronan (HA) (4). In many cancers, binding of HA to CD44
activates multiple receptor tyrosine kinases, including epidermal
growth factor receptor (EGFR) and ERBB2 (5). HA binding to CD44 leads to activation
of the MAPK and PI3K/AKT pathways, resulting in increased
proliferation and survival (4).
Inhibition of CD44-HA binding has been shown to prevent tumor
formation in colorectal cancer (6).
CD44, either alone or in combination with other surface markers,
has been used to identify and isolate cells with stem cell
properties from colon cancer tissue (7).
Aldefluor is a new flow cytometric methodology that
assesses ALDH activity in viable cells. This method was initially
used to sort hematopoietic cells (8,9). ALDH1
activity was combined with CD34 expression to identify distinct
hematopoietic stem and progenitor cell subpopulations (10). Based on its role in the
identification of haematopoietic stem cells, it has been
hypothesised that ALDH1 is plausibly useful to identify CSCs from
multiple myeloma and leukaemia patients. This was supported by the
fact that isolated ALDH bright cells exhibit high tumorigenicity
when inoculated into non-obese diabetic/severe combined
immunodeficiency (NOD/SCID) mice (11,12).
In addition, these ALDH bright cells exhibited the traditional
features of a stem cell such as a slow proliferation rate.
Dormancy is a process that CSCs use to evade the
immune system before or after metastasis. It is believed that
dormant metastatic cells originate from a pool of quiescent cells.
Being non-proliferative, resting in the quiescent state and losing
apoptotic potential, as well as retaining LRC markers or expressing
stem-like markers are defining features of these cells (13). Research hypotheses have been
developed to investigate the interchangeable state between slow and
fast proliferating cancer cells. One of the major research
questions was to address this process in dormant cancer cells after
metastasis, in order to determine if it was restricted to
metastasis or also occured in the primary tumor (14). CSCs survive extreme
micro-environmental conditions long-term by being dormant. However,
the decision to remain dormant or proliferate is coordinated by
extracellular conditions including immune reactions to antitumor
treatment and variations in angiogenic processes (15,16).
Key intracellular checkpoints controlling CSC entry and exit from
quiescence include modifications in the cell cycle (17). Understanding the behaviour of
‘dormant’ CSCs and unravelling approaches to identify them are
critical in the treatment of cancer.
Several studies have claimed that proliferation
markers like BrdU can be used as label-retaining markers for the
identification of slow dividing SCs (18). We exploited the properties of the
proliferation marker EdU, which is incorporated into the DNA of
actively dividing cells in the same manner as BrdU. Upon cell
division, it distributes equally to both daughter cells. Therefore,
EdU fluorescence decreases with each round of DNA synthesis and
cell division, however it is retained at original levels in non or
slow dividing cells (19).
The aim of the present study was to assess and
characterise the stemness properties of a new putative colorectal
adenocarcinoma cancer stem cell model. These cells were established
and promoted by Biomedicure (San Diego, CA, USA), however there was
no available information about their properties. However, the
methods used to generate the cells did not describe their
characteristics (20). Thus, the
need arose to verify them independently. As aforementioned, CSCs
were supposed to display defining characteristics and these were
investigated accordingly.
In the present study, we assessed the stemness
properties and proliferative capacity of CSC480 cells using in
vitro techniques. Furthermore, this study presented a new
approach for identifying different populations in heterogenous
cancers. Finally, we hypothesised that we could assess the utility
of EdU for the characterisation of an infrequently cycling (i.e.
EdU retaining), tumor-initiating, sub-population in human colon
cancer cells.
Materials and methods
Cell culture
Three colon cell lines were used. The normal fetal
human colon epithelial cell line FHC [American Type Culture
Collection (ATCC) Manassas, VA, USA], the grade 3–4 colon
adenocarcinoma cell line SW480 (ATCC) and colorectal cancer stem
cell line CSC480 (Biomedicure) (20). The SW480 and CSC480 cells were
maintained in Dulbecco's modified Eagle's medium (DMEM)-high
glucose media (Gibco; Thermo Fisher Scientific, Waltham, MA, USA)
supplemented with 10% (v/v) fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific) 100 g/ml streptomycin, 100 U/ml penicillin and
cultured in a humidified atmosphere of 5% CO2 at 37°C.
The FHC cell line was propagated using DMEM/F12 (Gibco; Thermo
Fisher Scientific) supplemented with 25 mM HEPES, 10 ng/ml cholera
toxin, 0.005 mg/ml insulin, 0.005 mg/ml transferrin, 100 ng/ml
hydrocortisone (Sigma-Aldrich Merck KGaA; Darmstadt, Germany) and
10% FBS (Gibco; Thermo Fisher Scientific).
Flow cytometry
ALDH activity assessment
ALDH activity was analysed using the Aldefluor assay
according to the manufacturer's instructions (StemCell
Technologies, Vancouver, BC, Canada). Approximately
1×106 cells were used for analysis. Firstly, the cells
were detached using Accutase dissociation reagent (Innovative Cell
Technologies, San Diego, CA, USA). The cells were then washed three
times with PBS and centrifuged at 250 × g for 2 min for each wash.
Cell concentration was determined through trypan blue exclusion.
The cells were then resuspended using Aldefluor-activated reagent
Bodipy-aminoacetaldehyde (BAAA; 1 µmol/l per 1×106
cells). Half of the cell suspension was transferred to another tube
containing diethylaminobenzaldehyde (DEAB) solution (50 mmol/l) and
incubated for one hour at 37°C to deactivate the
Adlefluor-activated reagent. The cells were then pelleted by
centrifugation at 250 × g for 2 min, the supernatant was removed
and the cells were resuspended in 200 µl of 1% BSA/Aldefluor
buffer.
CD44 flow cytometric analysis
For the analysis of the CD44 expression, the cells
were detached using Accutase dissociation reagent (Innovative Cell
Technologies). The cell suspension was washed twice with PBS and
centrifuged at 250 × g for 2 min for each wash. The cells were
incubated with mouse anti-human CD44-FITC primary antibody (cat.
no. MHCD4401; Invitrogen; Thermo Fisher Scientific) or mouse
isotype matched control (cat. no. 11-4724-42; eBioscience; Thermo
Fisher Scientific; the dilution for both antibodies is 5
µl/1×106 cells suspended in 400 µl of 1% BSA/PBS
solution) on ice, in the dark for 30 min. Subsequently, the cell
suspensions were centrifuged for 10 min at 100 × g at 4°C, the
supernatants were removed, the cells were resuspended in 1% BSA in
PBS and incubated with 7-AAD nuclear stain (eBioscience) on ice for
5 min. The cells were then analysed via flow cytometry.
Flow cytometric analysis
The flow cytometric analysis was performed on a
FACSAria flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
The rate of acquisition was set to <10,000 events per second.
The logarithmic amplification was used for EdU fluorescence. For
the detection of EdU with Alexa Fluor 647, 633/635 nm excitation
with the red emission filter (660/20 nm) was used, while for the
detection of CD44 with FITC, 488 nm excitation with a green filter
emission filter (530/30 nm) was used. A whole cell gate on forward
vs. side scatter was constructed to exclude debris, doublets and
aggregates. This gate was used for all subsequent analyses.
Immunofluorescence staining
SW480 and CSC480 cells were seeded into 8-well
chamber slides at a density of 10,000 cells/well. The cells were
pulsed with 10 µM EdU for 2 h (Invitrogen Life Technologies).
Subsequently, they were fixed with 4% formaldehyde and stained
using Click-iT EdU Imaging kit (Invitrogen Life Technologies)
following the manufacturer's protocol. The cells were washed three
times with 1X phosphate buffered saline (PBS) and were blocked in
1% bovine serum albumin (BSA) in 1X PBS. Subsequently, the cells
were incubated with mouse anti-human CD44 monoclonal antibody
(1:100 dilution; cat. no. 3570; Cell Signaling Technology, Inc.,
Danvers, MA, USA) or mouse isotype matched control (cat. no.
12-4724-42; eBioscience) at room temperature, in the dark for 30
min. The primary antibody was withdrawn and cells were washed three
times as above-mentioned. Then, the cells were incubated with
donkey anti-mouse Alexa Fluor 594 secondary antibody (1:1,000; cat.
no. AF594; Abcam, Cambridge, MA, USA). They were washed as
above-mentioned and counterstained with Vectashield/DAPI (cat. no.
H-1200; Vector Laboratories, Burlingame, CA, USA).
Image acquisition and analysis
Immunofluorescence images were acquired using an
AxioImager Z1 fluorescence microscope (Carl Zeiss, Inc., Thornwood,
NY, USA) and an Axiocam camera (Carl Zeiss, Inc.). Images were
processed using AxioVision 4.6.3 (Carl Zeiss, Inc.) digital image
editing software and adjusted for brightness and contrast only
using Adobe Photoshop software.
RNA extraction
The cells in T75 flasks (Corning Incorporated,
Corning, NY, USA) were washed twice with 4 ml Dulbecco's
phosphate-buffered saline (DPBS) and incubated with 3–4 ml trypsin
(Gibco; Thermo Fisher Scientific) for 3–5 min at 37°C. Once
detached, the trypsin was inactivated by the addition of an equal
amount of fresh media containing serum and the cells were pelleted
by centrifugation at 250 × g for 5 min. The cells were washed with
2–3 ml of PBS (depending on the pellet size) and pelleted in a 1.5
ml Eppendorf tube by centrifugation at 300 × g for 5 min at 4°C.
The cell pellets were stored at −80°C until processing.
Total RNA was extracted using the RNeasy kit
(Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's
instructions. The cell pellets (typically ~1×106 cells)
were resuspended in 350 µl lysis buffer and disrupted by vortexing
vigorously for 15 sec. Subsequently, 350 µl of 70% ethanol was
added before each sample was transferred to an RNeasy spin column.
Following wash steps, the RNA was eluted in 30 µl RNase/DNase-free
water. The amount and purity of the RNA was determined by measuring
absorbance at 260 nm (A260) and 280 nm (A280) on a NanoDrop
spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
An A260/A280 ratio of 1.8–2.0, indicating RNA free of contaminating
protein or phenol, was obtained for all samples. RNA was stored at
−80°C.
cDNA synthesis
cDNA was synthesized with miSript reverse
transcription kit (Qiagen) according to the manufacturer's
instructions. Briefly, to perform cDNA synthesis, up to 1 µg of RNA
was mixed with 5X miScript RT buffer, 1 µl of miScript Reverse
Transcriptase Mix and adjusted with water to a total of 20 µl. The
reaction was mixed and incubated at 37°C for 60 min and then
incubated at 95°C for 5 min to inactivate the miScript Reverse
Transcriptase Mix. cDNA samples were then stored at −20°C.
Real-time quantitative PCR
Quantitative real-time PCR was performed using the
iCycler iQ5 Real-Time PCR system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). Quantitative gene expression was determined for
ABCG2, CD44, Ki67, EpCAM, ALDH1A and SOX2 (OriGene Technologies,
Inc., Rockville, MD, USA). The primer sequences are listed in
Table I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Gene | Forward | Reverse |
|---|
| ABCG2 |
GTTCTCAGCAGCTCTTCGGCTT |
TCCTCCAGACACACCACGGATA |
| CD44 |
CCAGAAGGAACAGTGGTTTGGC |
ACTGTCCTCTGGGCTTGGTGTT |
| Ki67 |
GAAAGAGTGGCAACCTGCCTTC |
GCACCAAGTTTTACTACATCTGCC |
| EpCAM |
GCCAGTGTACTTCAGTTGGTGC |
CCCTTCAGGTTTTGCTCTTCTCC |
| ALDH1A |
CGGGAAAAGCAATCTGAAGAGGG |
GATGCGGCTATACAACACTGGC |
| SOX2 |
GCTACAGCATGATGCAGGACCA |
TCTGCGAGCTGGTCATGGAGTT |
| NANOG |
CTCCAACATCCTGAACCTCAGC |
CGTCACACCATTGCTATTCTTCG |
| GAPDH |
GTCTCCTCTGACTTCAACAGCG |
ACCACCCTGTTGCTGTAGCCAA |
The results were normalized to the housekeeping gene
GAPDH (OriGene Technologies, Inc.). Real-time PCR was performed
according to the following cycling protocol: 1 cycle at 95°C for 10
min, followed by 42 cycles at 94°C for 10 sec and 60°C for 10
sec.
EdU incorporation
The cells were exposed to a 2 h pulse of 10 µM EdU
(Invitrogen Life Technologies). Retention was assessed in SW480 and
CSC480 cell lines at different time-point post-labelling (24, 48,
72 and 96 h) after a 24 h EdU exposure (10 µM). Labelling was
performed according to the manufacturer's instructions
(Click-iT™ EdU; Invitrogen Life Technologies).
MTT proliferation assay
Three hours before each of the time-points, 20 µl of
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
solution (5 mg/ml in PBS) (Sigma-Aldrich; Merck KGaA) was added
into each well and the cells were incubated at 37°C for a further 3
h. The medium was removed and 100 µl of DMSO was added into each
well. The plate was gently rotated on an orbital shaker for 10 min
to completely dissolve the precipitate. The absorbance was detected
at 570 nm with a microplate reader (POLARstar Omega; BMG Labtech,
Offenburg, Germany).
Statistical analysis
GraphPad Prism 6 software suite (GraphPad Software,
Inc., La Jolla, CA, USA) was used to perform statistical analysis
using Student's t-test or one-way ANOVA. The experiments were
conducted in triplicate and data are presented as the means ±
standard error of mean (SEM). P<0.05 was considered to indicate
a statistically significant difference.
Results
Phenotypic and molecular assessment of
the CSC480 cells compared to SW480 cells
Using well-characterized biomarker assays, the stem
cell-like characteristics of the CSC480 cell line were assessed
compared to the parental cell line, SW480. It was observed that
these markers vary in their expression between the two cell lines.
CSC480 cells expressed significantly higher CD44 protein (90.5%)
than SW480 cells (42.4%) (Fig. 1A).
Furthermore, CSC480 cells expressed twice the amount of CD44 mRNA
than SW480 cells while FHC ‘normal’ colon epithelial line expressed
little if any, CD44 (P<0.001; Fig.
1B). CSC480 cells also expressed higher ALDH1 activity (45.54%)
compared with SW480 cells (10.08%) (Fig. 1C). We also found that CSC480
expressed four times more ALDH1 mRNA (P<0.05) (Fig. 1D) and three times more ABCG2
(P<0.01) (Fig. 1E) than SW480
cells. CSC480 cells expressed an ~1.2-fold increase in Sox2 mRNA
than SW480 cells (P<0.001) (Fig.
1F).
 | Figure 1.Phenotypic and molecular
characterisation of the CSC480 cancer cell line. (A) Flow
cytometric analysis of CD44 in CSC480 cell line. The scatter plots
on the top row represent isotype-FITC labelled control group and on
the bottom row represent CD44-FITC group. There was considerable
increase in CD44-FITC intensity in CSC480 cell lines as compared to
the control cancer cell line. (B) The expression of CD44 mRNA
levels in CSC480, SW480 and FHC cells. Data are mean ± SEM, n=3;
statistical analysis, one-way ANOVA; ***P<0.001 to FHC. (C)
Analysis of ALDH1 in CSC480 cells. Scatter plots on the left show
flow cytometric analysis of ALDH1 using Aldefluor assay. The area
under the dotted line represents Aldefluor-treated cells while the
area under the solid line represents ALDH1 Aldefluor-treated cells.
(D) The bar graph on the right displays ALDH1 mRNA expression
levels in CSC480 cell line, compared with SW480 control cell line.
Data are mean ± SEM, n=3; statistical analysis, Student's t-test;
*P<0.05. (E) The expression of ABCG2 mRNA levels in CSC480
cells. Data are mean ± SEM, n=3; statistical analysis, Student's
t-test; **P<0.01. (F) Bar graph on the right represents the
expression of SOX2 mRNA levels in CSC480 cell lines compared to
cancer cell line, SW480 and normal colon cell line, FHC used as
controls. Data are mean ± SEM, n=3; statistical analysis, one-way
ANNOVA; ***P<0.001 compared to FHC. |
Notably, EpCAM, a marker for isolating self-renewing
colorectal cancer cells, demonstrated ~1.5-fold higher expression
in SW480 cells compared to CSC480 cells (Fig. 2). The embryonic stem cell gene,
NANOG, was highly expressed in both cell types compared to FHC
cells. Both were significantly different from FHC cells
(P<0.001) (Fig. 2).
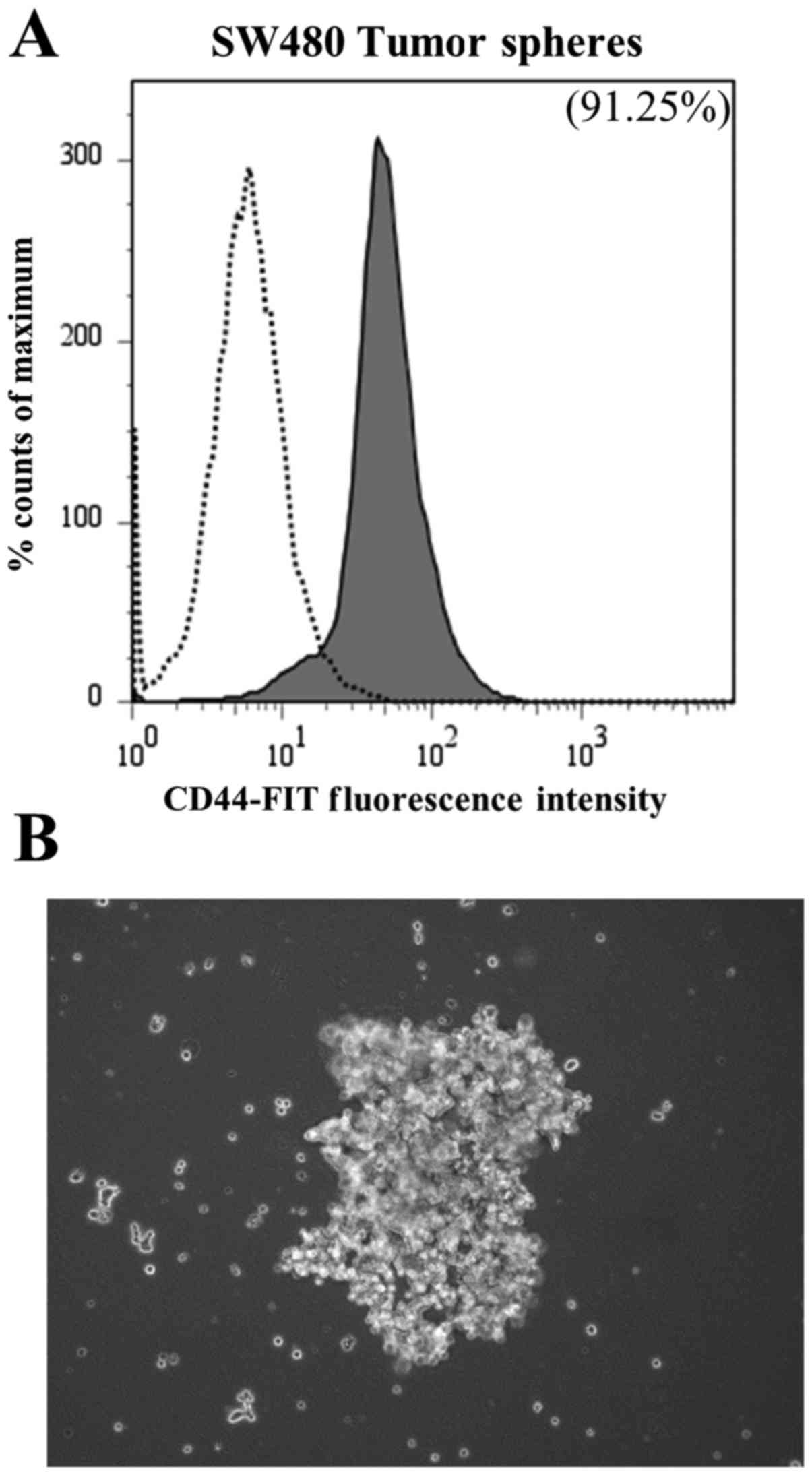
Comparing CSC-480 cells with SW480
tumor-sphere cells
To determine if the established cell line, CSC480,
presented similar expression of the CD44 marker compared to the
conventional method, we used an established method for enriching
cancer cells into a stem-like phenotype (21). SW480 cells were exposed to an
enrichment medium in low adherent culture to form tumor spheres and
analyzed for CD44 expression. Flow cytometric analysis revealed
that following tumor-sphere formation, SW480 cells expressed CD44
to a similar degree as CSC480 cells (91.25%) (Fig. 3; to be compared with Fig. 1A).
Assessing the proliferation of CSC480
cells compared to SW480 cells
CSC480 cells demonstrated enhanced proliferative
capacity compared to SW480 cells. Ki67, a well-established
proliferation marker, was used to assess the proliferative capacity
of the cell lines. It was found that CSC480 cells expressed Ki67 at
160% of SW480 levels (P<0.001) (Fig.
4A). Furthermore, MTT assays used to assess the cell growth
rate over a five-day period revealed that CSC480 cells grew 10–15%
faster than SW480 cells (Fig. 4B).
There was significant increase in the cell growth rate of CSC480
cells on day 3, 4 and 5 (P<0.001).
EdU can identify different populations
of cells in the CSC480 cell line
EdU is a well-studied marker for labelling cells in
the S phase. Research has revealed different uses for this marker,
including the use of EdU incorporation to identify cells
subsequently residing in a quiescent state (dormancy) (22). Exposing CSC480 cells to EdU for 2 h
revealed that the cells were divided into five different
populations according to their EdU fluorescent intensity and CD44
expression. Four CD44 stained populations were labelled with EdU,
while only a minor population was positive for CD44 alone. This
EdU-negative and CD44-positive population may represent the ‘true’
stem-like cell population. We observed that EdU was able to divide
SW480 cells into similar populations ratios as well (Fig. 5A). Immunofluorescence staining
demonstrated a similar trend when EdU was combined with CD44. It
has demonstrated that EdU stained CD44-positive population with
variable intensities. The highly expressing EdU population were
lower in CD44 expression. Conversely, highly CD44 expressing cells
revealed low to diminished EdU labeling intensity (Fig. 5B). This indicated that EdU may be
used to identify different populations in a heterogeneous cancer
population according to their division state.
 | Figure 5.Identifying CD44-positive populations
in SW480 and CSC480 cancer cell lines. (A) Flow cytometric analysis
of CD44-positive populations using thymidine analogue EdU in SW480
(scatter plot on left) and CSC480 cells (scatter plot on right) is
presented. Q1, EdUhigh/CD44low (19.1% for
SW480 and 1.2% for CSC480); Q2,
EdUhigh/CD44high (4.3% for SW480 and 13% for
CSC480); Q3, EdUlow-mid/CD44high (73.8% for
SW480 and 30.8% for CSC480); Q4,
EdUlow/CD44high (2.8% for SW480 and 54.9% for
CSC480); Q5, CD44high (1.5% for both SW480 and CSC480).
(B) Identification of CD44 and EdU-positive cell populations.
Immunofluorescence images of CD44-positive and EdU-positive cell
populations in CSC480 and SW480 cell lines are shown. Anti-CD44
(red), EdU-positive (green) and cell nucleus (DAPI, blue), right
panel, merged images. Top row, SW480 cells; bottom row, CSC480
cells, images captured at ×63 magnification. |
FHC cell line for assessing quiescence
markers and the resistance to therapeutic regimens in cancer
compared to normal cells
We used FHC gene expression to calibrate stem
cell-like characteristics in CSC480 cells compared to SW480 cell
line. FHC cells were very slow in division, which may be an
indicator that they were resting at the quiescence stage. The gene
expression of markers normally associated with stem cell-like
properties was analysed in FHC cells.
Notably, we found that the FHC cells expressed high
levels of the known stem cell marker ALDH1, whereas the
CSC480 and SW480 cells expressed low levels. FHC cells expressed
1,400-fold more ALDH1 than SW480 cells (P<0.01) and
~400-fold more than the CSC480 cells (P<0.001) (Fig. 6). This indicated that FHC could be
used for assessing markers of slow-cycling cancer cells and that
ALDH1 may be a marker of quiescence. Furthermore,
ABCG2, a cancer-resistance marker, was upregulated in FHC
compared to both CSC480 and SW480 cells. FHC cells were associated
with 80-fold more ABCG2 mRNA than SW480 cells (P<0.001)
and 30-fold more AGCG-2 mRNA than CSC480 cells (P<0.01)
(Fig. 7). Combining this
observation with data from other studies indicated that this
approach may be used for identifying quiescent cells.
To further confirm the slow division of FHC cells,
we treated them with the EdU proliferation marker to track their
division rate and we observed them under fluorescent microscopy.
Our observation of FHC cells revealed no EdU incorporation, which
indicated that these cells had slow dividing nature.
CD44-positive/EdU label retaining
cells are slow-dividing colon cancer cells
To identify the actively dividing cells from
non-dividing cells, CSC480 and SW480 cells were exposed to EdU for
24 h and harvested at different time-points. Four distinct
populations of cells were observed (Fig. 8). CSC480 cells collected at 24 and
48 h revealed a high retention of EdU and more CD44 expression
(85.84 and 85.28%) than SW480 cells (74 and 77%). At both
time-points, CSC480 cells expressed more CD44 than SW480 cells
(±91.41 and ±78.98%, respectively). Conversely, EdU was retained in
the cells harvested at two days, showing similar ratios of labeled
cells in both cell lines (SW480 cells, 92.51% and CSC480 cells,
93.88%). Both cell types harvested after 72 h demonstrated a
significant loss of EdU in the CD44-positive population (SW480
cells, 60.80% and CSC480 cells, 66.36%). SW480 cells collected
after 96 h in culture revealed that CD44-positive cells had
retained more EdU than CSC480 cells at the same time-point (65.48
and 38.89%, respectively).
Discussion
In recent years, it has become apparent that CSCs
are one of the main factors contributing to tumor development and
metastasis and there has been considerable effort invested in
understanding and characterising this enigmatic cell population.
However, these efforts have been restrained by several factors. The
lack of a cancer stem-cell model that recapitulates the actual CSC
population present in cancer tissues is considered one of the main
obstacles. Furthermore, the absence of a specific biomarker that
unarguably identifies a pure CSC population has rendered in
vivo characterisation difficult. The method of identifying and
purifying CSCs in different organs depends largely on their surface
phenotype and use of flow cytometry.
Adequate number of CSCs that have stable phenotypes
and similar backgrounds are required to perform reliable functional
assays. However, CSCs isolated from patients are generally rare and
readily differentiate in culture and as a consequence, a shortage
in material for functional assessment or for screening new agents
specific to CSCs persists (23).
Most CSC assays depend on the enrichment of CSCs from freshly
isolated tumors and the efficiency of cell sorting and varied
genetic background can also hinder CSC research. Furthermore,
freshly isolated cancer stem-like cells can be contaminated with
lymphocytes or stromal cells that affect the downstream assessment.
Thus, the establishment of human CSC lines is a desirable strategy
to investigate the mechanisms of tumor initiation, resistance to
novel treatment regimens, metastasis and recurrence (23).
Therefore, efforts to establish CSC lines have been
a landmark in cancer research. These efforts varied from transient
enrichment using specific growth factors as non-adherent spheres,
to using induced pluripotent stem cell (iPSC) technology to
reprogram gastrointestinal cancer cells by introducing embryonic
stem (ES) cells transcription factors (24).
The main aim of the present study was to analyse and
characterise the novel putative cancer ‘stem’ cell line CSC480.
These cells have not been previously studied and very limited
information was available concerning their behaviour. Furthermore,
it was not clear how these cells were enriched and no information
was available about the conformation of their stemness. To address
these gaps in knowledge and to unravel the nature of CSC480 cells,
they were subjected to thorough analyses using cellular and
molecular assays. We used CD44 and ALDH1A1 markers to assess the
stemness/progenitor-like properties of these cells. CSC480 cells
revealed increased expression of CD44 and ALDH1 compared to SW480
cancer cell line. CD44, a highly heterogeneous glycoprotein
encrypted by a single gene, is involved in multiple cellular
mechanisms including cellular migration, adhesion and
proliferation. Furthermore, it is pro-oncogenic and acts as a
regulator of multiple pathways as demonstrated in breast cancer
(25). CD44 is represented by two
isoforms. The standard isoform, which is expressed by the
mesenchymal and hematopoietic cells, while epithelial cells express
the variant isoform. CD44 variant overexpression in head and neck
squamous cell carcinoma is associated with invasion, clinical
stage, therapeutic resistance and relapse (26). Thus, CD44 may be a reliable marker
for tumor progression assessment. CD44 has also been used to mark
and isolate CSCs from a range of malignant tumors, including breast
(27) and colon (28) cancers. However, it is insufficient
to use CD44 as a stand-alone marker of CSCs as sometimes
CD44-positive populations are heterogeneous (29).
The present study demonstrated that almost all
CSC480 cells express CD44 marker. Thus, we used EdU to identify
different CD44-positive populations by flow cytometry. It was also
used to assess their proliferation capacity and their quiescence
status. Assessing the proliferation and identifying cells resting
in dormant state were previously reported (22,30).
EdU is a nucleoside analogue of thymidine that would be
incorporated into DNA during DNA synthesis phase (30). We employed this feature in the
present study to identify different populations present in CSC480
cells based on the DNA division state. This technique revealed that
EdU labels CSC480 CD44-positive cells with different intensities
according to DNA division rate. Four CD44 stained populations were
labelled with EdU, while only a minor subset of cells was positive
for CD44 alone. This EdU-negative and CD44-positive population may
represent the non or slow-dividing cells. We observed that EdU was
able to segregate SW480 cells into similar populations as well.
This indicated that EdU may be used to identify different
populations in a heterogeneous cancer population. This may have an
impact on identifying cancer cell populations based on their
division status, which may help in deciding therapeutic regimens
and defining the percentage of actively dividing cells compared to
dormant cells. Furthermore, based on the reported findings by
Deleyrolle et al (22), we
hypothesised that EdU could be used as a LRC marker. The cells that
are actively dividing will dilute the label during multiple rounds
of divisions. After a certain period of time (chase period), the
cells will end up having no detectable label. Conversely, cells in
slow division retain the label (31). Thus, the CSC480 dormant population
was analysed by combining CD44 with EdU after exposing cells to EdU
for 24 h. Our hypothesis implied that cells retaining EdU would be
CD44-positive. Subsequently, we subjected pulsed cells for the
analysis after collecting them at different time-points and we
found that cells lost their EdU content according to their number
of divisions (Fig. 8). However, not
all cells that retained EdU were CD44-positive. Furthermore, more
SW480 cells retained EdU on day 4 than CSC480 cells. This raised
the question whether label-retaining assays were suitable to
identify dormant stem cell populations, even though they provided a
valuable tool to delineate the cycling properties within a given
population. However, caution should be taken in consideration when
designing and interpreting these experimental results. Firstly,
although many stem cells are slow cycling, label-retention on its
own does not indicate ‘stemness’. In some cases, cells dividing
during the pulse period, withdrawn from the cell cycle and
differentiated will also appear as LRCs (31). The more ‘differentiated’ cell line,
SW480 has been found to have some LRC that were not CD44-positive.
Furthermore, the extremely slow cycling cells may not incorporate
EdU during pulse window and as a result may be missed during the
assessment process (31). To
accurately assess the cycling status of a given cell, EdU should be
combined with a cell cycle kinetic marker in addition to CD44.
CSC480 cells demonstrated increased expression of
ABCG2, a cancer-resistance protein, compared to SW480 cells.
Resistance to chemotherapy has been the most important
characteristic feature of CSCs and the main reason for cancer
metastasis and recurrence (32).
The enrichment of breast CSCs post-chemotherapy, has been reported
as an indicator of resistance to chemotherapy (33).
CSC480 cells were found to equally express NANOG
compared to the SW480 parental cell line. This may be an indicator
that CSC480 cell line represented partly self-renewing progenitor
cells. NANOG has been revealed as an indispensable component in
transforming gastrointestinal cancer cells into pluripotent stem
cells (24).
Epithelial cell adhesion molecule (EpCAM) showed up
to 2,500 and 1,500-fold increased expression in SW480 and CSC480
cells, respectively compared to FHC normal epithelial cells. This
marker has been revealed to be widely expressed in epithelial
tissue and all colorectal cancer cells (34,35). A
previous study (36) demonstrated
that EpCAM regulated self-renewal in colon initiating cells. It was
also delineated in the same study, that the persistence of EpCAM
expression was linked with increased invasiveness in vitro
and tumor initiating capacity in vivo. Furthermore, its
increased expression was correlated with increased expression of
stemness markers like OCT4, NANOG and c-Myc in colon cancer
(36).
The present study has also revealed that the fetal
human colon cell line, FHC, highly expressed ALDH1 in comparison to
SW480 and CSC480 cancer cell lines. FHC cells are featured by their
extremely slow division as observed in the present study. Thus, the
increased expression of ALDH1 in FHC cells may provide evidence
that it marked slow (dividing) cycling cells. Furthermore, elevated
expression of ABCG2 was also observed in FHC cells compared to
SW480 and CSC480 cells. This may confirm the reported feature that
dormant cells resist chemotherapy (37). FHC cells exhibited resistance to
apoptosis after genotoxic treatment (37). When FHC cells were pulsed with EdU,
none of the cells demonstrated EdU labelling (data not shown). This
finding indicated that many FHC cells may reside in a dormant
state. Assessing FHC cells in terms of tumorigenicity, revealed
that these cells are capable of growing in semisolid media under
anchorage-independent growth conditions in vitro and have
exhibited a capacity to form solid tumors in vivo (37).
Based on these findings, we analysed SW480 and
CSC480 cell gene expression using ALDH1 and ABCG2 and observed that
the CSC480 cell line exhibited elevated expression compared to
SW480 cells. This implied that CSC480 cells contained more cancer
stem-like cells that were resistant to chemotherapy in comparison
to SW480 cells, which needed to be determined experimentally.
Collectively, FHC cells could be considered as an indispensable
experimental model for assessing quiescence markers for identifying
dormant cancer stem-like cells and as a positive control for
examining chemotherapeutic drugs.
In the present study, we examined a number of
putative CSC characteristics in CSC480 cells compared to other
well-established cell lines. Since CSC480 is a newly reported cell
line and the present study was the first, further characterization
at the in vitro level should be considered to define these
cells. Assessing the molecular and phenotypic profile of certain
known colorectal stemness signature like Lrg5, BIM-1 and c-MYC
should be undertaken. Furthermore, identifying the self-renewal
capacity using clonal limited dilution assay should also be
pursued, as should in vivo growth assessment by inoculating
a limited number of cells subcutaneously compared with other cancer
cell lines. Cancer resistance to ABCG2 could be examined using a
range of drugs that are shown to be effluxed by ABCG2 such as
mitoxantrone and methotrexate (38).
In conclusion, CSC480 cells were composed of a high
percentage of transiently dividing and self-renewing cells and CD44
appeared to be a marker for these partially self-renewing precursor
cells.
Acknowledgements
Not applicable.
Funding
The present study was supported by a PhD scholarship
awarded to F.A. by King Saud University. In addition, the present
study was supported by the ‘College of Medicine Research Centre,
Deanship of Scientific Research, King Saud University’.
Availability of data and materials
The datasets used during the present study are
available from the corresponding authors upon reasonable
request.
Authors' contributions
FA contributed to the study design, experimental
work and manuscript write up; SMH contributed to the study design,
experimental work and manuscript revision; NA contributed to
experimental work and the manuscript; RA contributed to the
experimental work; SI contributed to the revision of the
manuscript; BB contributed to the experimental work; AM and AL
contributed to the experimental design; SW contributed to the
design and revision of the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
FHC
|
fetal human colon cell line
|
|
ALDH1
|
aldehyde dehydrogenase family 1 member
A1
|
|
ABCG2
|
ATP-binding cassette sub-family G
member 2
|
|
EpCAM
|
epithelial cell adhesion molecule
|
References
|
1
|
Dittmar T, Nagler C, Schwitalla S, Reith
G, Niggemann B and Zänker KS: Recurrence cancer stem cells-made by
cell fusion? Med Hypotheses. 73:542–557. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang ZJ and Wechsler-Reya R: Hit'em where
they live: Targeting the cancer stem cell niche. Cancer Cell.
11:3–5. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shen C, Xiang M, Nie C, Hu H, Ma Y and Wu
H: CD44 as a molecular marker to screen cancer stem cells in
hypopharyngeal cancer. Acta Otolaryngol. 133:1219–1226. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Misra S, Toole BP and Ghatak S: Hyaluronan
constitutively regulates activation of multiple receptor tyrosine
kinases in epithelial and carcinoma cells. J Biol Chem.
281:34936–34941. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ghatak S, Misra S and Toole BP: Hyaluronan
constitutively regulates ErbB2 phosphorylation and signaling
complex formation in carcinoma cells. J Biol Chem. 280:8875–8883.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim HR, Wheeler MA, Wilson CM, Iida J, Eng
D, Simpson MA, McCarthy JB and Bullard KM: Hyaluronan facilitates
invasion of colon carcinoma cells in vitro via interaction
with CD44. Cancer Res. 64:4569–4576. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Du L, Wang H, He L, Zhang J, Ni B, Wang X,
Jin H, Cahuzac N, Mehrpour M, Lu Y and Chen Q: CD44 is of
functional importance for colorectal cancer stem cells. Clin Cancer
Res. 14:6751–6760. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jones RJ, Barber JP, Vala MS, Collector
MI, Kaufmann SH, Ludeman SM, Colvin OM and Hilton J: Assessment of
aldehyde dehydrogenase in viable cells. Blood. 85:2742–2746.
1995.PubMed/NCBI
|
|
9
|
Armstrong L, Stojkovic M, Dimmick I, Ahmad
S, Stojkovic P, Hole N and Lako M: Phenotypic characterization of
murine primitive hematopoietic progenitor cells isolated on basis
of aldehyde dehydrogenase activity. Stem Cells. 22:1142–1151. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Storms RW, Green PD, Safford KM,
Niedzwiecki D, Cogle CR, Colvin OM, Chao NJ, Rice HE and Smith CA:
Distinct hematopoietic progenitor compartments are delineated by
the expression of aldehyde dehydrogenase and CD34. Blood.
106:95–102. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Matsui W, Huff CA, Wang Q, Malehorn MT,
Barber J, Tanhehco Y, Smith BD, Civin CI and Jones RJ:
Characterization of clonogenic multiple myeloma cells. Blood.
103:2332–2336. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pearce DJ, Taussig D, Simpson C, Allen K,
Rohatiner AZ, Lister TA and Bonnet D: Characterization of cells
with a high aldehyde dehydrogenase activity from cord blood and
acute myeloid leukemia samples. Stem Cells. 23:752–760. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Naumov GN, Bender E, Zurakowski D, Kang
SY, Sampson D, Flynn E, Watnick RS, Straume O, Akslen LA, Folkman J
and Almog N: A model of human tumor dormancy: An angiogenic switch
from the nonangiogenic phenotype. J Natl Cancer Inst. 98:316–325.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu Y and Zhu Z: Cell dormancy and tumor
refractory. Anticancer Agents Med Chem. 13:199–202. 2013.
View Article : Google Scholar
|
|
15
|
Quesnel B: Tumor dormancy: Long-term
survival in a hostile environment. Adv Exp Med Biol. 734:181–200.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wells A, Griffith L, Wells JZ and Taylor
DP: The dormancy dilemma: Quiescence versus balanced proliferation.
Cancer Res. 73:3811–3816. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kleffel S and Schatton T: Tumor dormancy
and cancer stem cells: Two sides of the same coin? Adv Exp Med
Biol. 734:145–179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Duvillié B, Attali M, Aiello V, Quemeneur
E and Scharfmann R: Label-retaining cells in the rat pancreas:
Location and differentiation potential in vitro. Diabetes.
52:2035–2042. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Diermeier-Daucher S, Clarke ST, Hill D,
Vollmann-Zwerenz A, Bradford JA and Brockhoff G: Cell type specific
applicability of 5-ethynyl-2′-deoxyuridine (EdU) for dynamic
proliferation assessment in flow cytometry. Cytometry A.
75:535–546. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qian Y: Tumorigenic cancerstemcells,
methods of isolating and using the same. Google Patents,
US20110206735. 2011.
|
|
21
|
Wang S, Kanojia D, Lo P, Chandrashekaran
V, Duan X, Berger FG, Wang Q and Hexin C: Enrichment and selective
targeting of cancer stem cells in colorectal cancer cell lines. Hum
Genet Embryol. S2:0062012.
|
|
22
|
Deleyrolle LP, Harding A, Cato K,
Siebzehnrubl FA, Rahman M, Azari H, Olson S, Gabrielli B, Osborne
G, Vescovi A and Reynolds BA: Evidence for label-retaining
tumour-initiating cells in human glioblastoma. Brain.
134:1331–1343. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rao GH, Liu HM, Li BW, Hao JJ, Yang YL,
Wang MR, Wang XH, Wang J, Jin HJ, Du L and Chen Q: Establishment of
a human colorectal cancer cell line P6C with stem cell properties
and resistance to chemotherapeutic drugs. Acta Pharmacol Sin.
36:793–804. 2013. View Article : Google Scholar
|
|
24
|
Miyoshi N, Ishii H, Nagai K, Hoshino H,
Mimori K, Tanaka F, Nagano H, Sekimoto M, Doki Y and Mori M:
Defined factors induce reprogramming of gastrointestinal cancer
cells. Proc Natl Acad Sci USA. 107:40–45. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Smith SM and Cai L: Cell specific CD44
expression in breast cancer requires the interaction of AP-1 and
NFκB with a novel cis-element. PLoS One. 7:e508672012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang SJ, Wong G, de Heer AM, Xia W and
Bourguignon LY: CD44 variant isoforms in head and neck squamous
cell carcinoma progression. Laryngoscope. 119:1518–1530. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dalerba P, Dylla SJ, Park IK, Liu R, Wang
X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al:
Phenotypic characterization of human colorectal cancer stem cells.
Proc Natl Acad Sci USA. 104:10158–10163. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ricardo S, Vieira AF, Gerhard R, Leitão D,
Pinto R, Cameselle-Teijeiro JF, Milanezi F, Schmitt F and Paredes
J: Breast cancer stem cell markers CD44, CD24 and ALDH1: Expression
distribution within intrinsic molecular subtype. J Clin Pathol.
64:937–946. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Buck SB, Bradford J, Gee KR, Agnew BJ and
Clarke ST: Detection of S-phase cell cycle progression using
5-ethynyl-2′-deoxyuridine incorporation with click chemistry, an
alternative to using 5-bromo-2′-deoxyuridine antibodies.
Biotechniques. 44:927–929. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hsu YC and Fuchs E: A family business:
Stem cell progeny join the niche to regulate homeostasis. Nat Rev
Mol Cell Biol. 13:103–114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ding XW, Wu JH and Jiang CP: ABCG2: A
potential marker of stem cells and novel target in stem cell and
cancer therapy. Life Sci. 86:631–637. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee HE, Kim JH, Kim YJ, Choi SY, Kim SW,
Kang E, Chung IY, Kim IA, Kim EJ, Choi Y, et al: An increase in
cancer stem cell population after primary systemic therapy is a
poor prognostic factor in breast cancer. Br J Cancer.
104:1730–1738. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Butler SJ, Richardson L, Farias N,
Morrison J and Coomber BL: Characterization of cancer stem cell
drug resistance in the human colorectal cancer cell lines HCT116
and SW480. Biochem Biophys Res Commun. 490:29–35. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Trzpis M, McLaughlin PM, de Leij LM and
Harmsen MC: Epithelial cell adhesion molecule: More than a
carcinoma marker and adhesion molecule. Am J Pathol. 171:386–395.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lin CW, Liao MY, Lin WW, Wang YP, Lu TY
and Wu HC: Epithelial cell adhesion molecule regulates tumor
initiation and tumorigenesis via activating reprogramming factors
and epithelial-mesenchymal transition gene expression in colon
cancer. J Biol Chem. 287:39449–39459. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Souček K, Gajdušková P, Brázdová M,
Hýzd'alová M, Kocí L, Vydra D, Trojanec R, Pernicová Z, Lentvorská
L, Hajdúch M, et al: Fetal colon cell line FHC exhibits tumorigenic
phenotype, complex karyotype, and TP53 gene mutation. Cancer
Genet Cytogenet. 197:107–116. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Doyle L and Ross DD: Multidrug resistance
mediated by the breast cancer resistance protein BCRP (ABCG2).
Oncogene. 22:7340–7358. 2003. View Article : Google Scholar : PubMed/NCBI
|