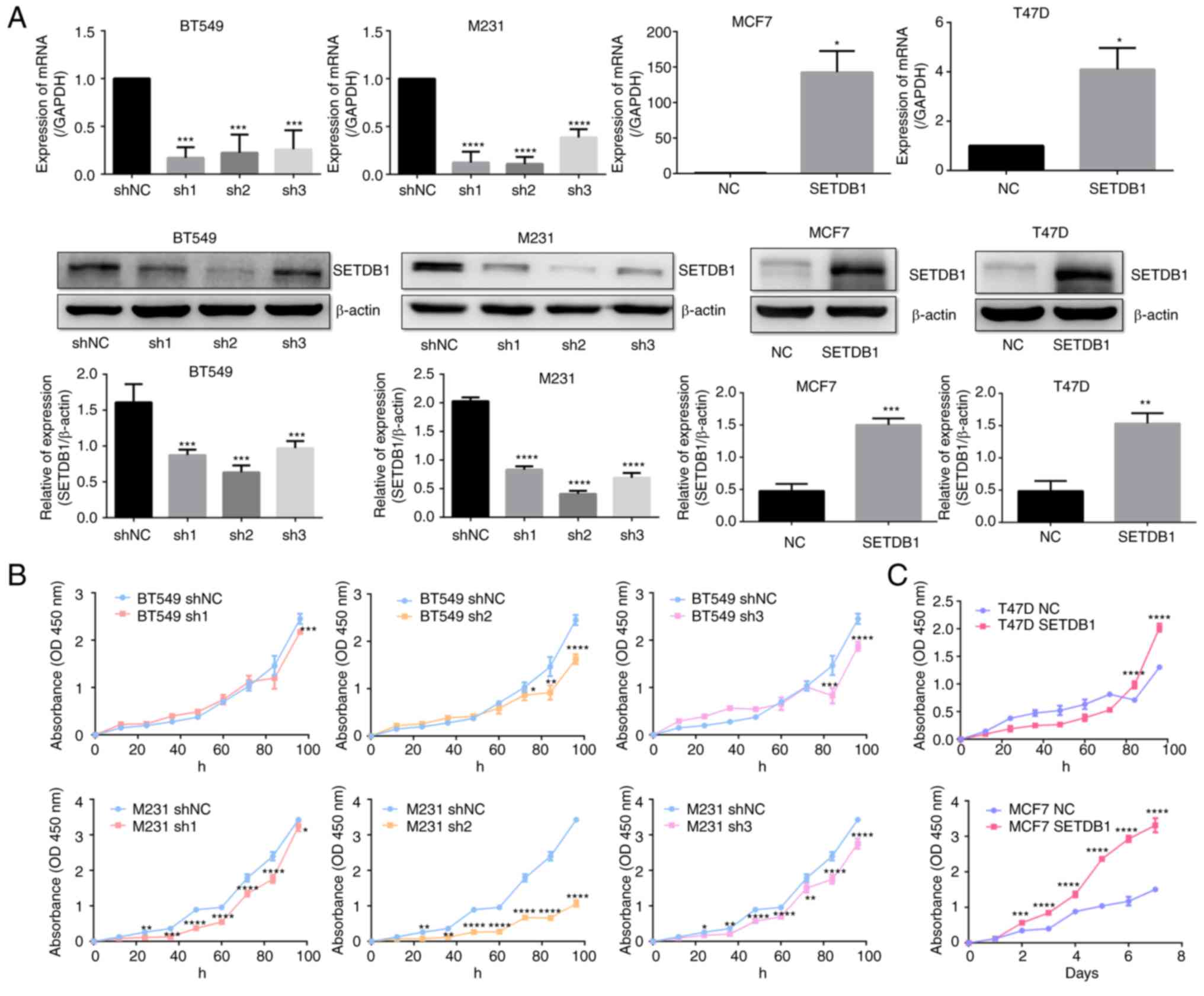
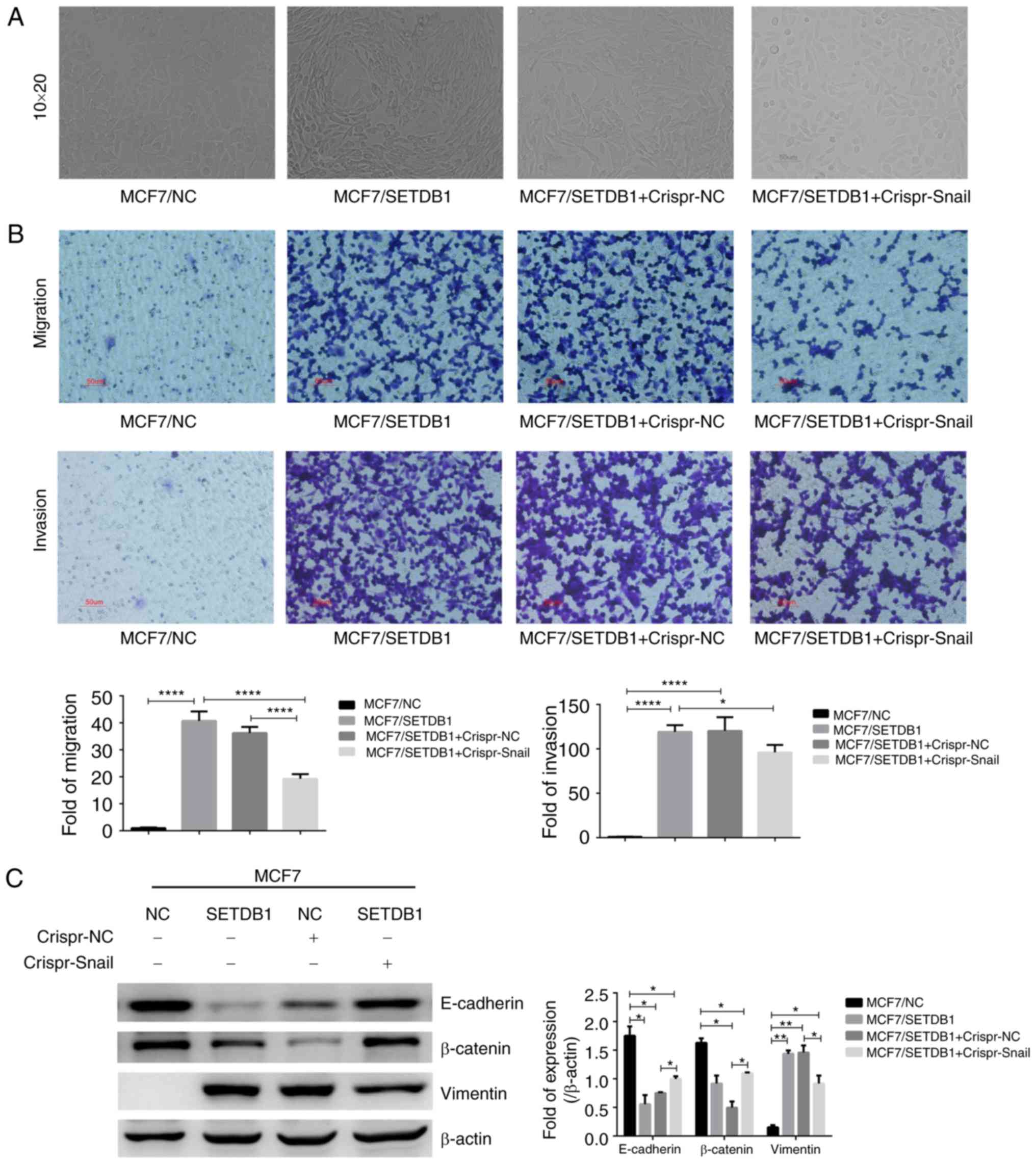
|
1
|
Benson JR and Jatoi I: The global breast
cancer burden. Future Oncol. 8:697–702. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gonzalez-Angulo AM, Morales-Vasquez F and
Hortobagyi GN: Overview of resistance to systemic therapy in
patients with breast cancer. Adv Exp Med Biol. 608:1–22. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Elsheikh SE, Green AR, Rakha EA, Powe DG,
Ahmed RA, Collins HM, Soria D, Garibaldi JM, Paish CE, Ammar AA, et
al: Global histone modifications in breast cancer correlate with
tumor phenotypes, prognostic factors, and patient outcome. Cancer
Res. 69:3802–3809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Albert M and Helin K: Histone
methyltransferases in cancer. Semin Cell Dev Biol. 21:209–220.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Harte PJ, Wu W, Carrasquillo MM and Matera
AG: Assignment of a novel bifurcated SET domain gene, SETDB1, to
human chromosome band 1q21 by in situ hybridization and radiation
hybrids. Cytogenet Cell Genet. 84:83–86. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schultz DC, Ayyanathan K, Negorev D, Maul
GG and Rauscher FJ III: SETDB1: A novel KAP-1-associated histone
H3, lysine 9-specific methyltransferase that contributes to
HP1-mediated silencing of euchromatic genes by KRAB zinc-finger
proteins. Genes Dev. 16:919–932. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Verschure PJ, van der Kraan I, de Leeuw W,
van der Vlag J, Carpenter AE, Belmont AS and van Driel R: In vivo
HP1 targeting causes large-scale chromatin condensation and
enhanced histone lysine methylation. Mol Cell Biol. 25:4552–4564.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang H, An W, Cao R, Xia L,
Erdjument-Bromage H, Chatton B, Tempst P, Roeder RG and Zhang Y:
mAM facilitates conversion by ESET of dimethyl to trimethyl lysine
9 of histone H3 to cause transcriptional repression. Mol Cell.
12:475–487. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sarraf SA and Stancheva I: Methyl-CpG
binding protein MBD1 couples histone H3 methylation at lysine 9 by
SETDB1 to DNA replication and chromatin assembly. Mol Cell.
15:595–605. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ichimura T, Watanabe S, Sakamoto Y, Aoto
T, Fujita N and Nakao M: Transcriptional repression and
heterochromatin formation by MBD1 and MCAF/AM family proteins. J
Biol Chem. 280:13928–13935. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bilodeau S, Kagey MH, Frampton GM, Rahl PB
and Young RA: SetDB1 contributes to repression of genes encoding
developmental regulators and maintenance of ES cell state. Genes
Dev. 23:2484–2489. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu S, Brind'Amour J, Karimi MM, Shirane
K, Bogutz A, Lefebvre L, Sasaki H, Shinkai Y and Lorincz MC:
Setdb1 is required for germline development and silencing of
H3K9me3-marked endogenous retroviruses in primordial germ cells.
Genes Dev. 29:1082015.
|
|
13
|
Koide S, Oshima M, Takubo K, Yamazaki S,
Nitta E, Saraya A, Aoyama K, Kato Y, Miyagi S, Nakajima-Takagi Y,
et al: Setdb1 maintains hematopoietic stem and progenitor cells by
restricting the ectopic activation of nonhematopoietic genes.
Blood. 128:638–649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thompson PJ, Dulberg V, Moon KM, Foster
LJ, Chen C, Karimi MM and Lorincz MC: hnRNP K coordinates
transcriptional silencing by SETDB1 in embryonic stem cells. PLoS
Genet. 11:e10049332015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fei Q, Yang X, Jiang H, Wang Q, Yu Y, Yu
Y, Yi W, Zhou S, Chen T, Lu C, et al: SETDB1 modulates PRC2
activity at developmental genes independently of H3K9
trimethylation in mouse ES cells. Genome Res. 25:1325–1335. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ceol CJ, Houvras Y, Jane-Valbuena J,
Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ,
Ferré F, et al: The histone methyltransferase SETDB1 is recurrently
amplified in melanoma and accelerates its onset. Nature.
471:513–517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Spyropoulou A, Gargalionis A, Dalagiorgou
G, Adamopoulos C, Papavassiliou KA, Lea RW, Piperi C and
Papavassiliou AG: Role of histone lysine methyltransferases SUV39H1
and SETDB1 in gliomagenesis: Modulation of cell proliferation,
migration, and colony formation. Neuromolecular Med. 16:70–82.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun Y, Wei M, Ren SC, Chen R, Xu WD, Wang
FB, Lu J, Shen J, Yu YW, Hou JG, et al: Histone methyltransferase
SETDB1 is required for prostate cancer cell proliferation,
migration and invasion. Asian J Androl. 16:319–324. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rodriguez-Paredes M, Martinez de Paz A,
Simó-Riudalbas L, Sayols S, Moutinho C, Moran S, Villanueva A,
Vázquez-Cedeira M, Lazo PA, Carneiro F, et al: Gene amplification
of the histone methyltransferase SETDB1 contributes to human lung
tumorigenesis. Oncogene. 33:2807–2813. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen K, Zhang F, Ding J, Liang Y, Zhan Z,
Zhan Y, Chen LH and Ding Y: Histone methyltransferase SETDB1
promotes the progression of colorectal cancer by inhibiting the
expression of TP53. J Cancer. 8:3318–3330. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ho Y, Lin YM, Huang YC, Chang J, Yeh KT,
Lin LI, Gong Z, Tzeng TY and Lu JW: Significance of histone
methyltransferase SETDB1 expression in colon adenocarcinoma. APMIS.
125:985–995. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Noh HJ, Kim KA and Kim KC: p53
Down-regulates SETDB1 gene expression during paclitaxel
induced-cell death. Biochem Biophys Res Commun. 446:43–48. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cicchini C, Battistelli C and Tripodi M:
SETDB1 is a new promising target in HCC therapy. Chin Clin Oncol.
5:732016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Karanth AV, Maniswami RR, Prashanth S,
Govindaraj H, Padmavathy R, Jegatheesan SK, Mullangi R and
Rajagopal S: Emerging role of SETDB1 as a therapeutic target.
Expert Opin Ther Targets. 21:319–331. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang W, Wang JG, Xu J, Zhou D, Ren K, Hou
C, Chen L and Liu X: HCRP1 inhibits TGF-β induced
epithelial-mesenchymal transition in hepatocellular carcinoma. Int
J Oncol. Mar 8–2017.(Epub ahead of print). doi:
10.3892/ijo.2017.3903. View Article : Google Scholar
|
|
26
|
Sun QY, Ding LW, Xiao JF, Chien W, Lim SL,
Hattori N, Goodglick L, Chia D, Mah V, Alavi M, et al: SETDB1
accelerates tumourigenesis by regulating the WNT signalling
pathway. J Pathol. 235:559–570. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu PC, Lu JW, Yang JY, Lin IH, Ou DL, Lin
YH, Chou KH, Huang WF, Wang WP, Huang YL, et al: H3K9 histone
methyltransferase, KMT1E/SETDB1, cooperates with the SMAD2/3
pathway to suppress lung cancer metastasis. Cancer Res.
74:7333–7343. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu L, Kimball S, Liu H, Holowatyj A and
Yang ZQ: Genetic alterations of histone lysine methyltransferases
and their significance in breast cancer. Oncotarget. 6:2466–2482.
2015.PubMed/NCBI
|
|
29
|
Regina C, Compagnone M, Peschiaroli A,
Lena A, Annicchiarico-Petruzzelli M, Piro MC, Melino G and Candi E:
Setdb1, a novel interactor of ΔNp63, is involved in breast
tumorigenesis. Oncotarget. 7:28836–28848. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang H, Cai K, Wang J, Wang X, Cheng K,
Shi F, Jiang L, Zhang Y and Dou J: MiR-7, inhibited indirectly by
lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of
breast cancer stem cells by downregulating the STAT3 pathway. Stem
Cells. 32:2858–2868. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Creighton CJ, Chang JC and Rosen JM:
Epithelial-mesenchymal transition (EMT) in tumor-initiating cells
and its clinical implications in breast cancer. J Mammary Gland
Biol Neoplasia. 15:253–260. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kahata K, Dadras MS and Moustakas A: TGF-β
family signaling in epithelial differentiation and
epithelial-mesenchymal transition. Cold Spring Harb Perspect Biol.
10:a0221942018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
McCormack N and O'Dea S: Regulation of
epithelial to mesenchymal transition by bone morphogenetic
proteins. Cell Signal. 25:2856–2862. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zardawi SJ, O'Toole SA, Sutherland RL and
Musgrove EA: Dysregulation of Hedgehog, Wnt and Notch signalling
pathways in breast cancer. Histol Histopathol. 24:385–398.
2009.PubMed/NCBI
|
|
35
|
Miyazono K: Transforming growth
factor-beta signaling in epithelial-mesenchymal transition and
progression of cancer. Proc Jpn Acad Ser B Phys Biol Sci.
85:314–323. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kataoka H, Murayama T, Yokode M, Mori S,
Sano H, Ozaki H, Yokota Y, Nishikawa S and Kita T: A novel
snail-related transcription factor Smuc regulates basic
helix-loop-helix transcription factor activities via specific E-box
motifs. Nucleic Acids Res. 28:626–633. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cano A, Pérez-Moreno MA, Rodrigo I,
Locascio A, Blanco MJ, del Barrio MG, Portillo F and Nieto MA: The
transcription factor snail controls epithelial-mesenchymal
transitions by repressing E-cadherin expression. Nat Cell Biol.
2:76–83. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Frietze S, O'Geen H, Blahnik KR, Jin VX
and Farnham PJ: ZNF274 recruits the histone methyltransferase
SETDB1 to the 3′ ends of ZNF genes. PLoS One. 5:e150822010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Du D, Katsuno Y, Meyer D, Budi EH, Chen
SH, Koeppen H, Wang H, Akhurst RJ and Derynck R: Smad3-mediated
recruitment of the methyltransferase SETDB1/ESET controls Snail1
expression and epithelial-mesenchymal transition. EMBO Rep.
19:135–155. 2018. View Article : Google Scholar : PubMed/NCBI
|