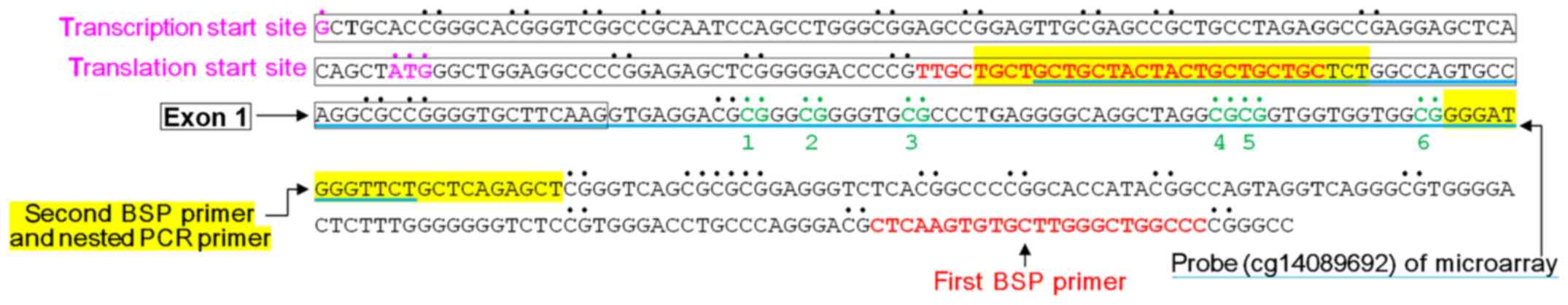
|
1
|
Stewart BW and Wild C: World Cancer Report
2014. Lyon, France: International Agency for Research on Cancer.
World Health Organization. 630:2014.
|
|
2
|
Halden RU: Plastics and health risks. Annu
Rev Public Health. 31:179–194. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Erkekoglu P and Kocer-Gumusel B:
Environmental effects of endocrine-disrupting chemicals: A special
focus on phthalates and bisphenol A. Environ Health. 6:1–36.
2016.
|
|
4
|
Solecki R, Kortenkamp A, Bergman Å,
Chahoud I, Degen GH, Dietrich D, Greim H, Håkansson H, Hass U,
Husoy T, et al: Scientific principles for the identification of
endocrine-disrupting chemicals: A consensus statement. Arch
Toxicol. 91:1001–1006. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Diamanti-Kandarakis E, Bourguignon JP,
Giudice LC, Hauser R, Prins GS, Soto AM, Zoeller RT and Gore AC:
Endocrine-disrupting chemicals: An Endocrine Society scientific
statement. Endocr Rev. 30:293–342. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schug TT, Janesick A, Blumberg B and
Heindel JJ: Endocrine disrupting chemicals and disease
susceptibility. J Steroid Biochem Mol Biol. 127:204–215. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Buluş AD, Aşci A, Erkekoglu P, Balci A,
Andiran N and Koçer-Gümüşel B: The evaluation of possible role of
endocrine disruptors in central and peripheral precocious puberty.
Toxicol Mech Methods. 26:493–500. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Orth P, Reichert P, Wang W, Prosise WW,
Yarosh-Tomaine T, Hammond G, Ingram RN, Xiao L, Mirza UA, Zou J, et
al: Crystal structure of the catalytic domain of human ADAM33. J
Mol Biol. 335:129–137. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Seniski GG, Camargo AA, Ierardi DF, Ramos
EA, Grochoski M, Ribeiro ES, Cavalli IJ, Pedrosa FO, de Souza EM,
Zanata SM, et al: ADAM33 gene silencing by promoter
hypermethylation as a molecular marker in breast invasive lobular
carcinoma. BMC Cancer. 9:802009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Topal O, Erinanc H, Ozer C, Canpolat ET,
Celik SB and Erbek SS: Expression of ‘a disintegrin and
metalloproteinase-33’ (ADAM-33) protein in laryngeal squamous cell
carcinoma. J Laryngol Otol. 126:511–515. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim KE, Song H, Hahm C, Yoon SY, Park S,
Lee HR, Hur DY, Kim T, Kim CH, Bang SI, et al: Expression of ADAM33
is a novel regulatory mechanism in IL-18-secreted process in
gastric cancer. J Immunol. 182:3548–3555. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Treviño LS, Wang Q and Walker CL:
Hypothesis: Activation of rapid signaling by environmental
estrogens and epigenetic reprogramming in breast cancer. Reprod
Toxicol. 54:136–140. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nephew KP and Huang TH: Epigenetic gene
silencing in cancer initiation and progression. Cancer Lett.
190:125–133. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gong C, Fujino K, Monteiro LJ, Gomes AR,
Drost R, Davidson-Smith H, Takeda S, Khoo US, Jonkers J, Sproul D,
et al: FOXA1 repression is associated with loss of BRCA1 and
increased promoter methylation and chromatin silencing in breast
cancer. Oncogene. 34:5012–5024. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Manica GC, Ribeiro CF, Oliveira MA,
Pereira IT, Chequin A, Ramos EA, Klassen LM, Sebastião AP,
Alvarenga LM, Zanata SM, et al: Down regulation of ADAM33 as
a predictive biomarker of aggressive breast cancer. Sci Rep.
7:444142017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Costa FF, Verbisck NV, Salim AC, Ierardi
DF, Pires LC, Sasahara RM, Sogayar MC, Zanata SM, Mackay A, O'Hare
M, et al: Epigenetic silencing of the adhesion molecule ADAM23 is
highly frequent in breast tumors. Oncogene. 23:1481–1488. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Singh S and Li SS: Epigenetic effects of
environmental chemicals bisphenol A and phthalates. Int J Mol Sci.
13:10143–10153. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Edge SB; and Cancer AJCo, : AJCC Cancer
Staging Handbook: From The AJCC Cancer Staging Manual. Springer;
New York, NY: 2010
|
|
19
|
Anjum S, Fourkala EO, Zikan M, Wong A,
Gentry-Maharaj A, Jones A, Hardy R, Cibula D, Kuh D, Jacobs IJ, et
al: A BRCA1-mutation associated DNA methylation signature in
blood cells predicts sporadic breast cancer incidence and survival.
Genome Med. 6:472014. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fackler MS, Bujanda ZL, Umbricht C, Teo W,
Zhang Z, Visvanathan K, Jeter S, Argani P, Wang C, Lyman JP, et al:
Detection of hypermethylated circulating serum DNA in metastatic
breast cancer and confirmation by the cMethDNA assay. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE52621March
1–2014
|
|
21
|
Zhuang J, Jones A, Lee SH, Ng E, Fiegl H,
Zikan M, Cibula D, Sargent A, Salvesen HB, Jacobs IJ, et al: The
dynamics and prognostic potential of DNA methylation changes at
stem cell gene loci in women's cancer. PLoS Genet. 8:e10025172012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dedeurwaerder S, Desmedt C, Calonne E,
Singhal SK, Haibe-Kains B, Defrance M, Michiels S, Volkmar M,
Deplus R, Luciani J, et al: DNA methylation profiling reveals a
predominant immune component in breast cancers. EMBO Mol Med.
3:726–741. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fackler MJ, Umbricht CB, Williams D,
Argani P, Cruz LA, Merino VF, Teo WW, Zhang Z, Huang P,
Visvananthan K, et al: Genome-wide methylation analysis identifies
genes specific to breast cancer hormone receptor status and risk of
recurrence. Cancer Res. 71:6195–6207. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bibikova M, Lin Z, Zhou L, Chudin E,
Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, et al:
High-throughput DNA methylation profiling using universal bead
arrays. Genome Res. 16:383–393. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen X, Li S, Zhang L, Li H, Hong G, Zhou
X, Zheng T, Zhang W, Hao C, Shi T, et al: An integrated approach to
uncover driver genes in breast cancer methylation genomes. PLoS
One. 8:e612142013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takai D and Jones PA: The CpG island
searcher: A new WWW resource. In Silico Biol. 3:235–240.
2003.PubMed/NCBI
|
|
27
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rohde C, Zhang Y, Reinhardt R and Jeltsch
A: BISMA - fast and accurate bisulfite sequencing data analysis of
individual clones from unique and repetitive sequences. BMC
Bioinformatics. 11:2302010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bremnes RM, Dønnem T, Al-Saad S, Al-Shibli
K, Andersen S, Sirera R, Camps C, Marinez I and Busund LT: The role
of tumor stroma in cancer progression and prognosis: Emphasis on
carcinoma-associated fibroblasts and non-small cell lung cancer. J
Thorac Oncol. 6:209–217. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang Y, Haitchi HM, Cakebread J, Sammut D,
Harvey A, Powell RM, Holloway JW, Howarth P, Holgate ST and Davies
DE: Epigenetic mechanisms silence a disintegrin and metalloprotease
33 expression in bronchial epithelial cells. J Allergy Clin
Immunol. 121:1391–1399. 2008. View Article : Google Scholar
|
|
32
|
Yoshinaka T, Nishii K, Yamada K, Sawada H,
Nishiwaki E, Smith K, Yoshino K, Ishiguro H and Higashiyama S:
Identification and characterization of novel mouse and human
ADAM33s with potential metalloprotease activity. Gene. 282:227–236.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Unoki M and Nakamura Y: Methylation at CpG
islands in intron 1 of EGR2 confers enhancer-like activity. FEBS
Lett. 554:67–72. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mueller E, Sarraf P, Tontonoz P, Evans RM,
Martin KJ, Zhang M, Fletcher C, Singer S and Spiegelman BM:
Terminal differentiation of human breast cancer through PPAR gamma.
Mol Cell. 1:465–470. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Elstner E, Müller C, Koshizuka K,
Williamson EA, Park D, Asou H, Shintaku P, Said JW, Heber D and
Koeffler HP: Ligands for peroxisome proliferator-activated
receptorgamma and retinoic acid receptor inhibit growth and induce
apoptosis of human breast cancer cells in vitro and in BNX
mice. Proc Natl Acad Sci USA. 95:8806–8811. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hurst CH and Waxman DJ: Activation of
PPARalpha and PPARgamma by environmental phthalate monoesters.
Toxicol Sci. 74:297–308. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Venkata NG, Robinson JA, Cabot PJ, Davis
B, Monteith GR and Roberts-Thomson SJ: Mono(2-ethylhexyl)phthalate
and mono-n-butyl phthalate activation of peroxisome
proliferator activated-receptors α and γ in breast. Toxicol Lett.
163:224–234. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
López-Carrillo L, Hernández-Ramírez RU,
Calafat AM, Torres-Sánchez L, Galván-Portillo M, Needham LL,
Ruiz-Ramos R and Cebrián ME: Exposure to phthalates and breast
cancer risk in northern Mexico. Environ Health Perspect.
118:539–544. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Urriola-Muñoz P, Li X, Maretzky T,
McIlwain DR, Mak TW, Reyes JG, Blobel CP and Moreno RD: The
xenoestrogens biphenol-A and nonylphenol differentially regulate
metalloprotease-mediated shedding of EGFR ligands. J Cell Physiol.
233:2247–2256. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Urriola-Muñoz P, Lagos-Cabré R and Moreno
RD: A mechanism of male germ cell apoptosis induced by bisphenol-A
and nonylphenol involving ADAM17 and p38 MAPK activation. PLoS One.
9:e1137932014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Romagnolo DF, Daniels KD, Grunwald JT,
Ramos SA, Propper CR and Selmin OI: Epigenetics of breast cancer:
Modifying role of environmental and bioactive food compounds. Mol
Nutr Food Res. 60:1310–1329. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fernandez SV, Huang Y, Snider KE, Zhou Y,
Pogash TJ and Russo J: Expression and DNA methylation changes in
human breast epithelial cells after bisphenol A exposure. Int J
Oncol. 41:369–377. 2012.PubMed/NCBI
|
|
43
|
Hsieh TH, Tsai CF, Hsu CY, Kuo PL, Lee JN,
Chai CY, Wang SC and Tsai EM: Phthalates induce proliferation and
invasiveness of estrogen receptor-negative breast cancer through
the AhR/HDAC6/c-Myc signaling pathway. FASEB J. 26:778–787. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dhimolea E, Wadia PR, Murray TJ, Settles
ML, Treitman JD, Sonnenschein C, Shioda T and Soto AM: Prenatal
exposure to BPA alters the epigenome of the rat mammary gland and
increases the propensity to neoplastic development. PLoS One.
9:e998002014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kang SC and Lee BM: DNA methylation of
estrogen receptor alpha gene by phthalates. J Toxicol Environ
Health A. 68:1995–2003. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ehrlich M: DNA methylation in cancer: Too
much, but also too little. Oncogene. 21:5400–5413. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Völkel W, Colnot T, Csanády GA, Filser JG
and Dekant W: Metabolism and kinetics of bisphenol A in humans at
low doses following oral administration. Chem Res Toxicol.
15:1281–1287. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Calafat AM, Kuklenyik Z, Reidy JA, Caudill
SP, Ekong J and Needham LL: Urinary concentrations of bisphenol A
and 4-nonylphenol in a human reference population. Environ Health
Perspect. 113:391–395. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wittassek M, Koch HM, Angerer J and
Brüning T: Assessing exposure to phthalates - the human
biomonitoring approach. Mol Nutr Food Res. 55:7–31. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kay VR, Chambers C and Foster WG:
Reproductive and developmental effects of phthalate diesters in
females. Crit Rev Toxicol. 43:200–219. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Melzer D, Harries L, Cipelli R, Henley W,
Money C, McCormack P, Young A, Guralnik J, Ferrucci L, Bandinelli
S, et al: Bisphenol A exposure is associated with in vivo
estrogenic gene expression in adults. Environ Health Perspect.
119:1788–1793. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shenker NS, Polidoro S, van Veldhoven K,
Sacerdote C, Ricceri F, Birrell MA, Belvisi MG, Brown R, Vineis P
and Flanagan JM: Epigenome-wide association study in the European
Prospective Investigation into Cancer and Nutrition (EPIC-Turin)
identifies novel genetic loci associated with smoking. Hum Mol
Genet. 22:843–851. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hoppin JA, Brock JW, Davis BJ and Baird
DD: Reproducibility of urinary phthalate metabolites in first
morning urine samples. Environ Health Perspect. 110:515–518. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nepomnaschy PA, Baird DD, Weinberg CR,
Hoppin JA, Longnecker MP and Wilcox AJ: Within-person variability
in urinary bisphenol A concentrations: Measurements from specimens
after long-term frozen storage. Environ Res. 109:734–737. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sharma P, Sahni NS, Tibshirani R, Skaane
P, Urdal P, Berghagen H, Jensen M, Kristiansen L, Moen C, Sharma P,
et al: Early detection of breast cancer based on gene-expression
patterns in peripheral blood cells. Breast Cancer Res. 7:R634–R644.
2005. View Article : Google Scholar : PubMed/NCBI
|