Introduction
Osteosarcoma is one of the most common types of
serious malignant bone tumor with high mortality rates in children
and adolescents (1,2). With neoadjuvant chemotherapy and
surgery widely used in clinical treatment, the 5-year survival rate
of osteosarcoma is able to reach 50–60% (3). However, the survival rate remains 30%
in patients with distant metastatic tumors and ~60% of patients
with osteosarcoma are diagnosed with small metastases, which
suggests a poor prognosis (4,5).
Therefore, it is important to identify the molecular mechanism
underlying osteosarcoma invasion and metastasis to explore novel
therapeutic targets of osteosarcoma.
MicroRNAs (miRNAs) are endogenous small noncoding
RNAs beTween-20 and 24 nucleotides in length that bind to the
3′untranslated region (UTR) of target mRNAs to silence target gene
expression (6). miRNAs are
post-transcriptional regulators, serving important roles in a
variety of physiological and pathological processes, including
morphogenesis, differentiation and carcinogenesis (7–9). In a
previous study, various miRNAs have been demonstrated to be
dysregulated in multiple types of cancer, including osteosarcoma
(10). Certain studies have also
highlighted the association between miRNAs, and abnormal regulation
of proliferation, invasion, apoptosis and cell cycle distribution
(11–13). Certain miRNAs have been identified
to be dysregulated in osteosarcoma (14); however, to the best of our
knowledge, the role of miR-106a has not previously been
investigated.
Vascular non-inflammatory molecule 2 (VNN2) protein
is a novel glycosylphosphatidyl inositol-anchored protein member of
the VNN family that serves an important role in transendothelial
migration of cells (15). The VNN
family includes membrane-associated proteins, a few of which have
been reported to participate in regulating neutrophil trafficking
and adherence (16). It is
generally accepted that neutrophils are present in numerous
different types of cancer and are eventually recruited to the tumor
microenvironment (17).
Infiltrating inflammatory cells are highly prevalent within the
tumor microenvironment and mediate numerous processes involved in
tumor progression in vivo (18). In addition, the VNN family belongs
to a wider pantetheinase family and serve a role in redox
regulation, which may associate with tumor progression in
vitro (19). Given that
miR-106a is upregulated in human osteosarcoma cells, and associated
with cell proliferation, invasion and apoptosis, we hypothesized
that the knockdown of miR-106a may result in VNN2 gene
overexpression. The present study aimed to determine the expression
and function of miR-106a in human U2OS cell lines and tumor tissues
obtained from patients with osteosarcoma. The results revealed that
the knockdown of miR-106a mediated tumor progression at least
partially by targeting VNN2 in osteosarcoma.
Materials and methods
Patients and tissue samples
Osteosarcoma tissues and matched non-cancerous
adjacent tissues (NATs) were obtained from 18 patients (9 males and
9 females; mean age, 22.7 years; age range, 15–35 years)
histopathologically diagnosed with osteosarcoma at the First
Affiliated Hospital of China Medical University (Shenyang, China).
No patients in the study received any preoperative treatment, and
all patients underwent resection of osteosarcoma at the time of
diagnosis between June 2013 and June 2016. Patients with
histological grade IIB/III osteosarcoma were included, and patients
with any other primary disease were excluded. The paired and
osteosarcoma tissues were immediately preserved in liquid nitrogen
and maintained at −80°C. All associated clinical data, including
age, sex and tumor node metastasis stage were obtained from the
medical records of patients. The present study received approval by
the Ethics Committee of the First Affiliated Hospital of China
Medical University and written informed consent was obtained from
all patients.
Cell lines and culture
Human osteosarcoma U2OS (cat. no. TCHu88), Saos-2
(cat. no. TCHu114), MG63 (cat. no. TCHu124), 293T (cat. no. GNHu17)
and human osteoblast hFOB1.19 (cat. no. GNHu14) cells were
purchased from the Institute of Biochemistry and Cell Research
Center at the Chinese Academy of Sciences (Shanghai, China). The
U2OS, Saos-2 and MG63 cells were cultured in high-glucose
Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine
serum (both from HyClone; GE Healthcare Life Sciences, Logan City,
UT, USA) and hFOB1.19 cells were cultured in DMEM/F12 medium
(HyClone; GE Healthcare Life Sciences) containing 10% fetal bovine
serum. All cells were cultured at 37°C in a humidified atmosphere
with 5% CO2.
Lentivirus infection and plasmid
transfection
miR-106a gene-expression plasmid (miR-106a),
miR-106a gene-inhibition plasmid packaged into letivirus core
vector (hU6-MCS-Ubiquitin-EGFP-IRES-puromycin)
(miR-106a-inhibitor), mock negative control (miR-NC), VNN2 wild
type gene over-expression plasmid (GV272/VNN2), VNN2 mutant type
gene over-expression plasmid (GV272/VNN2-mut), VNN2 negative
control plasmid (GV272/VNN2-NC) were all chemically synthesized by
Shanghai GeneChem Co., Ltd. (Shanghai, China). miR-106a-inhibitor
was used as the experimental group and miR-NC as the control group.
The experimental group and control group were treated with
lentivirus, which was inactivated and served as a carrier.
Therefore, the blank control group was removed (20). The sequences were as follows:
miR-106a-inhibitor, 5′-GTAAGAAGTGCTTACATTGCAG-3′; miR-NC,
5′-TTCTCCGAACGTGTCACGT-3′; and miR-106a,
5′-ACGGGCCCTCTAGACTCGAGTGTTTTAACCAGGTGAGTC-3′. The sequence of
miR-106a binding site in GV272/VNN2 3′UTR was 5′-GCCATTGCAAA-3′,
and GV272/VNN2-mut 3′UTR was 5′-GCACGGTACAA-3′. All plasmid DNA was
extracted using an EndoFree Mini Plasmid kit (Tiangen Biotech Co.,
Ltd., Beijing, China). U2OS cells were transfected with the plasmid
using X-tremeGENE HP (Roche Diagnostics, Basel, Switzerland) and
infected at a multiplicity of infection of 10 for 12 h in the
incubator, then incubated for 72 h to perform the subsequent
trials.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from the paired specimens, and U2OS,
Saos-2, MG63, human hFOB1.19 cells and infected U2OS cells were
isolated using the QIAzol reagent (Qiagen, Inc., Valencia, CA, USA)
according to the manufacturer's protocol. The stem-loop
reverse-transcription primers were designed as follows: miR-106a,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCT-3′ and U6
small nuclear RNA (U6),
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGGGACAAA-3′. The oligo dT
primers were used for VNN2 and GAPDH genes. Reverse transcription
was performed using a PrimeScript RT reagent kit (Takara Bio, Inc.,
Otsu, Japan) according to the manufacturer's protocol. The
sequences of the PCR primers were designed as follows: miR-106a
forward, 5′-CGCAAAAGTGCTTACAGTGCA-3′ and reverse,
5′-GTGCAGGGTCCGAGGT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′; VNN2 forward,
5′-CCATAAGGTGGGCAAGAGTCA-3′ and reverse,
5′-CTCCGGCTTTTCAGGGACAT-3′; GAPDH forward,
5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse, 5′-TGGTGAAGACGCCAGTGGA-3′.
All PCR reactions were performed using an Applied Biosystems 7900HT
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) using the SYBR PrimeScript RT-PCR kit
(Takara Bio, Inc.) according to the manufacturer's protocol. The
thermocycling conditions maintained were as follows: 95°C for 30
sec; 95°C for 5 sec; and 60°C for 32 sec, for 40 cycles. To ensure
the fidelity of PCR reactions results, the expression levels of
miR-106a were normalized to the expression of U6, while the
expression levels of VNN2 were normalized to the expression of
GAPDH. The relative expression levels were calculated and
normalized using the 2−ΔΔCq method (21).
Cell proliferation assay
The proliferation ability of infected
miR-106a-inhibitor and miR-106a-NC U2OS cells was measured using an
MTS assay kit (Promega Corporation, Madison, WI, USA) according to
the manufacturer's protocol. Cells were plated (2×103
cells/well) in 96-well plates, and cultured for 24, 48, 72 and 96
h. In a 96-well plate, 20 µl MTS reagent was added for every 100 µl
medium/well and then incubated at 37°C in 5% CO2 for 4
h. The optical density value of each sample was recorded at the
wavelength of 490 nm via a microplate reader.
Cell migration and invasion
assays
The migration and invasion ability of infected U2OS
cells were detected using Transwell assays. The upper and lower
chambers of a 24-well plate (Corning Incorporated, Corning, NY,
USA) were washed with serum-free DMEM, and then 40 µl Matrigel (BD
Biosciences, San Jose, CA, USA) diluted 1:8 with serum-free medium
was added to the upper chamber to evenly cover the surface of the
polycarbonate membrane for the invasion assay, whereas no Matrigel
was added for migration assay. The 24-well plate was put into an
incubator for 4 h at 37°C to allow the Matrigel to solidify. A
total of 200 µl DMEM containing 1×105 cells was seeded
on the top of the upper chamber in the Transwell invasion chamber
while 600 µl DMEM with 10% FBS was added into the lower chamber.
The Transwell invasion assay was put in a cell incubator at 37°C
with 5% CO2. After 48 h, the cells on the upper surface
were wiped away using a wet cotton swab. For the adherent cells,
the migrated cells attach to the other side of the membrane in a
Transwell assay (22). Cells that
passed through the upper chamber and attach to the lower surface
were fixed using absolute ethanol for 15 min at room temperature
and stained with 0.1% crystal violet for 15 min at room
temperature. Visible cells were counted using an inverted phase
contrast microscope at a magnification of ×400.
Analysis of cell apoptosis
After infection for 72 h, cells were detached using
EDTA-free trypsin and washed three times with ice-cold PBS. The
cell apoptosis rate was detected using the FITC Annexin V Apoptosis
Detection kit (BD Biosciences) according to the manufacturer's
protocol using a flow cytometer.
Analysis of cell cycle
Cells in the logarithmic phase were harvested
following infection for 72 h and washed twice with ice-cold PBS,
then fixed with 5 ml 70% ethanol overnight at −20°C. Fixed cells
were washed twice with ice-cold PBS and then centrifuged at 800 × g
for 15 min at 4°C. Cells were resuspended in 0.4 ml ice-cold PBS
and subjected to 1 ml propidium iodide (PI)/Triton X-100 containing
RNase staining for 30 min at 4°C followed by flow cytometry. Data
were analyzed using the CellQuest software (version 7.5.3; BD
Biosciences).
Dual-luciferase reporter assay
Cells were seeded into a 24-well plate
(1×105 cells/well) and co-transfected with 0.4 µg of
miR-106a or miR-NC vectors, and 0.1 µg of GV272/VNN2 or
GV272/VNN2-mut vectors containing 3′-UTR as well as GV272/VNN2-NC
vector. The firefly luciferase reporter gene was constructed into
the vectors to quantitatively reflect the inhibitory effect. Cells
were harvested 48 h after transfection for luciferase activity
assays using the Reporter Assay system (Promega Corporation)
according to the manufacturer's protocol.
Western blot analysis
Cells were harvested 72 h after infection, and total
protein was extracted using radio immunoprecipitation assay lysis
buffer and phenylmethanesulfonyl fluoride (both from Beyotime
Institute of Biotechnology, Shanghai, China) according to the
manufacturer's protocol. Total protein (30 µg/lane) were separated
using 10% SDS-PAGE and electroblotted onto a 0.2-µm pore size
polyvinylidene fluoride membrane (Beyotime Institute of
Biotechnology). The membrane was blocked by 5% skim milk at room
temperature for 2 h and then incubated overnight at 4°C with rabbit
anti-human VNN2 antibody (1:1,000; cat. no., 25643-1-AP) or β-actin
antibody (1:1,000; cat. no., 20536-1-AP) (both from ProteinTech
Group, Inc., Chicago, IL, USA) as a control. Following washing of
the membrane three times with Tris-buffered saline containing
Tween-20 (1X TBST), the membrane was incubated for 2 h at room
temperature with horseradish peroxidase-conjugated affinipure goat
anti-rabbit IgG secondary antibody (1:5,000; cat. no., SA00001-2;
ProteinTech Group, Inc.). Following washing of the membrane three
times with TBST again, an ultrasensitive chemiluminescence solution
was used from the BeyoECL Plus kit (Beyotime Biotechnology,
Shanghai, China) to detect the protein bands according to the
manufacturer's protocol (23). The
bands were observed using MF-Chemisis 2.0 and GelCapture software
(DNR Bio-Imaging Systems, Ltd., Neve Yamin, Israel).
Bioinformatic and statistical
analysis
Open access miRNA databases (TargetScan, http://www.targetscan.org/vert_72/; PicTarget,
https://pictar.mdc-berlin.de/; and
MicroCosm, http://www.ebi.ac.uk/enright-srv/microcosm/) were used
for prediction of miR-106a target genes. SPSS software (version 22;
IBM Corp., Armonk, NY, USA) was used to analyze all results. All
statistical data are presented as mean ± standard deviation
following three independent experiments. Student's t-tests were
used when only two groups were present and one-way analysis of
variance followed by the Scheffe post hoc test was used when more
than two groups were present. The spearman's rank correlation test
was used to measure the correlation between the expression of
miR-106a and VNN2. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-106a is overexpressed in human
osteosarcoma cell lines and patient tissues
To quantify the expression of miR-106a in 18
osteosarcoma and non-cancerous adjacent tissues samples, RT-qPCR
was performed. The results indicated that the miR-106a expression
level was significantly increased in osteosarcoma tissues compared
with NTAs (Fig. 1A). In the human
U2OS, Saos-2 and MG63 cells, levels of miR-106a were significantly
upregulated, compared with that in normal hFOB1.19 osteoblast cells
(Fig. 1B). Since U2OS cells
exhibited the highest miR-106a expression level among the three
osteosarcoma cell lines, this cell line was chosen for further
studies.
Decreased miR-106a expression in human
U2OS cells affects cell proliferation and invasion
To explore the association of miR-106a expression
with human U2OS cells, the cells were infected with
lentivirus-mediated small interfering RNA (miR-106a-inhibitor).
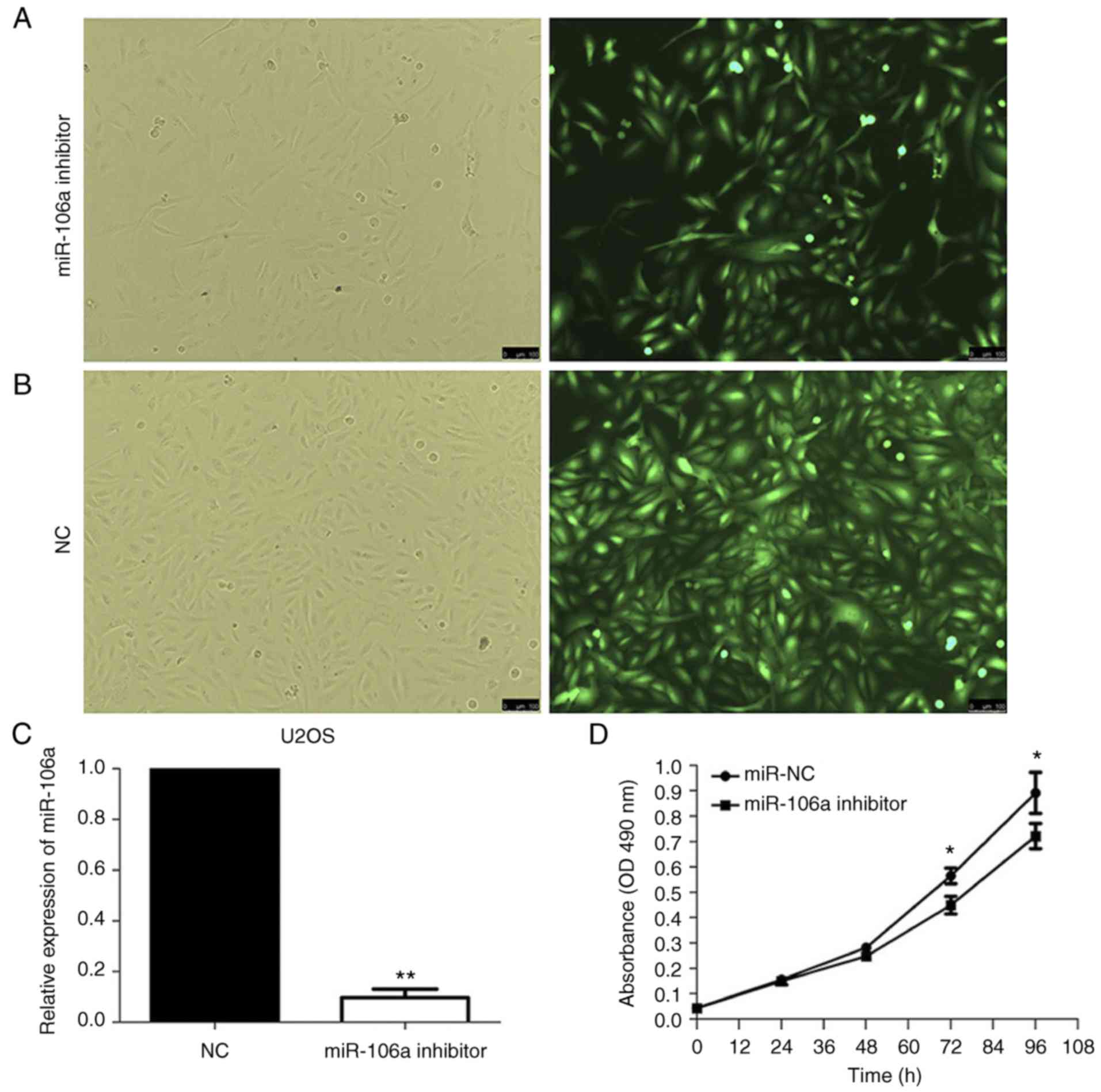
Infected cells labeled with green fluorescent protein were observed
under a fluorescent microscope, and the infection efficiency was
>90% when comparing the number of cells using the green
fluorescent view in the same field (Fig. 2A and B). Infection efficiency was
also determined by RT-qPCR at 72 h after infection (Fig. 2C).
The MTS assay used to investigate human U2OS cell
proliferation following infection with miR-106a-inhibitor or NC was
then performed. The MTS assay was performed at 24, 48, 72 and 96 h.
No significant differences were identified in absorbance at 490 nm
in the first 48 h. At 72 and 96 h, a significant decrease in the
absorbance at 490 nm was identified in human U2OS cells infected
with miR-106a-inhibitor compared with the NC-infected cells
(P<0.05; Fig. 2D). This
indicated that the knockdown of miR-106a expression may have
suppressed cell proliferation in human U2OS cells.
Next, an invasion assay was performed to investigate
the influence of knockdown of miR-106a of human U2OS cells on the
migration and invasion ability. Cells that passed through the upper
chamber and attached to the lower surface of the membrane were
fixed (Fig. 2E) and quantified
(Fig. 2F). The number of
miR-106a-inhibitor-infected U2OS cells that passed through the
upper chamber was significantly decreased compared with that of
miR-NC-infected cells.
Decreased miR-106a expression in human
U2OS cells influences the cell apoptosis ratio and cycle
distribution
In order to explore the mechanisms underlying the
inhibitory effect of miR-106a-inhibitor infection on cell
proliferation and invasion, flow cytometric analysis of apoptosis
and the cell cycle were applied (Fig.
3). As the Annexin V and PI staining results demonstrated,
knockdown of miR-106a resulted in a significant increase in the
apoptosis rate of human U2OS cells from 2.7 to 6.7% compared with
miR-NC-infected cells (P<0.05; Fig.
3A). The cell cycle analysis demonstrated that knockdown of
miR-106a significantly increased the percentage of cells in the
G2/M phase from 12.80 to 22.54% and decreased the percentage in the
S phase from 30.97 to 18.04% (both P<0.05; Fig. 3B).
Knockdown of miR-106a expression in
human U2OS cells leads to an increase in VNN2 expression
To explore whether VNN2 is a candidate target gene
of miR-106a, VNN2 protein expression levels were detected by
western blot analysis in human U2OS cells following the knockdown
of miR-106a (Fig. 2C). The results
revealed a significant upregulation in VNN2 protein expression in
knocked down U2OS cells compared with miR-NC-infected cells
(Fig. 4A).
RT-qPCR was performed to analyze the effect of
downregulating miR-106a expression on VNN2 expression at the
transcriptional level. The expression level of VNN2 mRNA was
significantly increased in miR-106a inhibitor-infected U2OS cells
compared with that in NC-infected U2OS cells (P<0.05; Fig. 4B). An inverse association between
miR-106a expression and VNN2 expression levels was verified via
RT-qPCR and western blot analysis.
miR-106a directly targets VNN2
By using open access online databases (TargetScan,
PicTarget and miRBase Targets), VNN2 was initially identified as a
candidate target gene of miR-106a. A complementary site of miR-106a
was identified in the VNN2 3′-UTR (Fig.
5A).
To confirm the possibility that miR-106a directly
targets VNN2, the miR-106a binding region at the 3′-UTR of VNN2 or
VNN2-mut mRNA were cloned downstream of the firefly luciferase
reporter gene in GV272 vectors. miR-106a or miR-NC vectors and
GV272 vectors were then co-transfected into 293T cells. The
relative luciferase activity of the reporter containing VNN2 3′-UTR
was significantly suppressed by 54±0.03% (P<0.05) when miR-106a
vectors were co-transfected, whereas the relative luciferase
activity of 3′-UTR-NC reporter was not affected (Fig. 5B). The results of luciferase
activity assay indicated that miR-106a may target and suppress VNN2
gene expression by binding the 3′-UTR of VNN2 mRNA.
VNN2 expression level is inversely
correlated with expression of miR-106a in human osteosarcoma
tissues
To quantify the expression of VNN2 in 18
osteosarcoma tissue and NTA samples, RT-qPCR was performed. The
results indicated that the VNN2 expression level was significantly
decreased in osteosarcoma tissues, compared with NTAs (Fig. 6A). Finally, the spearman's rank
correlation test was used to analyze the expression of VNN2 and
miR-106a in 18 osteosarcoma with non-cancerous adjacent tissues.
The result indicated a negative correlation between them (R=−0.640,
P<0.01; Fig. 6B).
Discussion
MicroRNAs emerged as novel gene regulators and have
been extensively studied in various types of human cancer (24,25).
Up to a third of the protein-coding genes in the human genome are
regulated by >2,000 microRNAs that have been discovered in human
so far (26). The function of
microRNA is primarily ascribed to the dynamic regulation of human
cancer as oncogenes or tumor suppressors (27). Based on previous studies, the same
type of microRNA exhibit similar functions in different types of
human cancer. For example, microRNA-21 has been detected to be
significantly upregulated in liver cancer, breast cancer and
malignant glioma (11,28,29).
MicroRNA-21 is able to promote the proliferation and invasion of
hepatocellular carcinoma cells by downregulating the expression of
tumor suppressor phosphatase and tensin homolog, while the
knockdown of microRNA-21 in malignant glioma results in caspase
activation, inducing an increase in apoptosis (29). It has also been reported that the
tumorigenic ability of the microRNA-21-knockdown MCF-7 breast
cancer cell line is significantly reduced in nude mice, and further
studies have demonstrated that microRNA-21 may promote cell
proliferation by inhibiting the target gene tropomyosin 1 (30). Furthermore, the upregulation of
miR-21 indicates a positive correlation with advanced
clinicopathological features and poor prognosis in patients of
renal cell carcinoma (31). These
studies indicate the proto-oncogene activity of microRNA-21 and
suggest that the same microRNA in different types of human cancer
has similar functions.
Recent several studies have reported that
microRNA-106a is frequently upregulated in different types of human
cancer, and is involved in tumor development, initiation,
progression, invasion and metastasis (32,33).
Shen et al (34)
demonstrated that the expression of microRNA-106a was increased in
thyroid cancer, and miRNA-106a directly targeting retinoic acid
receptor β was associated with the viability, apoptosis,
differentiation and the iodine uptake function of thyroid cancer
cell lines by regulating mitogen activated protein kinase signaling
pathway in vitro. Espinosa-Parrilla et al (35) identified that genetic variation in
microRNA-106a had an essential role in genetic susceptibility to
gastric cancer and contributed to the molecular mechanisms of
gastric carcinogenesis. These results demonstrated the important
role of microRNA-106a in liver and gastric cancer. However, there
is limited data available regarding the functional role and
underlying mechanism of microRNA-106a in osteosarcoma. Therefore,
the aim of the present study was to detect the expression level and
to explore the possible target of microRNA-106a in
osteosarcoma.
In the present study, it was revealed that
microRNA-106a was significantly upregulated in osteosarcoma tissue
compared with the adjacent normal tissue as well as in osteosarcoma
cell lines, which was contrary to another study whereby miR-106a-5p
was downregulated in osteosarcoma tissues (36). In the present study, knockdown of
microRNA-106a significantly the inhibited malignant phenotypic
processes of osteosarcoma, including proliferation, migration and
invasion. Cell cycle and apoptosis analysis were subsequently
applied to reveal the mechanisms underlying these phenotypic
changes. The cytometric results demonstrated that the knockdown of
microRNA-106a resulted in significantly increased apoptosis rates,
decreased the percentage of cells in the S phase and increased the
cell population in the G2/M phase. To determine the potential
target of microRNA-106a, online databases were used, which
identified VNN2 as a candidate gene. This result supports the
hypothesis that microRNA-106a acts as an oncogene by targeting
VNN2. To verify this hypothesis and the prediction of online
databases, RT-qPCR and western blotting were performed to determine
the expression of VNN2 in U2OS cells following knockdown of
microRNA-106a compared with NC. There was reasonable concordance
whereby the expression of VNN2 was significantly upregulated in
both cases. Additional, the expression of VNN2 in osteosarcoma
tissue compared with the adjacent normal tissue was detected by
RT-qPCR. It was demonstrated that the VNN2 expression level was
significantly decreased in osteosarcoma tissues compared with NTAs,
and an inverse correlation between miR-106a and VNN2 expression was
observed. Additionally, dual-luciferase was performed to further
confirm the possibility that microRNA-106a targets VNN2. The
relative luciferase activity of VNN2 3′-UTR group was significantly
suppressed, which indicated that microRNA-106a was able to bind to
the 3′-UTR of VNN2 mRNA, thereby affecting proliferation, invasion
and apoptosis in osteosarcoma.
It is generally known that oxidative phosphorylation
is replaced by aerobic glycolysis in tumor cells, which only makes
use of metabolic recombination to maintain intracellular ATP and
NADH levels instead of obtaining ATP via mitochondria (37). An enormous amount of ATP and NADH
are necessary for tumor cell proliferation and metastasis, and also
to maintain the base metabolism of tumor cells. Destruction of
tumor cell energy and redox substances may be associated with the
molecular mechanisms that underlie metabolic reprogramming of
cancer cells (38). VNN2 as a
pantetheine hydrolase serves an important role in the pantothenate
and coenzyme A biosynthesis pathway (39). Considering the cell cycle
dysregulation and apoptosis rate changes observed in the present
study, we hypothesized that VNN2 regulates the aerobic glycolysis
of tumor cells and alter metabolite-driven gene regulation of
osteosarcoma cells to induce apoptosis and cell cycle changes.
Verification of this hypothesis is planned in future studies.
In conclusion, the overexpression of microRNA-106a
in human osteosarcoma tissues and U2OS cell line was observed, and
the knockdown of microRNA-106a was demonstrated to inhibit
osteosarcoma cell proliferation and invasion and induce apoptosis
by targeting VNN2. The current study identified microRNA-106a may
be a potential novel diagnostic marker and targeting
microRNA-106a-VNN2 interaction may be a potential therapeutic
strategy in human osteosarcoma. However, the function of VNN2 in
osteosarcoma remains unclear, and further studies are required to
identify the underlying mechanisms of VNN2 effects on osteosarcoma
cells.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Science and Technology Department of Liaoning
Province (grant no. 201302106) and the Science and Technology
Department of Shenyang City (grant no. F14-231-1-48).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HT conceived and designed the experiments. CY, YX
and LC performed the experiments. CY, LC and ZS wrote the
manuscript. CY, LP, WZ and ZS performed the data analysis. All
authors read and approved the manuscript.
Ethics approval and consent to
participate
The present study received approval by the Ethics
Committee of the First Affiliated Hospital of China Medical
University (Shenyang, China) and written informed consent was
obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ward E, DeSantis C, Robbins A, Kohler B
and Jemal A: Childhood and adolescent cancer statistics, 2014. CA
Cancer J Clin. 64:83–103. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the Surveillance, Epidemiology, and End Results program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bielack SS, Kempf-Bielack B, Delling G,
Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M,
Winkelmann W, et al: Prognostic factors in high-grade osteosarcoma
of the extremities or trunk: An analysis of 1,702 patients treated
on neoadjuvant cooperative osteosarcoma study group protocols. J
Clin Oncol. 20:776–790. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chou AJ, Kleinerman ES, Krailo MD, Chen Z,
Betcher DL, Healey JH, Conrad EU III, Nieder ML, Weiner MA, Wells
RJ, et al: Addition of muramyl tripeptide to chemotherapy for
patients with newly diagnosed metastatic osteosarcoma: A report
from the Children's Oncology Group. Cancer. 115:5339–5348. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Aung L, Tin AS, Quah TC and Pho RW:
Osteogenic sarcoma in children and young adults. Ann Acad Med
Singapore. 43:305–313. 2014.PubMed/NCBI
|
|
6
|
Behm-Ansmant I, Rehwinkel J and Izaurralde
E: MicroRNAs silence gene expression by repressing protein
expression and/or by promoting mRNA decay. Cold Spring Harb Symp
Quant Biol. 71:523–530. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambros V: MicroRNA pathways in flies and
worms: Growth, death, fat, stress, and timing. Cell. 113:673–676.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Song L and Tuan RS: MicroRNAs and cell
differentiation in mammalian development. Birth Defects Res C
Embryo Today. 78:140–149. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Skaftnesmo KO, Prestegarden L, Micklem DR
and Lorens JB: MicroRNAs in tumorigenesis. Curr Pharm Biotechnol.
8:320–325. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lulla RR, Costa FF, Bischof JM, Chou PM,
de F Bonaldo M, Vanin EF and Soares MB: Identification of
differentially expressed MicroRNAs in osteosarcoma. Sarcoma.
2011:7326902011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He L, He X, Lim LP, de Stanchina E, Xuan
Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting
BCL2. Proc Natl Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Palmini G, Marini F and Brandi ML: What is
new in the miRNAWorld regarding osteosarcoma and chondrosarcoma?
Molecule. 22:pii: E417. 2017. View Article : Google Scholar
|
|
15
|
Huang J, Takeda Y, Watanabe T and Sendo F:
A sandwich ELISA for detection of soluble GPI-80, a
glycosylphosphatidyl-inositol (GPI)-anchored protein on human
leukocytes involved in regulation of neutrophil adherence and
migration-its release from activated neutrophils and presence in
synovial fluid of rheumatoid arthritis patients. Microbiol Immunol.
45:467–471. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Suzuki K, Watanabe T, Sakurai S, Ohtake K,
Kinoshita T, Araki A, Fujita T, Takei H, Takeda Y, Sato Y, et al: A
novel glycosylphosphatidyl inositol-anchored protein on human
leukocytes: A possible role for regulation of neutrophil adherence
and migration. J Immunol. 162:4277–4284. 1999.PubMed/NCBI
|
|
17
|
Singel KL and Segal BH: Neutrophils in the
tumor microenvironment: Trying to heal the wound that cannot heal.
Immunol Rev. 273:329–343. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Eruslanov EB, Bhojnagarwala PS, Quatromoni
JG, Stephen TL, Ranganathan A, Deshpande C, Akimova T, Vachani A,
Litzky L, Hancock WW, et al: Tumor-associated neutrophils stimulate
T cell responses in early-stage human lung cancer. J Clin Invest.
124:5466–5480. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nitto T, Araki Y, Takeda Y and Sendo F:
Pharmacological analysis for mechanisms of GPI-80 release from
tumour necrosis factor-alpha-stimulated human neutrophils. Br J
Pharmacol. 137:353–360. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li P, Liu H, Zhang Z, Lv X, Wang H, Ma J,
Ma Z, Qu X and Teng YE: Expression and comparison of Cbl-b in lung
squamous cell carcinoma and adenocarcinoma. Med Sci Monit.
24:623–635. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Justus CR, Leffler N, Ruiz-Echevarria M
and Yang LV: In vitro cell migration and invasion assays. J Vis
Exp. Jun 1–2014.doi: 10.3791/51046. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mahmood T and Yang PC: Western blot:
Technique, theory, and trouble shooting. N Am J Med Sci. 4:429–434.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu S, Si ML, Wu H and Mo YY: MicroRNA-21
targets the tumor suppressor gene tropomyosin 1 (TPM1). J
Biol Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu Z and Lu Y, Xiao Y and Lu Y:
Upregulation of miR-21 expression is a valuable predicator of
advanced clinicopathological features and poor prognosis in
patients with renal cell carcinoma through the p53/p21-cyclin
E2-Bax/caspase-3 signaling pathway. Oncol Rep. 37:1437–1444. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Edatt L, Maurya AK, Raji G, Kunhiraman H
and Kumar SV: MicroRNA106a regulates matrix metalloprotease 9 in a
sirtuin-1 dependent mechanism. J Cell Physiol. 233:238–248. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pan YJ, Zhuang Y, Zheng JN and Pei DS:
MiR-106a: Promising biomarker for cancer. Bioorg Med Chem Lett.
26:5373–5377. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shen CT, Qiu ZL, Song HJ, Wei WJ and Luo
QY: miRNA-106a directly targeting RARB associates with the
expression of Na+/I− symporter in thyroid
cancer by regulating MAPK signaling pathway. J Exp Clin Cancer Res.
35:1012016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Espinosa-Parrilla Y, Munoz X, Bonet C,
Garcia N, Venceslá A, Yiannakouris N, Naccarati A, Sieri S, Panico
S, Huerta JM, et al: Genetic association of gastric cancer with
miRNA clusters including the cancer-related genes MIR29, MIR25,
MIR93 and MIR106: Results from the EPIC-EURGAST study.
Int J Cancer. 135:2065–2076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
He QY, Wang GC, Zhang H, Tong DK, Ding C,
Liu K, Ji F, Zhu X and Yang S: miR-106a-5p suppresses the
proliferation, migration, and invasion of osteosarcoma cells by
targeting HMGA2. DNA Cell Biol. 35:506–520. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kroemer G and Pouyssegur J: Tumor cell
metabolism: Cancer's Achilles' heel. Cancer Cell. 13:472–482. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lopez-Rios F, Sanchez-Arago M,
Garcia-Garcia E, Ortega AD, Berrendero JR, Pozo-Rodríguez F,
López-Encuentra A, Ballestín C and Cuezva JM: Loss of the
mitochondrial bioenergetic capacity underlies the glucose avidity
of carcinomas. Cancer Res. 67:9013–9017. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nitto T and Onodera K: Linkage between
coenzyme a metabolism and inflammation: Roles of pantetheinase. J
Pharmacol Sci. 123:1–8. 2013. View Article : Google Scholar : PubMed/NCBI
|