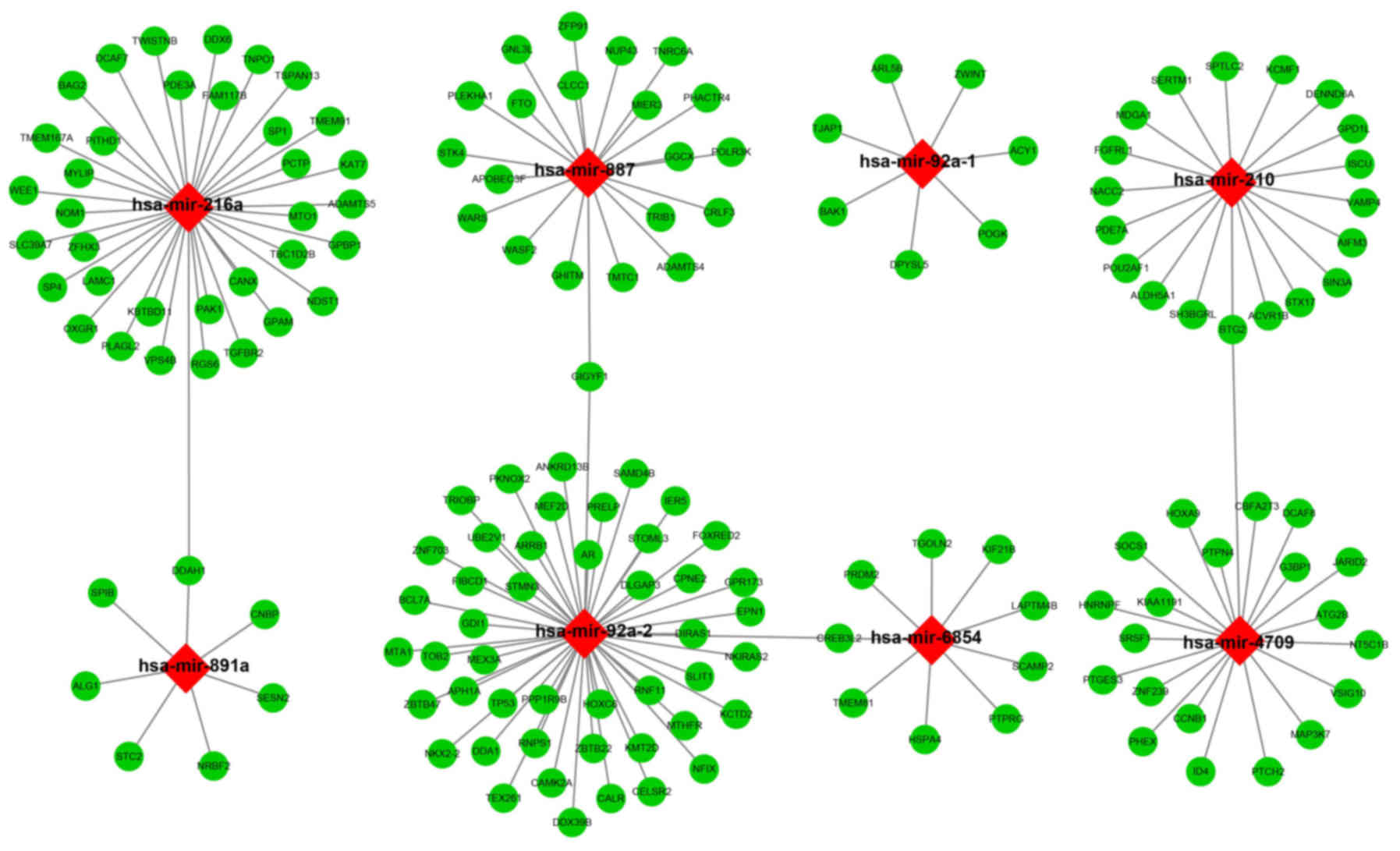
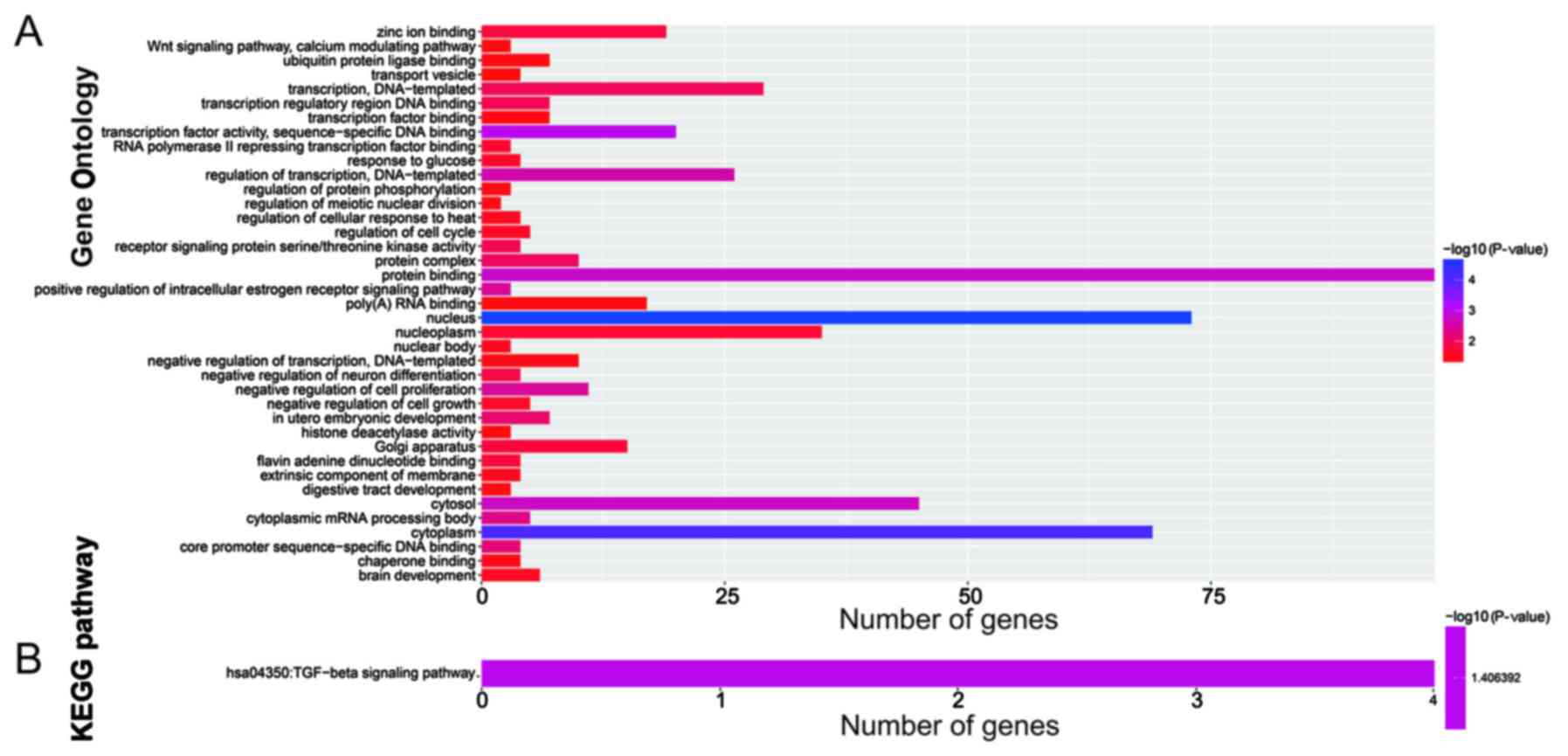
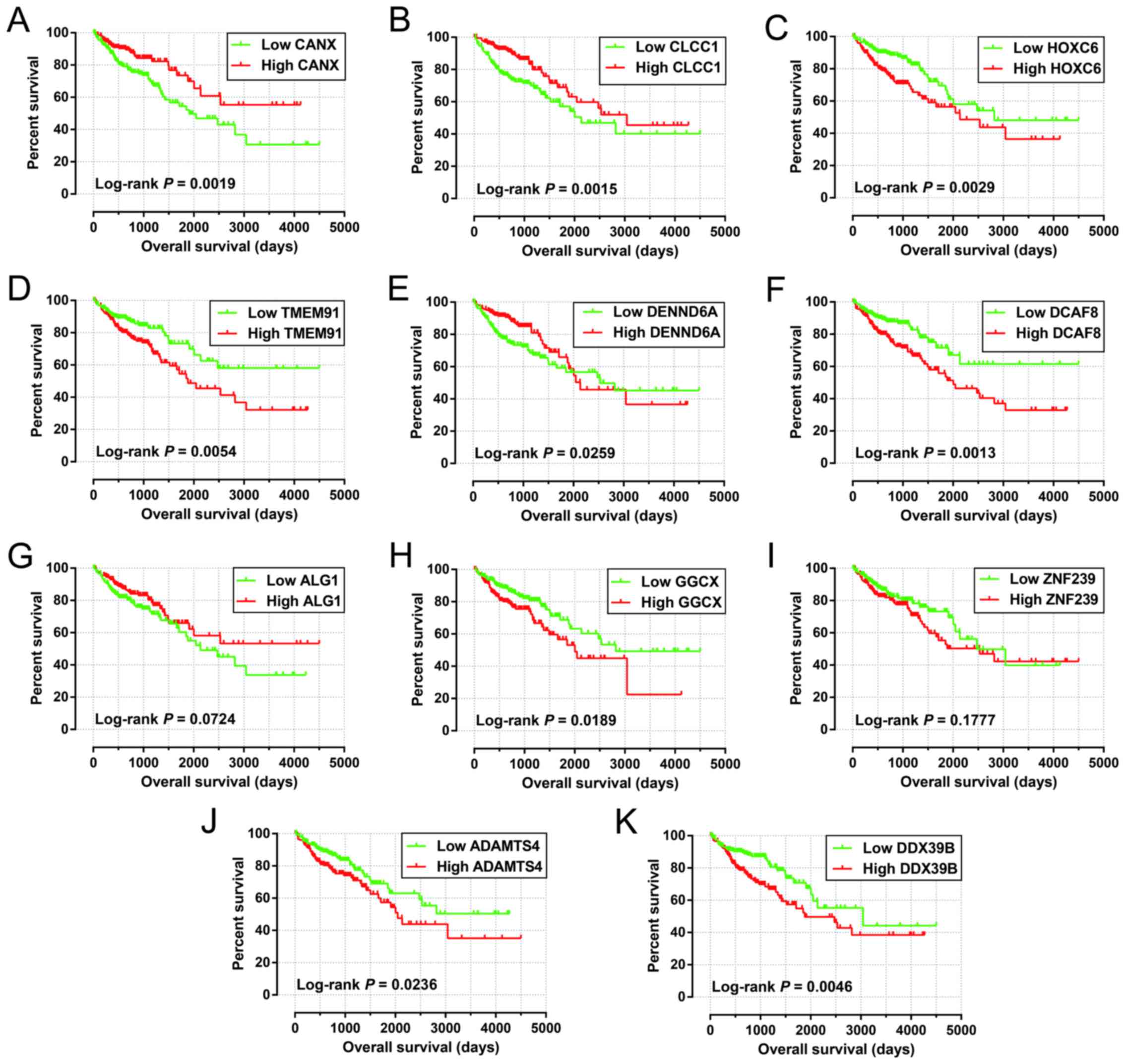
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA:
Cancer J Clin. 65:87–108. 2015.PubMed/NCBI
|
|
2
|
McGuire S: World cancer report 2014.
Geneva, Switzerland: World Health Organization, International
Agency for Research on Cancer, WHO press, 2015. Adv Nutr.
7:418–419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in china,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zeng H, Zheng R, Guo Y, Zhang S, Zou X,
Wang N, Zhang L, Tang J, Chen J, Wei K, et al: Cancer survival in
China, 2003–2005: A population-based study. Int J Cancer.
136:1921–1930. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Towler BP, Jones CI and Newbury SF:
Mechanisms of regulation of mature miRNAs. Biochem Soc Trans.
43:1208–1214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Strubberg AM and Madison BB: MicroRNAs in
the etiology of colorectal cancer: Pathways and clinical
implications. Dis Model Mech. 10:197–214. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tomczak K, Czerwinska P and Wiznerowicz M:
The Cancer Genome Atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol. 19:A68–A77. 2015.
|
|
9
|
Cancer Genome Atlas Research Network, .
Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA,
Ellrott K, Shmulevich I, Sander C and Stuart JM: The cancer genome
atlas pan-cancer analysis project. Nat Genet. 45:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cancer Genome Atlas Network: Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11:R1062010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liao X, Huang K, Huang R, Liu X, Han C, Yu
L, Yu T, Yang C, Wang X and Peng T: Genome-scale analysis to
identify prognostic markers in patients with early-stage pancreatic
ductal adenocarcinoma after pancreaticoduodenectomy. OncoTargets
Ther. 10:4493–4506. 2017. View Article : Google Scholar
|
|
13
|
Zhou M, Zhao H, Wang Z, Cheng L, Yang L,
Shi H, Yang H and Sun J: Identification and validation of potential
prognostic lncRNA biomarkers for predicting survival in patients
with multiple myeloma. J Exp Clin Cancer Res. 34:1022015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang R, Liao X and Li Q: Identification
and validation of potential prognostic gene biomarkers for
predicting survival in patients with acute myeloid leukemia.
OncoTargets Ther. 10:5243–5254. 2017. View Article : Google Scholar
|
|
15
|
Lossos IS, Czerwinski DK, Alizadeh AA,
Wechser MA, Tibshirani R, Botstein D and Levy R: Prediction of
survival in diffuse large-B-cell lymphoma based on the expression
of six genes. N Engl J Med. 350:1828–1837. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Heagerty PJ and Zheng Y: Survival model
predictive accuracy and ROC curves. Biometrics. 61:92–105. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:doi: 10.7554/eLife.05005. 2015. View Article : Google Scholar
|
|
18
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang X: Improving microRNA target
prediction by modeling with unambiguously identified
microRNA-target pairs from CLIP-ligation studies. Bioinformatics.
32:1316–1322. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:D146–D152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chou CH, Shrestha S, Yang CD, Chang NW,
Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al: miRTarBase
update 2018: A resource for experimentally validated
microRNA-target interactions. Nucleic Acids Res. 46:D296–D302.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hsu SD, Lin FM, Wu WY, Liang C, Huang WC,
Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al: miRTarBase: A
database curates experimentally validated microRNA-target
interactions. Nucleic Acids Res. 39:D163–D169. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
da Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cancer Genome Atlas Research Network.
Electronic address simplewheeler@bcm.edu; Cancer
Genome Atlas Research Network: Comprehensive and integrative
genomic characterization of hepatocellular carcinoma. Cell.
169(1327–1341): e13232017.
|
|
26
|
Wang JY, Wang CL, Wang XM and Liu FJ:
Comprehensive analysis of microRNA/mRNA signature in colon
adenocarcinoma. Eur Rev Med Pharmacol Sci. 21:2114–2129.
2017.PubMed/NCBI
|
|
27
|
Yang J, Ma D, Fesler A, Zhai H,
Leamniramit A, Li W, Wu S and Ju J: Expression analysis of microRNA
as prognostic biomarkers in colorectal cancer. Oncotarget.
8:52403–52412. 2016.PubMed/NCBI
|
|
28
|
Jacob H, Stanisavljevic L, Storli KE,
Hestetun KE, Dahl O and Myklebust MP: Identification of a
sixteen-microRNA signature as prognostic biomarker for stage II and
III colon cancer. Oncotarget. 8:87837–87847. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS,
Zhang XS, Cui J, Zeng YX and Li J: Tumor-derived exosomes promote
tumor progression and T-cell dysfunction through the regulation of
enriched exosomal microRNAs in human nasopharyngeal carcinoma.
Oncotarget. 5:5439–5452. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiaoli Z, Yawei W, Lianna L, Haifeng L and
Hui Z: Screening of target genes and regulatory function of mirnas
as prognostic indicators for prostate cancer. Med Sci Monit.
21:3748–3759. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhao JY, Wang F, Li Y, Zhang XB, Yang L,
Wang W, Xu H, Liu DZ and Zhang LY: Five mirnas considered as
molecular targets for predicting esophageal cancer. Med Sci Monit.
21:3222–3230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ranade AR, Cherba D, Sridhar S, Richardson
P, Webb C, Paripati A, Bowles B and Weiss GJ: MicroRNA 92a-2*: A
biomarker predictive for chemoresistance and prognostic for
survival in patients with small cell lung cancer. J Thorac Oncol.
5:1273–1278. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang Y, Wang N, Yin D, Li YK, Guo L, Shi
LP and Huang X: Changes in the expression of serum MiR-887-5p in
patients with endometrial cancer. Int J Gynecolo Cancer.
26:1143–1147. 2016. View Article : Google Scholar
|
|
34
|
Xia H, Ooi LL and Hui KM:
MicroRNA-216a/217-induced epithelial-mesenchymal transition targets
PTEN and SMAD7 to promote drug resistance and recurrence of liver
cancer. Hepatology. 58:629–641. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang D, Zhao L, Shen Q, Lv Q, Jin M, Ma
H, Nie X, Zheng X, Huang S, Zhou P, et al: Down-regulation of
KIAA1199/CEMIP by miR-216a suppresses tumor invasion and metastasis
in colorectal cancer. Int J Cancer. 140:2298–2309. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang RT, Xu M, Xu CX, Song ZG and Jin H:
Decreased expression of miR216a contributes to non-small-cell lung
cancer progression. Clin Cancer Res. 20:4705–4716. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li L and Ma HQ: MicroRNA-216a inhibits the
growth and metastasis of oral squamous cell carcinoma by targeting
eukaryotic translation initiation factor 4B. Mol Med Rep.
12:3156–3162. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lu J, Li X, Wang F, Guo Y, Huang Y, Zhu H,
Wang Y, Lu Y and Wang Z: YB-1 expression promotes pancreatic cancer
metastasis that is inhibited by microRNA-216a. Exp Cell Res.
359:319–326. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Y, Tang X, Shi M, Wen C and Shen B:
MiR-216a decreases MALAT1 expression, induces G2/M arrest and
apoptosis in pancreatic cancer cells. Biochem Biophys Res Commun.
483:816–822. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hou BH, Jian ZX, Cui P, Li SJ, Tian RQ and
Ou JR: miR-216a may inhibit pancreatic tumor growth by targeting
JAK2. FEBS Lett. 589:2224–2232. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang S, Chen X and Tang M: MicroRNA-216a
inhibits pancreatic cancer by directly targeting Janus kinase 2.
Oncol Rep. 32:2824–2830. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bavelloni A, Ramazzotti G, Poli A, Piazzi
M, Focaccia E, Blalock W and Faenza I: MiRNA-210: A current
overview. Anticancer Res. 37:6511–6521. 2017.PubMed/NCBI
|
|
43
|
Huang X and Zuo J: Emerging roles of
miR-210 and other non-coding RNAs in the hypoxic response. Acta
Biochim Biophys Sin. 46:220–232. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen J, Wang W, Zhang Y, Chen Y and Hu T:
Predicting distant metastasis and chemoresistance using plasma
miRNAs. Med Oncol. 31:7992014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ullmann P, Qureshi-Baig K, Rodriguez F,
Ginolhac A, Nonnenmacher Y, Ternes D, Weiler J, Gäbler K, Bahlawane
C, Hiller K, et al: Hypoxia-responsive miR-210 promotes
self-renewal capacity of colon tumor-initiating cells by repressing
ISCU and by inducing lactate production. Oncotarget. 7:65454–65470.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nijhuis A, Thompson H, Adam J, Parker A,
Gammon L, Lewis A, Bundy JG, Soga T, Jalaly A, Propper D, et al:
Remodelling of microRNAs in colorectal cancer by hypoxia alters
metabolism profiles and 5-fluorouracil resistance. Hum Mol Genet.
26:1552–1564. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sun Y, Xing X, Liu Q, Wang Z, Xin Y, Zhang
P, Hu C and Liu Y: Hypoxia-induced autophagy reduces
radiosensitivity by the HIF-1alpha/miR-210/Bcl-2 pathway in colon
cancer cells. Int J Oncol. 46:750–756. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang J, Zhao J, Shi M, Ding Y, Sun H, Yuan
F and Zou Z: Elevated expression of miR-210 predicts poor survival
of cancer patients: A systematic review and meta-analysis. PLoS
One. 9:e892232014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhan M, Li Y, Hu B, He X, Huang J, Zhao Y,
Fu S and Lu L: Serum microRNA-210 as a predictive biomarker for
treatment response and prognosis in patients with hepatocellular
carcinoma undergoing transarterial chemoembolization. J Vasc Interv
Radiol. 25(1279–1287): e12712014.
|
|
50
|
Li Y, Ma X, Zhao J, Zhang B, Jing Z and
Liu L: microRNA-210 as a prognostic factor in patients with breast
cancer: Meta-analysis. Cancer Biomark. 13:471–481. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hong L, Yang J, Han Y, Lu Q, Cao J and
Syed L: High expression of miR-210 predicts poor survival in
patients with breast cancer: A meta-analysis. Gene. 507:135–138.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Camps C, Buffa FM, Colella S, Moore J,
Sotiriou C, Sheldon H, Harris AL, Gleadle JM and Ragoussis J:
hsa-miR-210 is induced by hypoxia and is an independent prognostic
factor in breast cancer. Clin Cancer Res. 14:1340–1348. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Toyama T, Kondo N, Endo Y, Sugiura H,
Yoshimoto N, Iwasa M, Takahashi S, Fujii Y and Yamashita H: High
expression of microRNA-210 is an independent factor indicating a
poor prognosis in Japanese triple-negative breast cancer patients.
Jpn J Clin Oncol. 42:256–263. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lai NS, Dong QS, Ding H, Miao ZL and Lin
YC: MicroRNA-210 overexpression predicts poorer prognosis in glioma
patients. J Clin Neurosci. 21:755–760. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lai NS, Wu DG, Fang XG, Lin YC, Chen SS,
Li ZB and Xu SS: Serum microRNA-210 as a potential noninvasive
biomarker for the diagnosis and prognosis of glioma. Br J Cancer.
112:1241–1246. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Cai H, Lin L, Cai H, Tang M and Wang Z:
Prognostic evaluation of microRNA-210 expression in pediatric
osteosarcoma. Med Oncol. 30:4992013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Eilertsen M, Andersen S, Al-Saad S,
Richardsen E, Stenvold H, Hald SM, Al-Shibli K, Donnem T, Busund LT
and Bremnes RM: Positive prognostic impact of miR-210 in non-small
cell lung cancer. Lung Cancer. 83:272–278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
McCormick RI, Blick C, Ragoussis J,
Schoedel J, Mole DR, Young AC, Selby PJ, Banks RE and Harris AL:
miR-210 is a target of hypoxia-inducible factors 1 and 2 in renal
cancer, regulates ISCU and correlates with good prognosis. Br J
Cancer. 108:1133–1142. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ma J, Sun F, Li C, Zhang Y, Xiao W, Li Z,
Pan Q, Zeng H, Xiao G, Yao K, et al: Depletion of intermediate
filament protein Nestin, a target of microRNA-940, suppresses
tumorigenesis by inducing spontaneous DNA damage accumulation in
human nasopharyngeal carcinoma. Cell Death Dis. 5:e13772014.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Ding D, Zhang Y, Yang R, Wang X, Ji G, Huo
L, Shao Z and Li X: miR-940 Suppresses tumor cell invasion and
migration via regulation of CXCR2 in hepatocellular carcinoma.
Biomed Res Int. 2016:76183422016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yuan B, Liang Y, Wang D and Luo F: MiR-940
inhibits hepatocellular carcinoma growth and correlates with
prognosis of hepatocellular carcinoma patients. Cancer Sci.
106:819–824. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang F, Wang Z, Gu X and Cui J: miR-940
Upregulation suppresses cell proliferation and induces apoptosis by
targeting PKC-δ in ovarian cancer OVCAR3 cells. Oncol Res.
25:107–114. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Rashed MH, Kanlikilicer P,
Rodriguez-Aguayo C, Pichler M, Bayraktar R, Bayraktar E, Ivan C,
Filant J, Silva A, Aslan B, et al: Exosomal miR-940 maintains
SRC-mediated oncogenic activity in cancer cells: A possible role
for exosomal disposal of tumor suppressor miRNAs. Oncotarget.
8:20145–20164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Rajendiran S, Parwani AV, Hare RJ,
Dasgupta S, Roby RK and Vishwanatha JK: MicroRNA-940 suppresses
prostate cancer migration and invasion by regulating MIEN1. Mol
Cancer. 13:2502014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hou L, Chen M, Yang H, Xing T, Li J, Li G,
Zhang L, Deng S, Hu J, Zhao X, et al: MiR-940 inhibited cell growth
and migration in triple-negative breast cancer. Med Sci Monit.
22:3666–3672. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Song B, Zhang C, Li G, Jin G and Liu C:
MiR-940 inhibited pancreatic ductal adenocarcinoma growth by
targeting MyD88. Cell Physiol Biochem. 35:1167–1177. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu X, Ge X, Zhang Z, Zhang X, Chang J, Wu
Z, Tang W, Gan L, Sun M and Li J: MicroRNA-940 promotes tumor cell
invasion and metastasis by downregulating ZNF24 in gastric cancer.
Oncotarget. 6:25418–25428. 2015.PubMed/NCBI
|
|
68
|
Yang HW, Liu GH, Liu YQ, Zhao HC, Yang Z,
Zhao CL, Zhang XF and Ye H: Over-expression of microRNA-940
promotes cell proliferation by targeting GSK3beta and sFRP1 in
human pancreatic carcinoma. Biomed Pharmacother. 83:593–601. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Liu X, Kwong A, Sihoe A and Chu KM: Plasma
miR-940 may serve as a novel biomarker for gastric cancer. Tumour
Biol. 37:3589–3597. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Xie Z, Yin X, Gong B, Nie W, Wu B, Zhang
X, Huang J, Zhang P, Zhou Z and Li Z: Salivary microRNAs show
potential as a noninvasive biomarker for detecting resectable
pancreatic cancer. Cancer Prev Res. 8:165–173. 2015. View Article : Google Scholar
|