Introduction
Pituitary tumors are considered as one of the most
common intracranial neoplasms of the central nervous system
(1,2). Although the majority of pituitary
adenomas are benign tumors, ~25% of the tumors invade areas of the
intracranial region (3). The widely
used surgical resection, chemotherapy and radiotherapy techniques
are only partially effective or wholly ineffective for patients
harboring invasive pituitary tumors, which leads to high mortality
rates and a poor outcome (4).
Therefore, it is important and urgent to identify novel targets and
characterize the underlying molecular mechanisms to improve the
treatment of pituitary tumors.
MicroRNAs (miRNAs/miRs) are characterized as a class
of small non-coding RNAs with a length of ~22 nucleotides (5–7).
miRNAs function as key regulators of gene expression via binding to
the 3′-untranslated region (UTR) of the target mRNAs, and
consequently inducing the degradation or translation inhibition of
the mRNAs (5,7,8).
Notably, emerging evidence has illustrated the critical roles of
miRNAs in the pathogenesis of different cancer types (9–16). For
example, miR-106b targeted the tumor suppressor phosphatase and
tensin homolog and promoted the growth of pituitary cancer cells
(17). Another study reported that
miR-133 suppressed the migration and invasion of pituitary tumor
cells by downregulating the expression of forkhead box C1 (14). Recently, a growing body of evidence
has suggested that miRNAs regulate the progression of cancer by
affecting the metabolism of cancer cells, particularly aerobic
glycolysis (18–23).
It is well documented that the pentose phosphate
pathway (PPP) serves an important role in glucose metabolism and
biosynthesis (24). PPP generates
nicotinamide adenine dinucleotide phosphate (NADPH) to facilitate
the synthesis of lipid and ribose 5-phosphate for the biosynthesis
of nucleotides, which maintains the rapid growth of cancer cells
(25–27). The ribose-5-phosphate generated by
the PPP is converted into the intermediates of the glycolytic
pathways. As the hallmark of cancer, aerobic glycolysis triggers
the conversion of glucose into lactate and generates adenosine
triphosphate (ATP) in cancer cells (28–30).
Glucose-6-phosphate dehydrogenase (G6PD), the first and
rate-limiting enzyme of the PPP, is essential for nucleotide
precursor production and redox homeostasis maintenance (26,31).
Increasing evidence has demonstrated that G6PD is highly expressed
in human cancer and associated with a worse patient prognosis
(31–34). In previous studies, upregulated G6PD
promoted reactive oxygen species (ROS) production by facilitating
the NADPH-dependent activation of NADPH oxidase 4 (35–37).
Inhibiting the activity of G6PD resulted in tumor suppressive
functions. The negative regulation of G6PD by miRNAs has been
demonstrated in recent studies (38,39);
G6PD was modulated by miR-206 and shown to be critical for the
differentiation of rhabdomyosarcoma (38). Additionally, it has been reported
that miR-1 post-transcriptionally repressed the expression of G6PD,
increased the ROS level and aggravated the cardiac oxidative stress
(40). However, the regulatory
association between G6PD and miR-1 in cancer remains unknown.
In order to investigate the involvement of miR-1 in
pituitary tumor, the expression of miR-1 was detected in pituitary
cancer tissues and cell lines in the present study. The effect of
miR-1 on the growth of pituitary tumor cells was examined and the
underlying molecular mechanism was further investigated.
Materials and methods
Pituitary tumor tissues
Snap-frozen pituitary tumor tissues were obtained
from 50 pituitary tumor patients (age range, 45–68 years; mean age,
58.4 years; female:male ratio, 1:1.32) between October 2013 and
August 2015. Patients who had received chemotherapy or radiotherapy
were excluded. The pituitary samples from patients without a
diagnosis of pituitary cancer were included as the non-tumor
control group to compare the expression of miR-1 in cancer tissues
(n=50; mean age, 55.8 years; age range, 41–69 years; female:male
ratio, 1:1.27). These tissues were used for analyzing the
expression of miR-1 in cancer, and for evaluating the association
between the expression of miR-1 and the metastasis and tumor size
of the patients. Another 120 patients, who were used for analyzing
the association between the expression of miR-1 and the 5-year
overall survival rate, were enrolled between April 2008 and July
2011 (n=120; mean age, 60.2 years; female:male ratio, 1.18).
All the tissue samples were obtained from the
People's Hospital of Yichang City Center affiliated to China Three
Gorges University (Yichang, Hubei, China). Patients were not
subjected to radiotherapy or chemotherapy prior to the surgical
resection. All tissues were stored in liquid nitrogen until use.
Written informed consent was provided by all participants and the
study was approved by the Ethical Committee of China Three Gorges
University. The clinicopathological parameters, including age,
gender, differentiation, tumor size, lymph node metastasis and
clinical stage, of all 170 patients are summarized in Table I.
 | Table I.Clinical parameters of the pituitary
tumor patients. |
Table I.
Clinical parameters of the pituitary
tumor patients.
| Clinical
characteristics | n |
|---|
| Age, years |
|
|
≤60 | 63 |
|
>60 | 107 |
| Sex |
|
| Male | 77 |
|
Female | 93 |
|
Differentiation |
|
|
Poor | 81 |
|
High/moderate | 89 |
| Tumor size, cm |
|
|
<4 | 75 |
| ≥4 | 95 |
| Lymph node
metastasis |
|
|
Present | 74 |
|
Absent | 96 |
| Clinical stage |
|
|
I–II | 82 |
|
III–IV | 88 |
Cell culture and transfection
The human pituitary adenoma HP75 cell line and the
rat pituitary gland neoplasm MMQ cell line were purchased from the
American Type Culture Collection (ATCC, Manassas, VA, USA). Cells
were cultured in Dulbecco's modified Eagle's medium (DMEM)
containing 12.5% horse serum and 2.5% fetal bovine serum (all
Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), with
5 U/ml penicillin and 5 g/ml streptomycin (Beyotime Institute of
Biotechnology, Shanghai, China) in an atmosphere with 5%
CO2 at 37°C. For the cell transfection, MMQ and HP75
cells were seeded in 6-well plates and when the cell confluence
reached 70–80%, miR-1 mimics (5′-UGGAAUGUAAAGAACU-3′) or negative
control miRNA (5′-UUUGUACUACACAAAAGUACUG-3′) were transfected into
the cells at the final concentration of 20 nM for 48 h with
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols.
Plasmid construction
The full length of G6PD was amplified by PCR (as
described below) and inserted into the pcDNA3.0-Flag vector. To
investigate the effect of G6PD on the growth of pituitary cancer
cells, Flag-G6PD was transfected into the cells with Lipofectamine
2000.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) for gene expression
Total RNA was extracted from pituitary tissues and
cell lines with TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The purity and quality of RNA were determined
with the NanoDrop-2000 (NanoDrop Technologies; Thermo Fisher
Scientific, Inc.). cDNA was obtained with the Omniscript Reverse
Transcription kit (Qiagen GmbH, Hilden, Germany) by reverse
transcription with 0.5 µg RNA. Using the cDNA as template, the
relative expression of miR-1 was determined with SYBR green mix
(Tiangen Biotech Co., Ltd., Beijing, China) on the ABI7500
quantitative PCR instrument (Applied Biosystems; Thermo Fisher
Scientific, Inc.) in a 10-µl reaction system. The primers of miR-1
and U6 were designed as below: miR-1 forward,
5′-CAGTGCGTGTCGTGGAGT-3′ and reverse, 5′-GGCCTGGATGTAAAGAAGT-3′;
and U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′. The reaction conditions were set as:
95°C for 5 min, followed by 40 cycles at 95°C for 15 sec and 60°C
for 1 min. Data were analyzed with the 2−ΔΔCq method
(41). The level of miR-1 was
normalized using U6 RNA as the internal reference.
Luciferase reporter assay
The wild-type or mutant 3′-UTR sequences of G6PD
containing the putative binding sites of miR-1 were cloned into the
pmirGLO reporter vector (Promega Corporation, Madison, WI, USA),
respectively. MMQ and HP75 cells were co-transfected with this
luciferase reporter vector and the indicated miRNA using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocols. Subsequent to
transfection for 48 h, the cells were harvested and the luciferase
activity was determined with the Dual-luciferase Reporter assay
system (cat. no. E1910; Promega Corporation). Renilla
luciferase activity was measured as the method of endogenous
normalization.
Western blot analysis
MMQ and HP75 cells were transfected with miR-1
mimics or control miRNA using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocols. Subsequent to transfection for 48 h, the cells were
collected and lyzed with the NP-40 lysis buffer (Beyotime Institute
of Biotechnology). The protein concentration was determined with
the bicinchoninic acid assay (BCA) kit (Beyotime Institute of
Biotechnology). Protein from each sample (20 µg) was loaded and
separated by 15% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and then transferred onto nitrocellulose
membranes. The membranes were blocked with 5% skimmed milk in PBS
at room temperature for 1 h and then incubated with anti-G6PD
antibody (1:3,000 dilution; cat. no. PA5-18734; Thermo Fisher
Scientific, Inc.) at room temperature for 2 h. The membranes were
washed twice with Tris-buffered saline plus Tween-20 and incubated
with goat anti-mouse horseradish-peroxidase-conjugated secondary
antibody (1:5,000 dilution; cat. no. AS003; ABclonal Biotech Co.,
Ltd., Woburn, MA, USA) for 1 h at room temperature. The expression
of β-actin (1:3,000; cat. no. AA128; Beyotime Institute of
Biotechnology) was detected as the loading control. The protein
bands were visualized with electrochemiluminescence reagent
(Pierce; Thermo Fisher Scientific, Inc.) using the typhoon scanner
(GE Healthcare, Chicago, IL, USA).
Cell proliferation analysis
The growth of the cells was evaluated using the
CellTiter 96 AQueous One Solution Cell Proliferation assay kit
(Promega Corporation) according to the manufacturer's protocols.
Briefly, the cells transfected with miR-1 mimics or control miRNA
were re-seeded into a 96-well plate at a density of 1,000
cells/well. Subsequent to being cultured for 24 h, 20 µl MTT was
added once into the DMEM at the time point of 1, 2, 3, 4 or 5 days
and incubated at 37°C for 3 h. The absorbance (formazan was
dissolved in DMSO) of each well at 480 nm was measured with a
microplate reader. The experiment was performed in triplicate.
Cell apoptosis
The cell apoptosis percentage of the pituitary tumor
cells was determined with the Annexin V-Fluorescein Isothiocyanate
Apoptosis Detection kit (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocols. Briefly, cells
were transfected with miR-1 mimics or control miRNA. Subsequent to
transfection for 48 h, the cells were stained with Annexin V-FITC
and propidium iodide working solution for 15 min in the dark at
room temperature. The cell apoptosis rate was analyzed using flow
cytometry (FACSCalibur; BD Biosciences, San Jose, CA, USA).
Colony formation assay
Pituitary tumor cells transfected with the indicated
miRNA were seeded in the 6-well plate at 1,000 cells/well.
Subsequent to being cultured for 2 weeks, the cells were washed
twice with PBS and fixed with methanol for 15 min at room
temperature. The cell colonies were stained with 1% crystal violet
for 10 min at room temperature. The colonies were counted by light
microscopy.
Prediction of the targets of
miR-1
The downstream targets of miR-1 were found using the
TargetScan database (http://www.targetscan.org/mamm_31/), with the species
set as human and the conserved microRNA family as miR-1/206 HMRDC.
Upon submitting the search request, the putative downstream targets
of miR-1 were summarized in a table, which included the gene name
and the binding sites in the 3′-UTR.
Oxidative PPP flux analysis
Pituitary tumor cells were transfected with miR-1 or
control miRNA using Lipofectamine 2000 (Invitrogen: Thermo Fisher
Scientific, Inc.). When the cell confluence reached 60–70%, the
cells were washed with fresh DMEM without glucose and incubated
with medium containing 10 mM [2-13C] glucose for 10 h.
The medium from the experimental and control groups was analyzed in
a 20-nm nuclear magnetic resonance tube with a 9.4T spectrometer at
100.66 MHz. Incorporation of 13C in the carbon 2 and 3
of the lactate represented the glucose metabolism from glycolysis
and the PPP, respectively. The oxidative PPP flux was determined by
the ratio of 13C in carbon 2 (glycolysis) and carbon 3
(oxidative PPP flux) of lactate, as well as the rate of glucose
uptake. The number of cells that were transfected with miR-1 or
control miRNA was compared to eliminate the influence caused by the
variation of cell amounts.
NADPH level determination
Pituitary tumor cells were transfected with miR-1
mimics or control miRNA. Subsequent to transfection for 48 h, the
cells were lysed with buffer containing 0.1 M Tris-HCl (pH 8.0),
0.05% Triton X-100 and 0.01 M EDTA. The lysates were sonicated and
centrifuged at 3,000 × g for 10 min at 4°C. The level of NADPH was
detected for the absorbance at the wavelength of 341 nm by
spectrometry. The protein concentration was determined using the
BCA kit (Beyotime Institute of Biotechnology) to normalize the
influence of the cell number.
Statistical analysis
The data from three independent experiments are
presented as the mean ± standard error. The statistical analysis
was performed using the SPSS software (13.0, IBM Corp., Armonk, NY,
USA). The difference between two groups was analyzed with Student's
t-test. The differences between more than two groups were detected
by one-way analysis of variance followed by Dunnett's post hoc
test. Kaplan-Meier survival analysis was performed to compare the
overall survival rates of patients with pituitary tumors grouped by
high and low miR-1 expression levels (the mean value of
miR-1expression in pituitary cancer tissues was considered as the
cut-off). The log-rank test was used to analyze the statistical
significance. Spearman's rank correlation test was performed to
analyze the correlation between the expression of miR-1 and G6PD in
the pituitary tumor tissues. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-1 is downregulated in pituitary
tumor tissues and associated with a poor prognosis in patients with
pituitary cancer
To understand the function of miR-1 in pituitary
tumors, the expression of miR-1 in pituitary tumor tissues and
adjacent normal pituitary tissues was evaluated by RT-qPCR
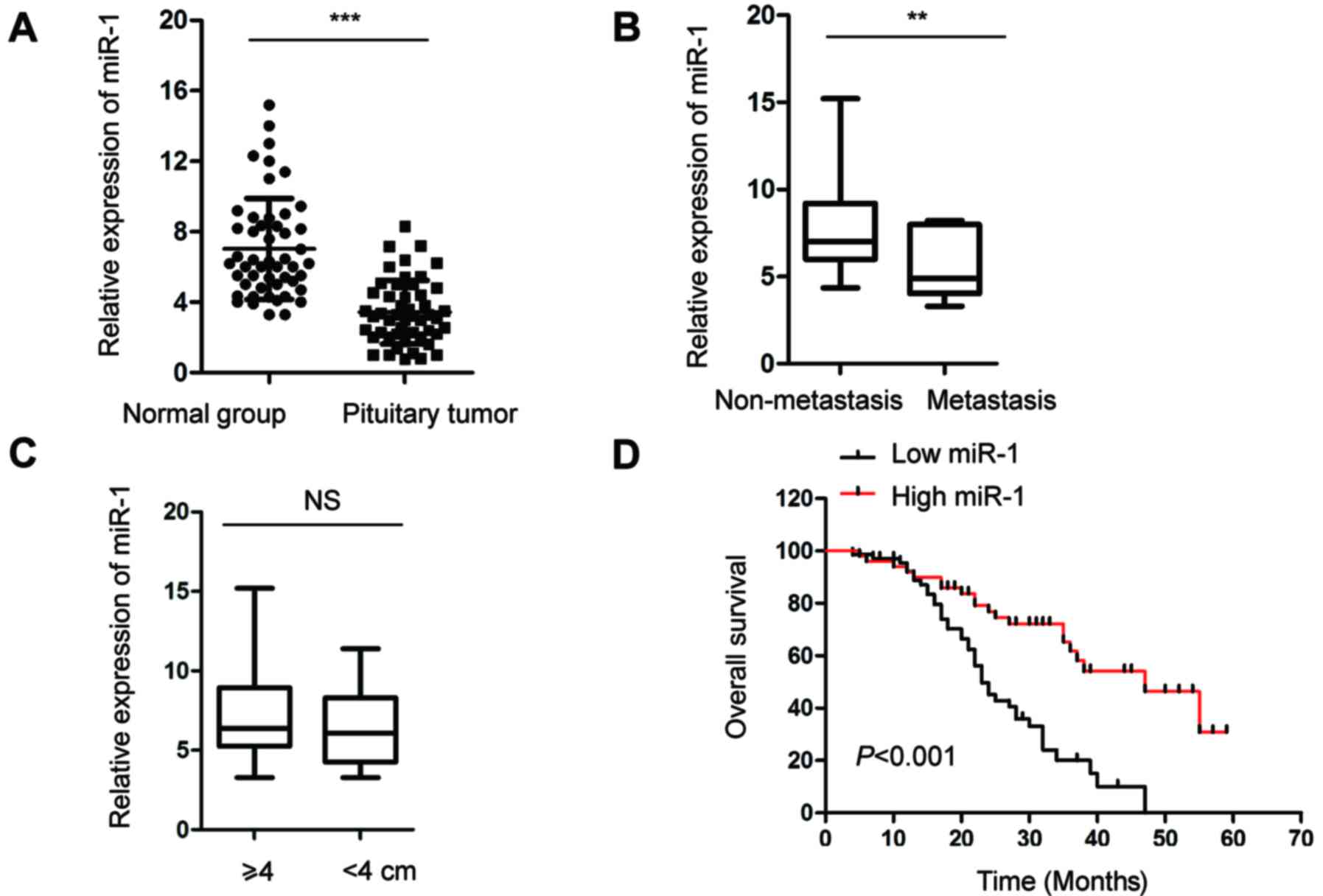
analysis. As shown in Fig. 1A, the
expression of miR-1 was significantly decreased in pituitary tumor
tissues compared with that in the control group. The decreased
expression of miR-1 in pituitary tumor tissues motivated
investigation of the association between the level of miR-1 and the
clinicopathological factors of patients with pituitary cancer. The
analysis data showed that among the 50 patients, the expression of
miR-1 was significantly lower in the patients with lymph metastasis
compared with that in the patients without metastasis (Fig. 1B). However, there was no significant
difference in the expression of miR-1 with regard to the size of
the tumors (Fig. 1C). Based on
these data, Kaplan-Meier survival analysis was performed to
investigate the association between the expression of miR-1 and the
5-year overall survival of patients was analyzed in another 120
pituitary tumor patients. As shown in Fig. 1D, low expression of miR-1 was
significantly associated with a worse prognosis in the patients.
These results suggested that decreased abundance of miR-1 was a
possible biomarker for predicting the prognosis of patients with
pituitary cancer.
Overexpression of miR-1 suppresses the
proliferation and induces the apoptosis of pituitary tumor
cells
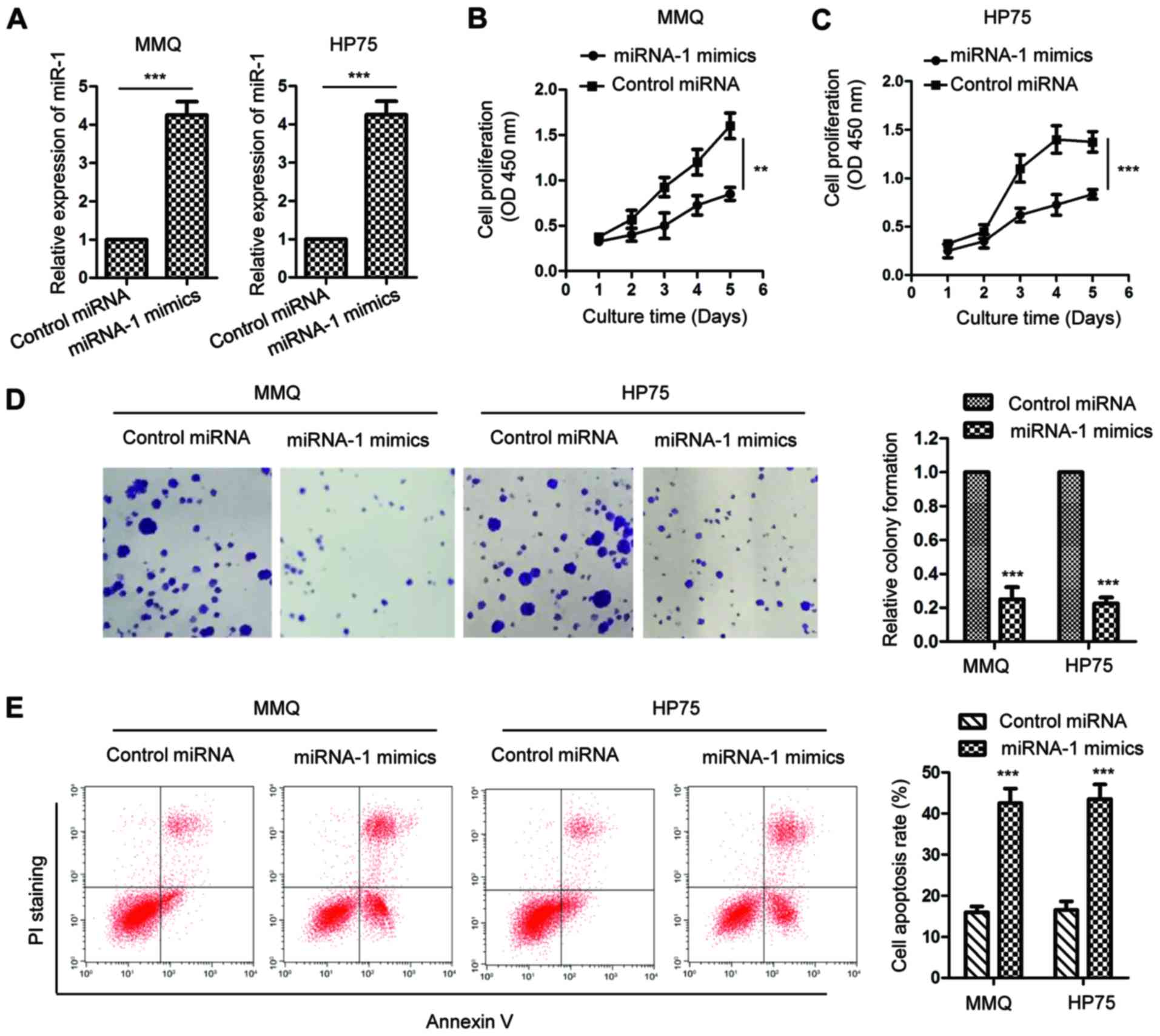
Given the decreased expression of miR-1 in pituitary
tumors, the influence of miR-1 on the growth of pituitary tumor
cells was then investigated. MMQ and HP75 cells were transfected
with miR-1 mimics or control miRNA. The expression of miR-1 was
confirmed by RT-qPCR analysis, as presented in Fig. 2A. An MTT assay was performed to
detect the proliferation rate of pituitary tumor cells expressing
miR-1 mimics or control miRNA. The results showed that upregulation
of miR-1 significantly decreased the proliferation of the MMQ and
HP75 cells (Fig. 2B and C). To
further characterize the inhibitory effect of miR-1 on pituitary
tumor cells, an in vitro colony formation assay was
performed with MMQ and HP75 cells harboring overexpressed miR-1 or
control miRNA. The data indicated that in the MMQ and HP75 cells,
highly expressed miR-1 significantly suppressed the colony
formation of the pituitary tumor cells (Fig. 2D). The cell apoptosis of MMQ and
HP75 cells with overexpressed miR-1 was also evaluated. As
illustrated in Fig. 2E, a
significantly increased cell apoptosis rate was observed in the
pituitary tumor cells with highly expressed miR-1. These results
demonstrated that overexpression of miR-1 inhibited the growth of
pituitary tumor cells.
G6PD is a downstream target of miR-1
in pituitary tumor cells
To understand the functional mechanism of miR-1 in
pituitary cancer, the downstream targets of miR-1 were predicted by
the TargetScan database, and ~440 possible conserved targets of
miR-1 were found. These targets mainly function in regulating the
proliferation and differentiation of cells. G6PD ranked top among
all the predicted target candidates of miR-1. The putative binding
sites of miR-1 at the 3′-UTR of G6PD are shown in Fig. 3A. To test this observation, a
luciferase reporter assay was performed by co-transfecting miR-1
mimics with the wild-type or mutant 3′-UTR of G6PD into the
pituitary tumor cells. The data showed that overexpression of miR-1
significantly decreased the luciferase activity of the wild-type
but not the mutant 3′-UTR of G6PD in the MMQ and HP75 cells
(Fig. 3B and C). To further confirm
this result, the protein level of G6PD in pituitary tumor cells
expressing miR-1 mimics or control miRNA was detected by western
blotting with anti-G6PD antibody. A decreased protein level of G6PD
was observed with the high expression of miR-1 in the MMQ and HP75
cells (Fig. 3D). At the same time,
the mRNA level of G6PD was also evaluated with overexpressed miR-1.
However, the results showed that no significance was obtained for
the mRNA abundance of G6PD in the presence of miR-1 compared with
that of the control cells (Fig.
3E). These results demonstrated that G6PD was a target of miR-1
and negatively regulated the protein expression of G6PD in
pituitary tumor cells.
miR-1 targets G6PD to suppress the
NADPH production and glycolysis of pituitary cancer cells
It has been well documented that the PPP generates
NADPH and ribose-5-phosphate to facilitate the biosynthesis of
nucleotides and glycolysis, respectively, which maintains the rapid
growth of cancer cells (42). G6PD
has been demonstrated as the first and rate-limiting enzyme of the
PPP (42). Due to the negative
regulation of miR-1 on G6PD, an assessment of whether
overexpression of miR-1 modulated the PPP in pituitary tumor cells
was performed. The oxidative PPP flux (flux through the oxidative
branch of PPP and glycolysis), glucose consumption and lactate
production of pituitary tumor cells transfected with overexpressed
miR-1 were detected. As shown in Fig.
4A, the ectopic expression of miR-1 resulted in a significant
decrease in the oxidative PPP flux. Since PPP serves important
roles in the production of cellular NADPH, the influence of miR-1
on the generation of NADPH was also measured. The data showed that
overexpression of miR-1 suppressed the level of NADPH compared with
that of the control group (Fig.
4B). The glucose uptake and lactate production were also
significantly decreased in the MMQ and HP75 cells expressing miR-1
mimics (Fig. 4C and D). These
results indicated that miR-1 suppressed the PPP and glucose
metabolism of the pituitary tumor cells.
Overexpression of G6PD reverses the
inhibitory effect of miR-1 on the growth of pituitary tumor
cells
To further characterize the association between
miR-1 and G6PD, the expression of G6PD in paired pituitary tumor
tissues and adjacent normal tissues was detected by RT-qPCR. The
data showed that the mRNA level of G6PD was significantly increased
in the pituitary tumor tissues compared with that in the
corresponding normal tissues (Fig.
5A). Spearman's rank correlation test indicated that the
expression of miR-1 and G6PD in pituitary tumor tissues was
significantly inversely correlated (Fig. 5B). To confirm that the inhibitory
effect of miR-1 on the growth of pituitary tumor cells was through
the regulation of G6PD, MMQ and HP75 cells expressing miR-1 mimics
were transfected to ectopically express G6PD. As shown in Fig. 5C and D, the MTT assay showed that
highly expressed miR-1 decreased the cell proliferation, while
restoring the expression of G6PD significantly inhibited the
suppressive function of miR-1 on the growth of pituitary tumor
cells. To further confirm this observation, the colony formation of
pituitary tumor cells that were transfected with miR-1 or control
miRNA with G6PD was performed. The data showed that in comparison
with the cells expressing miR-1, the overexpression of G6PD
significantly promoted the colony formation of the MMQ and HP75
cells (Fig. 5E). The
fluorescence-activated cell sorting (FACS) analysis showed that
overexpression of G6PD attenuated the cell apoptosis of pituitary
tumor cells that were induced with the transfection of miR-1
(Fig. 5F). These results suggested
that G6PD serves important roles for mediating the inhibitory
effect of miR-1 on the growth of pituitary tumor cells.
Discussion
Abnormal expression of miRNAs has been involved in
the initiation and progression of human cancer (10,11).
The tumor suppressive function of miR-1 in cancer has been revealed
in recent years (39,43–45).
Downregulation of miR-1 was shown to enhance the tumorigenesis and
invasiveness of oral squamous cell carcinoma (46). In ovarian cancer, miR-1 inhibited
cell proliferation and migration through regulation of the c-Met
pathway (47). The present study
revealed that miR-1 was downregulated in pituitary tumor tissues
and was associated with the poor prognosis of patients.
Overexpression of miR-1 suppressed the proliferation of pituitary
tumor cells, which was consistent with the previously described
tumor suppressive roles of miR-1 in other types of cancer.
It is well known that miRNAs exert their functions
by negatively regulating the expression of downstream target genes.
In gastric cancer, miR-1 was found to suppress the expression of
vascular endothelial growth factor A and endothelin-1, which
consequently inhibited the tube formation of endothelial cells
(48). Additionally, miR-1
suppressed the growth of esophageal carcinoma cells and enhanced
the sensitivity of cells to anticancer drugs (49). In the present study, the
bioinformatics and luciferase reporter assay revealed that miR-1
bound the 3′-UTR of G6PD. Overexpression of miR-1 decreased the
protein level of G6PD but exhibited no significant effect on the
mRNA level of G6PD. The binding of miR-1 with the 3′-UTR of G6PD
may affect the structure of G6PD mRNA and block the translation of
this mRNA into G6PD protein, which finally results in the decreased
protein abundance of G6PD in miR-1-overexpressing pituitary tumor
cells. Decreased expression of miR-1 was associated with the
metastasis of pituitary tumor patients in this study. Notably,
previous publications reported that the overexpression of G6PD was
associated with the high risk of recurrent metastasis in primary
breast carcinoma and esophageal squamous cell carcinoma (50,51).
Elevated G6PD level promoted the migration and invasion of
hepatocellular carcinoma cells by inducing the
epithelial-mesenchymal transition (52). All these results suggested the
critical involvement of G6PD in the metastasis of cancer. Thus, the
potential function of G6PD in regulating the metastasis of
pituitary tumor cells deserves further investigation.
As the hallmark of cancer, cancer cells specifically
reprogram the metabolism from oxidative phosphorylation to
glycolysis, which accelerates the generation of ATP and
intermediate materials for the biosynthesis of molecules in cells
(28–30). Notably, the PPP, as the major source
of NADPH, provides the reducing power for the biosynthesis of
cancer cells (25,27). As the rate-limiting factor of PPP,
the expression of G6PD was previously found to be highly
upregulated in several cancer types (31). Transcriptional activation of G6PD by
Tap73, the homolog of p53, enhanced the PPP and facilitated the
growth of cancer cells (53,54).
Due to its oncogenic function, decreasing the level of G6PD is a
novel strategy to suppress tumorigenesis. In the present study, the
downregulation of G6PD by miR-1 inhibited the generation of NADPH,
and decreased the glucose consumption and lactate production. These
results suggested that miR-1 was a novel regulator of the PPP in
pituitary tumor cells.
In conclusion, this study found that miR-1 was
downregulated in pituitary tumor cells and tissues. Overexpression
of miR-1 suppressed cell growth by targeting G6PD to inhibit the
metabolism of cancer cells. The downregulation of miR-1 was
significantly associated with a worse prognosis in patients with
pituitary tumors. These results suggested that miR-1 and G6PD may
be potential targets for the therapy of human pituitary cancer.
Acknowledgements
Not applicable.
Funding
The study was supported by the Natural Science
Foundation of Hubei Province in China (grant no. 2016CFB599) and
the National Natural Science Foundation of China (grant no.
81770360).
Availability of data and materials
All the data and materials in this study are
available from the corresponding author upon request.
Authors' contributions
CH designed the study and performed the majority of
the experiments. JuY and JD collected the clinical samples and
detected the expression of miR-1 in the tissues. SL and HW
performed the western blotting. YX and FZ performed the FACS
analysis. YJ and LT performed the statistical analysis. JiY
designed the study, wrote the manuscript, revised it critically for
important intellectual content and gave the final approval of the
version to be published. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethical Committee of
China Three Gorges University. Written informed consent was
provided by all participants.
Patient consent for publication
Consent for publication was obtained from all
patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Laws ER: Pituitary tumor apoplexy: A
review. J Intensive Care Med. 23:146–147. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nasi D, Perano D, Ghadirpour R, Iaccarino
C, Servadei F and Romano A: Primary pituitary neuroendocrine tumor:
Case report and literature review. Surg Neurol Int. 8:1012017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ezzat S and Asa SL: Mechanisms of disease:
The pathogenesis of pituitary tumors. Nat Clin Pract Endocrinol
Metab. 2:220–230. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jiang X and Zhang X: The molecular
pathogenesis of pituitary adenomas: An update. Endocrinol Metab.
28:245–254. 2013. View Article : Google Scholar
|
|
5
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Ann Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mohr AM and Mott JL: Overview of microRNA
biology. Semin Liver Dis. 35:3–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stilling G, Sun Z, Zhang S, Jin L, Righi
A, Kovācs G, Korbonits M, Scheithauer BW, Kovacs K and Lloyd RV:
MicroRNA expression in ACTH-producing pituitary tumors:
Up-regulation of microRNA-122 and −493 in pituitary carcinomas.
Endocrine. 38:67–75. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kwak PB, Iwasaki S and Tomari Y: The
microRNA pathway and cancer. Cancer Sci. 101:2309–2315. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qu H, Xu W, Huang Y and Yang S:
Circulating miRNAs: Promising biomarkers of human cancer. Asian Pac
J Cancer Prev. 12:1117–1125. 2011.PubMed/NCBI
|
|
13
|
Sapochnik M, Nieto LE, Fuertes M and Arzt
E: Molecular mechanisms underlying pituitary pathogenesis. Biochem
Genet. 54:107–119. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang DS, Zhang HQ, Zhang B, Yuan ZB, Yu
ZK, Yang T, Zhang SQ, Liu Y and Jia XX: miR-133 inhibits pituitary
tumor cell migration and invasion via down-regulating FOXC1
expression. Genet Mol Res. 15:2016.
|
|
15
|
Yu C, Li J, Sun F, Cui J, Fang H and Sui
G: Expression and clinical significance of miR-26a and pleomorphic
adenoma gene 1 (PLAG1) in invasive pituitary adenoma. Med Sci
Monit. 22:5101–5108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou K, Zhang T, Fan Y, Serick, Du G, Wu P
and Geng D: MicroRNA-106b promotes pituitary tumor cell
proliferation and invasion through PI3K/AKT signaling pathway by
targeting PTEN. Tumour Biol. 37:13469–13477. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng Z, Zhang Y, Zhang Z, Yang Y and Song
T: Effect of miR-106b on invasiveness of pituitary adenoma via
PTEN-PI3K/AKT. Med Sci Monit. 23:1277–1285. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wilfred BR, Wang WX and Nelson PT:
Energizing miRNA research: A review of the role of miRNAs in lipid
metabolism, with a prediction that miR-103/107 regulates human
metabolic pathways. Mol Genet Metab. 91:209–217. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Alamoudi AA, Alnoury A and Gad H: miRNA in
tumour metabolism and why could it be the preferred pathway for
energy reprograming. Brief Funct Genomics. 17:157–169. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Antoniali G, Serra F, Lirussi L, Tanaka M,
D'Ambrosio C, Zhang S, Radovic S, Dalla E, Ciani Y, Scaloni A, et
al: Mammalian APE1 controls miRNA processing and its interactome is
linked to cancer RNA metabolism. Nat Commun. 8:7972017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jordan SD, Krüger M, Willmes DM, Redemann
N, Wunderlich FT, Brönneke HS, Merkwirth C, Kashkar H, Olkkonen VM,
Böttger T, et al: Obesity-induced overexpression of miRNA-143
inhibits insulin-stimulated AKT activation and impairs glucose
metabolism. Nat Cell Biol. 13:434–446. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lynn FC: Meta-regulation: microRNA
regulation of glucose and lipid metabolism. Trends Endocrinol
Metab. 20:452–459. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ono K: MicroRNA links obesity and impaired
glucose metabolism. Cell Res. 21:864–866. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wamelink MM, Struys EA and Jakobs C: The
biochemistry, metabolism and inherited defects of the pentose
phosphate pathway: A review. J Inherit Metab Dis. 31:703–717. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang P, Du W and Wu M: Regulation of the
pentose phosphate pathway in cancer. Protein Cell. 5:592–602. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Patra KC and Hay N: The pentose phosphate
pathway and cancer. Trends Biochem Sci. 39:347–354. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cho ES, Cha YH, Kim HS, Kim NH and Yook
JI: The pentose phosphate pathway as a potential target for cancer
therapy. Biomol Ther. 26:29–38. 2018. View Article : Google Scholar
|
|
28
|
Zheng J: Energy metabolism of cancer:
Glycolysis versus oxidative phosphorylation (Review). Oncol Lett.
4:1151–1157. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Akram M: Mini-review on glycolysis and
cancer. J Cancer Educ. 28:454–457. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li XB, Gu JD and Zhou QH: Review of
aerobic glycolysis and its key enzymes-new targets for lung cancer
therapy. Thorac Cancer. 6:17–24. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang C, Zhang Z, Zhu Y and Qin S:
Glucose-6-phosphate dehydrogenase: A biomarker and potential
therapeutic target for cancer. Anticancer Agents Med Chem.
14:280–289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Naik SN and Anderson DE: The association
between glucose-6-phosphate dehydrogenase deficiency and cancer in
American Negroes. Oncology. 25:356–364. 1971. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Messeri G, Tozzi P, Boddi V and Ciatto S:
Glucose-6-phosphate dehydrogenase activity and estrogen receptors
in human breast cancer. J Steroid Biochem. 19:1647–1650. 1983.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pisano M, Cocco P, Cherchi R, Onnis R and
Cherchi P: Glucose-6-phosphate dehydrogenase deficiency and lung
cancer: A hospital based case-control study. Tumori. 77:12–15.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tho LL, Lee WH and Candlish JK:
Erythrocytic enzymes decomposing reactive oxygen species and
glucose 6-phosphate dehydrogenase deficiency. Singapore Med J.
29:60–62. 1988.PubMed/NCBI
|
|
36
|
Cai T, Kuang Y, Zhang C, Zhang Z, Chen L,
Li B, Li Y, Wang Y, Yang H, Han Q and Zhu Y: Glucose-6-phosphate
dehydrogenase and NADPH oxidase 4 control STAT3 activity in
melanoma cells through a pathway involving reactive oxygen species,
c-SRC and SHP2. Am J Cancer Res. 5:1610–1620. 2015.PubMed/NCBI
|
|
37
|
Nadeem A, Al-Harbi NO, Ahmad SF, Ibrahim
KE, Siddiqui N and Al-Harbi MM: Glucose-6-phosphate dehydrogenase
inhibition attenuates acute lung injury through reduction in NADPH
oxidase-derived reactive oxygen species. Clin Exp Immunol.
191:279–287. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Coda DM, Lingua MF, Morena D, Foglizzo V,
Bersani F, Ala U, Ponzetto C and Taulli R: SMYD1 and G6PD
modulation are critical events for miR-206-mediated differentiation
of rhabdomyosarcoma. Cell Cycle. 14:1389–1402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hu T, Chang YF, Xiao Z, Mao R, Tong J,
Chen B, Liu GC, Hong Y, Chen HL, Kong SY, et al: miR-1 inhibits
progression of high-risk papillomavirus-associated human cervical
cancer by targeting G6PD. Oncotarget. 7:86103–86116. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang L, Yuan Y, Li J, Ren H, Cai Q, Chen
X, Liang H, Shan H, Fu ZD, Gao X, et al: MicroRNA-1 aggravates
cardiac oxidative stress by post-transcriptional modification of
the antioxidant network. Cell Stress Chaperones. 20:411–420. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gelman SJ, Naser F, Mahieu NG, McKenzie
LD, Dunn GP, Chheda MG and Patti GJ: Consumption of NADPH for 2-HG
synthesis increases pentose phosphate pathway flux and sensitizes
cells to oxidative stress. Cell Rep. 22:512–522. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Han C, Zhou Y, An Q, Li F, Li D, Zhang X,
Yu Z, Zheng L, Duan Z and Kan Q: MicroRNA-1 (miR-1) inhibits
gastric cancer cell proliferation and migration by targeting MET.
Tumour Biol. 36:6715–6723. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu R, Li J, Lai Y, Liao Y, Liu R and Qiu
W: Hsa-miR-1 suppresses breast cancer development by
down-regulating K-ras and long non-coding RNA MALAT1. Int J Biol
Macromol. 81:491–497. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xu W, Zhang Z, Zou K, Cheng Y, Yang M,
Chen H, Wang H, Zhao J, Chen P, He L, et al: MiR-1 suppresses tumor
cell proliferation in colorectal cancer by inhibition of
Smad3-mediated tumor glycolysis. Cell Death Dis. 8:e27612017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Peng CY, Liao YW, Lu MY, Yu CH, Yu CC and
Chou MY: Downregulation of miR-1 enhances tumorigenicity and
invasiveness in oral squamous cell carcinomas. J Formos Med Assoc.
116:782–789. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Qu W, Chen X, Wang J, Lv J and Yan D:
MicroRNA-1 inhibits ovarian cancer cell proliferation and migration
through c-Met pathway. Clin Chim Acta. 473:237–244. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xie M, Dart DA, Guo T, Xing XF, Cheng XJ,
Du H, Jiang WG, Wen XZ and Ji JF: MicroRNA-1 acts as a tumor
suppressor microRNA by inhibiting angiogenesis-related growth
factors in human gastric cancer. Gastric Cancer. 21:41–54. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yu Q, Liu Y, Wen C, Zhao Y, Jin S, Hu Y,
Wang F, Chen L, Zhang B, Wang W, et al: MicroRNA-1 inhibits
tumorigenicity of esophageal squamous cell carcinoma and enhances
sensitivity to gefitinib. Oncol Lett. 15:963–971. 2018.PubMed/NCBI
|
|
50
|
Wang X, Li X, Zhang X, Fan R, Gu H, Shi Y
and Liu H: Glucose-6-phosphate dehydrogenase expression is
correlated with poor clinical prognosis in esophageal squamous cell
carcinoma. Eur J Surg Oncol. 41:1293–1299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Pu H, Zhang Q, Zhao C, Shi L, Wang Y, Wang
J and Zhang M: Overexpression of G6PD is associated with high risks
of recurrent metastasis and poor progression-free survival in
primary breast carcinoma. World J Surg Oncol. 13:3232015.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lu M, Lu L, Dong Q, Yu G, Chen J, Qin L,
Wang L, Zhu W and Jia H: Elevated G6PD expression contributes to
migration and invasion of hepatocellular carcinoma cells by
inducing epithelial-mesenchymal transition. Acta Biochim Biophys
Sin. 50:370–380. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang X, Wu G, Cao G, Yang L, Xu H, Huang J
and Hou J: Zoledronic acid inhibits the pentose phosphate pathway
through attenuating the Ras-TAp73-G6PD axis in bladder cancer
cells. Mol Med Rep. 12:4620–4625. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jiang P, Du W and Yang X: A critical role
of glucose-6-phosphate dehydrogenase in TAp73-mediated cell
proliferation. Cell Cycle. 12:3720–3726. 2013. View Article : Google Scholar : PubMed/NCBI
|