|
1
|
Ablain J, Rice K, Soilihi H, de Reynies A,
Minucci S and de Thé H: Activation of a promyelocytic
leukemia-tumor protein 53 axis underlies acute promyelocytic
leukemia cure. Nat Med. 20:167–174. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li K, Wang F, Cao WB, Lv XX, Hua F, Cui B,
Yu JJ, Zhang XW, Shang S, Liu SS, et al: TRIB3 promotes APL
progression through stabilization of the oncoprotein PML-RARα and
inhibition of p53-mediated senescence. Cancer Cell. 31:697–710
e697. 2017. View Article : Google Scholar : PubMed/NCBI
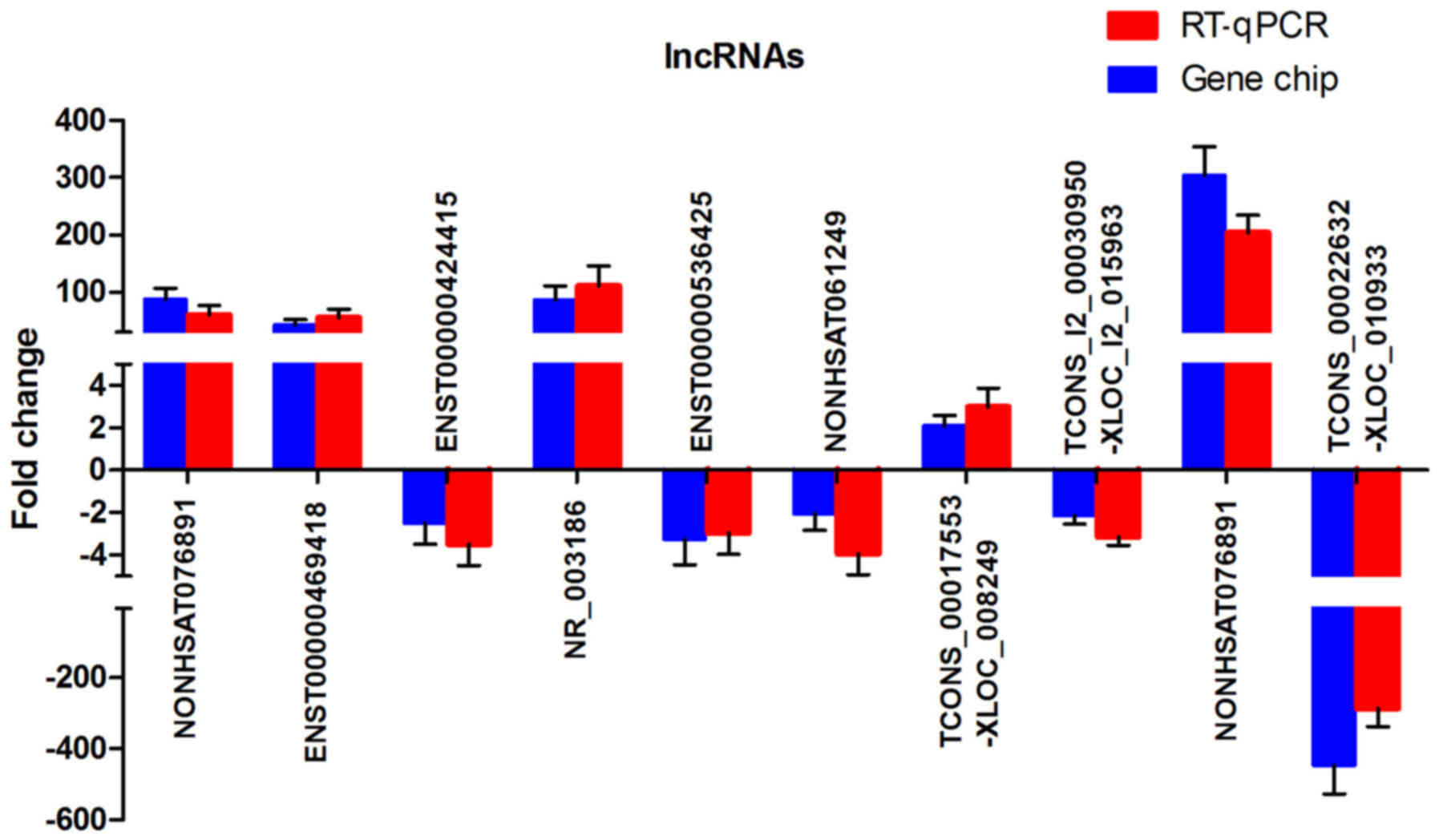
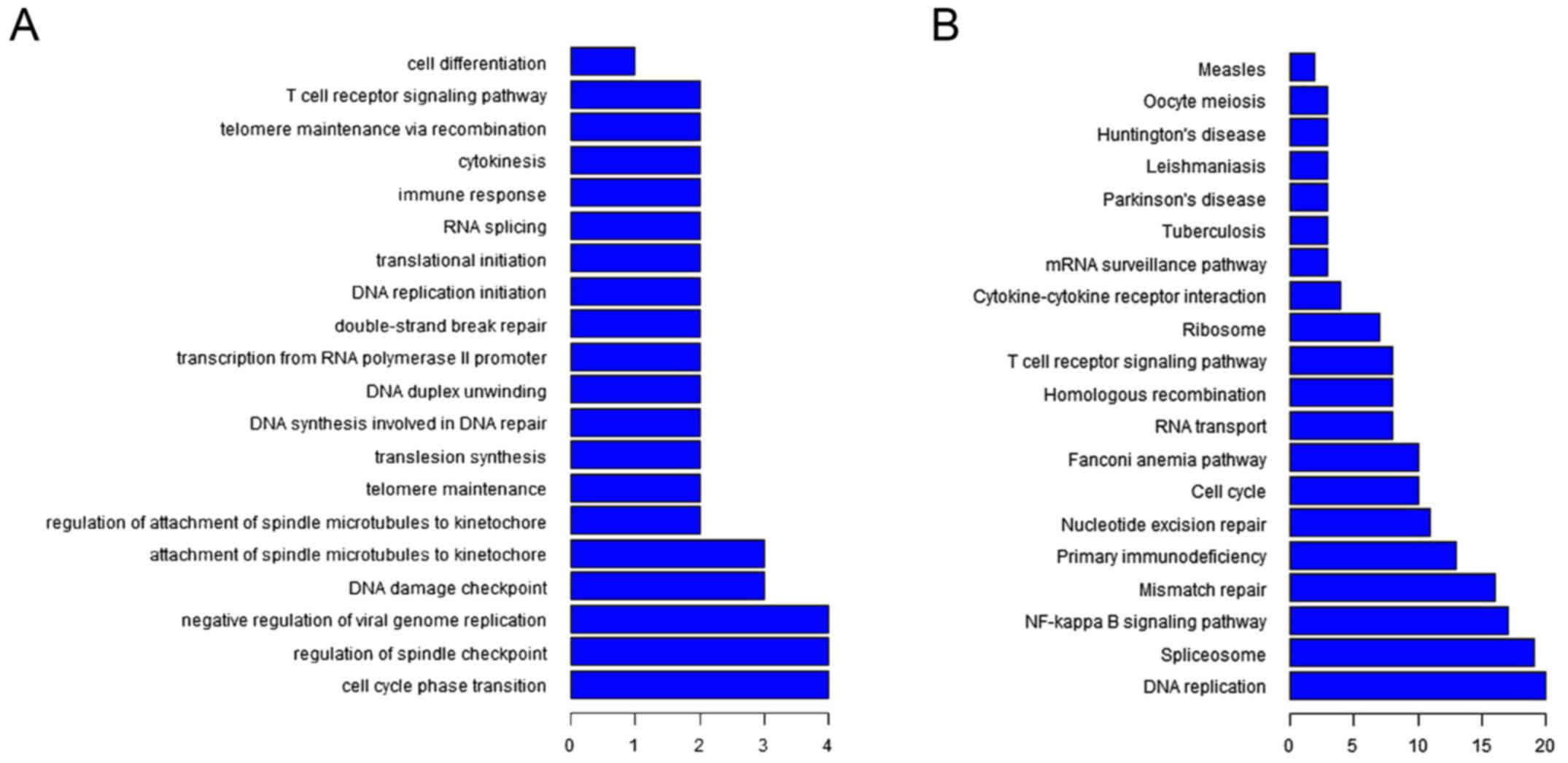
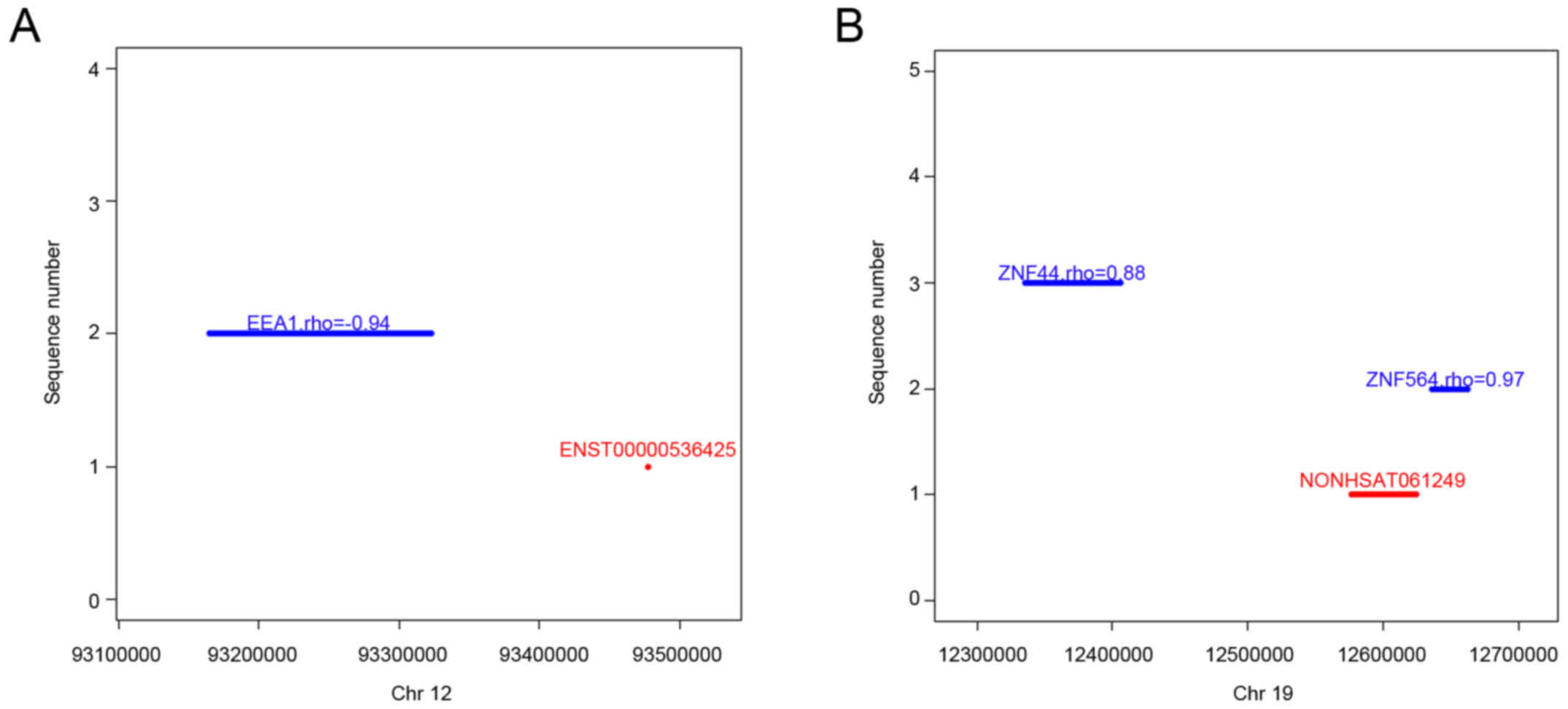
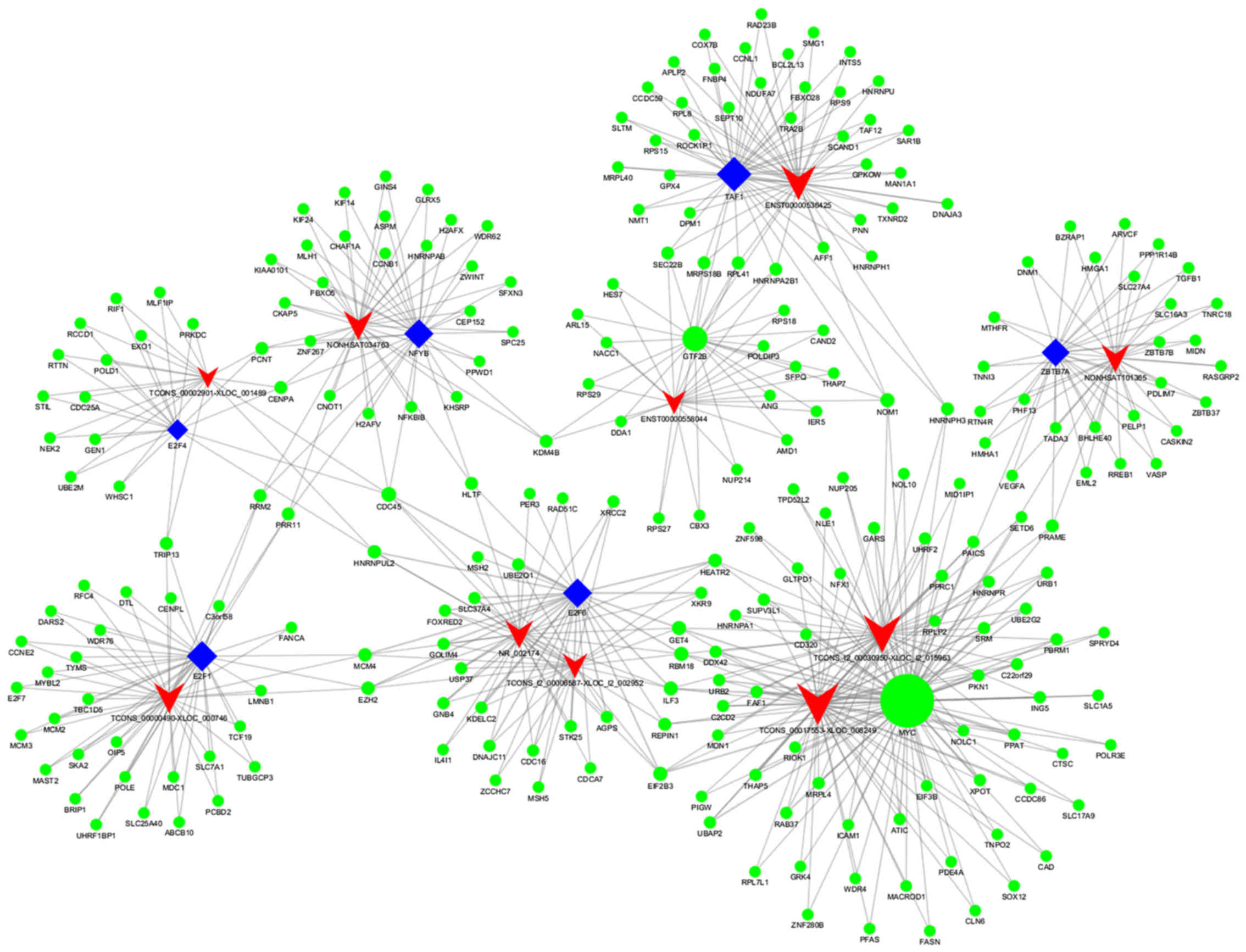
|
|
3
|
Wang ZY and Chen Z: Acute promyelocytic
leukemia: From highly fatal to highly curable. Blood.
111:2505–2515. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lo-Coco F, Avvisati G, Vignetti M, Thiede
C, Orlando SM, Iacobelli S, Ferrara F, Fazi P, Cicconi L, Di Bona
E, et al: Retinoic acid and arsenic trioxide for acute
promyelocytic leukemia. N Engl J Med. 369:111–121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lehmann-Che J, Bally C and de Thé H:
Resistance to therapy in acute promyelocytic leukemia. N Engl J
Med. 371:1170–1172. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu HH, Qin YZ and Huang XJ: Resistance to
arsenic therapy in acute promyelocytic leukemia. N Engl J Med.
370:1864–1866. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pan JQ, Zhang YQ, Wang JH, Xu P and Wang
W: lncRNA co-expression network model for the prognostic analysis
of acute myeloid leukemia. Int J Mol Med. 39:663–671. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cao L, Xiao PF, Tao YF, Hu SY, Lu J, Zhao
WL, Li ZH, Wang NN, Wang J, Feng X, et al: Microarray profiling of
bone marrow long non-coding RNA expression in Chinese pediatric
acute myeloid leukemia patients. Oncol Rep. 35:757–770. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen ZH, Wang WT, Huang W, Fang K, Sun YM,
Liu SR, Luo XQ and Chen YQ: The lncRNA HOTAIRM1 regulates the
degradation of PML-RARA oncoprotein and myeloid cell
differentiation by enhancing the autophagy pathway. Cell Death
Differ. 24:212–224. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei S, Zhao M, Wang X, Li Y and Wang K:
PU.1 controls the expression of long noncoding RNA HOTAIRM1 during
granulocytic differentiation. J Hematol Oncol. 9:442016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zeng C, Xu Y, Xu L, Yu X, Cheng J, Yang L,
Chen S and Li Y: Inhibition of long non-coding RNA NEAT1 impairs
myeloid differentiation in acute promyelocytic leukemia cells. BMC
Cancer. 14:6932014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang X, Weissman SM and Newburger PE:
Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle
progression during myeloid maturation in NB4 human promyelocytic
leukemia cells. RNA Biol. 11:777–787. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zheng PZ, Wang KK, Zhang QY, Huang QH, Du
YZ, Zhang QH, Xiao DK, Shen SH, Imbeaud S, Eveno E, et al: Systems
analysis of transcriptome and proteome in retinoic acid/arsenic
trioxide-induced cell differentiation/apoptosis of promyelocytic
leukemia. Proc Natl Acad Sci USA. 102:7653–7658. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sanz MA, Grimwade D, Tallman MS, Lowenberg
B, Fenaux P, Estey EH, Naoe T, Lengfelder E, Büchner T, Döhner H,
et al: Management of acute promyelocytic leukemia: Recommendations
from an expert panel on behalf of the European LeukemiaNet. Blood.
113:1875–1891. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guttman M and Rinn JL: Modular regulatory
principles of large non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gerstein MB, Kundaje A, Hariharan M, Landt
SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et
al: Architecture of the human regulatory network derived from
ENCODE data. Nature. 489:91–100. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guenzl PM and Barlow DP: Macro lncRNAs: A
new layer of cis-regulatory information in the mammalian genome.
RNA Biol. 9:731–741. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Y, Fu L, Sun A, Tang D, Xu Y, Li Z,
Chen M and Zhang G: C/EBPβ contributes to transcriptional
activation of long non-coding RNA NEAT1 during APL cell
differentiation. Biochem Biophys Res Commun. 499:99–104. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alvarez-Dominguez JR and Lodish HF:
Emerging mechanisms of long noncoding RNA function during normal
and malignant hematopoiesis. Blood. 130:1965–1975. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Garding A, Bhattacharya N, Claus R, Ruppel
M, Tschuch C, Filarsky K, Idler I, Zucknick M, Caudron-Herger M,
Oakes C, et al: Epigenetic upregulation of lncRNAs at 13q14.3 in
leukemia is linked to the In Cis downregulation of a gene cluster
that targets NF-κB. PLoS Genet. 9:e10033732013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zuo W, Huang F, Chiang YJ, Li M, Du J,
Ding Y, Zhang T, Lee HW, Jeong LS, Chen Y, et al: c-Cbl-mediated
neddylation antagonizes ubiquitination and degradation of the TGF-β
type II receptor. Mol Cell. 49:499–510. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Attwooll C, Lazzerini Denchi E and Helin
K: The E2F family: Specific functions and overlapping interests.
EMBO J. 23:4709–4716. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Leung JY and Nevins JR: E2F6 associates
with BRG1 in transcriptional regulation. PLoS One. 7:e479672012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rishi L, Hannon M, Salomè M, Hasemann M,
Frank AK, Campos J, Timoney J, O'Connor C, Cahill MR, Porse B and
Keeshan K: Regulation of Trib2 by an E2F1-C/EBPα feedback loop in
AML cell proliferation. Blood. 123:2389–2400. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Prasad MA, Ungerbäck J, Åhsberg J,
Somasundaram R, Strid T, Larsson M, Månsson R, De Paepe A,
Lilljebjörn H, Fioretos T, et al: Ebf1 heterozygosity results in
increased DNA damage in pro-B cells and their synergistic
transformation by Pax5 haploinsufficiency. Blood. 125:4052–4059.
2015. View Article : Google Scholar : PubMed/NCBI
|