Introduction
Glioblastoma (GBM; also known as grade IV
astrocytoma) is the most aggressive and lethal type of brain tumor,
according to the World Health Organization (WHO) criteria (1). Due to the high invasive potential of
GBM cells, they easily infiltrate into the healthy brain tissues
and ultimately result in tumor recurrence and patient mortality
(2). Despite continuous
developments in GBM treatment, the median survival time of GBM
patients is still only around 14 months (3). Therefore, assessment and
identification of the molecular events underlying the biological
behavior of invasive tumor cells may provide novel markers for GBM
treatment and improve patient prognosis.
Myristoylated alanine rich protein kinase C
substrate (MARCKS) was first identified over 20 years ago in brain
synaptosomes (4). It is involved in
cellular processes, such as motility through control of the actin
cytoskeleton, motility and membrane trafficking (5,6).
MARCKS is a well conserved protein that is ubiquitously expressed
in various tissues. Several studies have demonstrated that MARCKS
is involved in the pathological processes of various malignancies,
including tumor invasion, apoptosis and therapeutic resistance
(7–11). However, the role of MARCKS in glioma
tumorigenesis remains poorly understood. It has been reported that
MARCKS expression is inversely correlated with GBM cell
proliferation, suggesting that MARCKS may be regarded as a tumor
suppressor (12). However, another
study on MARCKS in glioma observed that higher MARCKS expression
leads to increased tumor invasion (13). These data suggest that the function
of MARCKS in glioma is multifaceted and complex.
In the present study, MARCKS expression in GBM
specimens was determined and its biological roles in glioma
tumorigenesis were characterized. It was shown that MARCKS
expression was upregulated in GBMs, and patients with high MARCKS
protein expression had shorter survival times. In addition, it was
demonstrated that inhibition of MARCKS in vitro suppressed
cell migration and invasion, resulting in the decreased expression
of its downstream epithelial-mesenchymal transition
(EMT)-associated genes. These data indicate that MARCKS may be a
prognostic biomarker and potential therapeutic target for GBM.
Materials and methods
Tissue samples
A total of 62 tumor samples and 30 normal brain
tissues from patients with GBM (38 males and 24 females; age range,
17–67; median, 46.3 years), confirmed by a pathologist according to
the WHO criteria (1), were
collected from the Neurosurgery Department of the Affiliated
Hospital of Southwest Medical University (Sichuan, China) between
January 2012 and January 2015. Normal brain tissue from the
peritumoral area was obtained during the tumor resection
procedures. These tissues were examined by a pathologist and
confirmed to be free of tumor cells. None of the included 62
patients with GBM had undergone preoperative chemotherapy or
radiotherapy, and complete follow-up data were collected. All
patients with GBM were followed up from 4 to 30 months. The overall
survival (OS) was defined as from the date of histological
diagnosis of GBM to the date of death or last known alive. The
present study was approved by the Ethics Committees of the
Affiliated Hospital of Southwest Medical University and informed
consent was obtained from all the patients whose clinical tissue
samples were used for research purposes.
Survival analysis
Survival data were collected for all 62 patients
with GBM, who were grouped into low or high expression groups,
according to the mean mRNA expression level of MARCKS (mean value,
1.148). The Kaplan-Meier method was used to evaluate patient
survival.
RNA extraction and reverse
transcription, quantitative polymerase chain reaction
(RT-qPCR)
Total RNA was isolated from GBM and normal brain
tissues using an RNA extraction kit (Tiangen Biotech Co., Ltd.,
Beijing, China), according to the manufacturer's instructions. The
concentration and purity of total RNA was measured on a Nanodrop
ND-100 spectrophotometer (Thermo Fisher Scientific, Inc., Waltham,
MA, USA). Total RNA was converted into cDNA using ReverTra
Ace® qPCR RT Master mix (Toyobo Life Science, Osaka,
Japan), according to the manufacturer's protocol. Subsequently,
qPCR was conducted using a SYBR-Green Realtime PCR Master mix
(Toyobo Life Science), according to the manufacturer's protocol.
The PCR primers for GAPDH and MARCKS were synthesized by Sangon
Biotech Co., Ltd. (Shanghai, China) and the sequences were as
follows: MARCKS sense primer, 5′-AGCCCGGTAGAGAAGGAGG-3′, and
antisense primer, 5′-TTGGGCGAAGAAGTCGAGGAG-3′; GAPDH sense primer,
5′-ATCATCAGCAATGCCTCCTG-3′ and antisense,
5′-ATGGACTGTGGTCATGAGTC-3′. GAPDH was used as an internal control.
The relative gene expression data was analyzed by the
2−∆∆Cq method (14).
Cell culture and treatment
Human GBM U87 MG and LN-229 cell lines (American
Type Culture Collection, Manassas, VA, USA) were provided by Dr
Y.W. Liu of the Nanfang Hospital of Southern Medical University
(Guangzhou, China). Cells were cultured in Dulbecco's modified
Eagle's medium (DMEM; Hyclone; GE Healthcare Life Sciences, Logan,
UT, USA) with 10% fetal bovine serum (FBS; Hyclone; GE Healthcare
Life Sciences) at 37°C with 5% CO2. For drug treatment,
U87 and LN229 glioma cells were treated with 50 µM phosphoinositide
3-kinase (PI3K) inhibitor LY294002 (Tocris Bioscience, Bristol, UK)
for 24 h at 37°C.
Cell transient transfection with small
interfering (si)RNAs
MARCKS siRNA (si-MARCKS) was designed and chemically
synthesized by Sangon Biotech Co., Ltd. The sequence of si-MARCKS
was as follows: Sense, 5′-GCCCAGTTCTCCAAGACCGTT-3′ and antisense,
5′-CGGUCUUGGAGAACUGGGCTT-3′. The sequence of the si-negative
control (si-NC) was also designed by Sangon Biotech Co., Ltd.
(sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′). U87 and LN229 cells were seeded onto
a 6-well plate (Corning Incorporated, Corning, NY, USA) and grown
to 60–70% confluence 24 h before transfection. si-MARCKS and si-NC
(50 nmol/l) were then transfected into cells using
Lipofectamine® 2000 siRNA transfection reagent
(Fermentas; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Lipofectamine® 2000 siRNA
transfection reagent alone was added to the culture medium of the
U87 and LN229 cells to serve as the control group (Blank). Cells
were collected after 24 h for further experiments.
Matrigel invasion assays
Cell invasion was assessed using a Transwell assay
with 24-well Transwell plates (8 µm pore size; BD Biosciences,
Franklin Lakes, NJ, USA); chamber membranes were precoated with 45
µg Matrigel (BD Biosciences) to form a matrix barrier. For the
invasion assay, 5×104 transfected cells were suspended
in 200 µl serum-free DMEM and added to the upper chambers. DMEM
(600 µl) with 10% FBS was placed in each of the lower chambers.
Following incubation for 8 h at 37°C in a 5% CO2
atmosphere, cells remaining on the upper membrane were removed
carefully with cotton wool. Cells that had invaded through the
membrane were fixed in pure methanol and stained with 0.5% crystal
violet (Beyotime Institute of Biotechnology, Haimen, China) for 8
min at room temperature, rinsed in PBS and subsequently counted in
10 microscopic fields (magnification, ×100) and photographed using
an inverted phase contrast microscope (Leica Microsystems GmbH,
Wetzlar, Germany). Experiments were independently repeated three
times.
Scratch migration assay
Transfected U87 and LN229 cells were cultured in
DMEM with 1% FBS in two 6-well plates until fully confluent. A
straight scratch was carefully made through the central axis of the
plate using a 20 µl micropipette tip. Images were acquired every 8
h of the same scratched region until the scratch closed completely.
Images were captured using an inverted phase contrast microscope
(magnification, ×50).
Western blot analysis
Western blot analysis was performed as previously
described (15,16) with primary rabbit polyclonal
antibodies including those against: MARCKS (cat. no. ab52616;
1:5,000; Abcam, Cambridge, UK), PI3K, phosphorylated (p)PI3K
(Tyr458; cat. no. 9655; 1:1,000; Cell Signaling Technology, Inc.,
Danvers, MA, USA), Akt (cat. no. 4691; 1:1,000, Cell Signaling
Technology, Inc.), pAkt (Ser473; cat. no. 4060; 1:2,000; Cell
Signaling Technology, Inc.), β-catenin (1:1,000; cat. no. 8480;
Cell Signaling Technology, Inc.), N-cadherin (1:1,000; cat. no.
13116; Cell Signaling Technology, Inc.), vimentin (1:1,000; cat.
no. 5741; Cell Signaling Technology, Inc.), E-cadherin (1:1,000;
cat. no. 3195; Cell Signaling Technology, Inc.), Zinc finger
protein SNAI1 (Snail; 1:1,000; cat. no. 3879; Cell Signaling
Technology, Inc.), Zinc finger protein SNAI2 (Slug; 1:1,000; cat.
no. 9585; Cell Signaling Technology, Inc.) and β-actin (1:1,000;
cat. no. 4970; Cell Signaling Technology, Inc.). The horseradish
peroxidase-conjugated anti-rabbit IgG secondary antibody (1:1,000;
cat. no. 7074; Cell Signaling Technology, Inc.) was used. Protein
signals were detected using an chemiluminescent detection system
(Pierce; Thermo Fisher Scientific, Inc.). The gray-scale value was
quantified by ImageJ software (version 1.8.0; National Institutes
of Health, Bethesda, MD, USA) to calculate relative protein
expression.
Statistical analysis
All experiments were repeated at least three times
and data were expressed as the mean ± standard deviation.
Differences in MARCKS expression between two groups were compared
using the Mann-Whitney U test. One-way analysis of variance or
Student's t-test was used for comparisons between groups, followed
by the Least Significant Difference test. P<0.05 was considered
to indicate a statistically significant difference. Statistical
analyses were performed using SPSS 20.0 (IBM Corp., Armonk, NY,
USA). The Kaplan-Meier estimate was used to evaluate and compare
the prognosis of patients with GBM using GraphPad Prism 6.0
(GraphPad Software, Inc., La Jolla, CA, USA).
Results
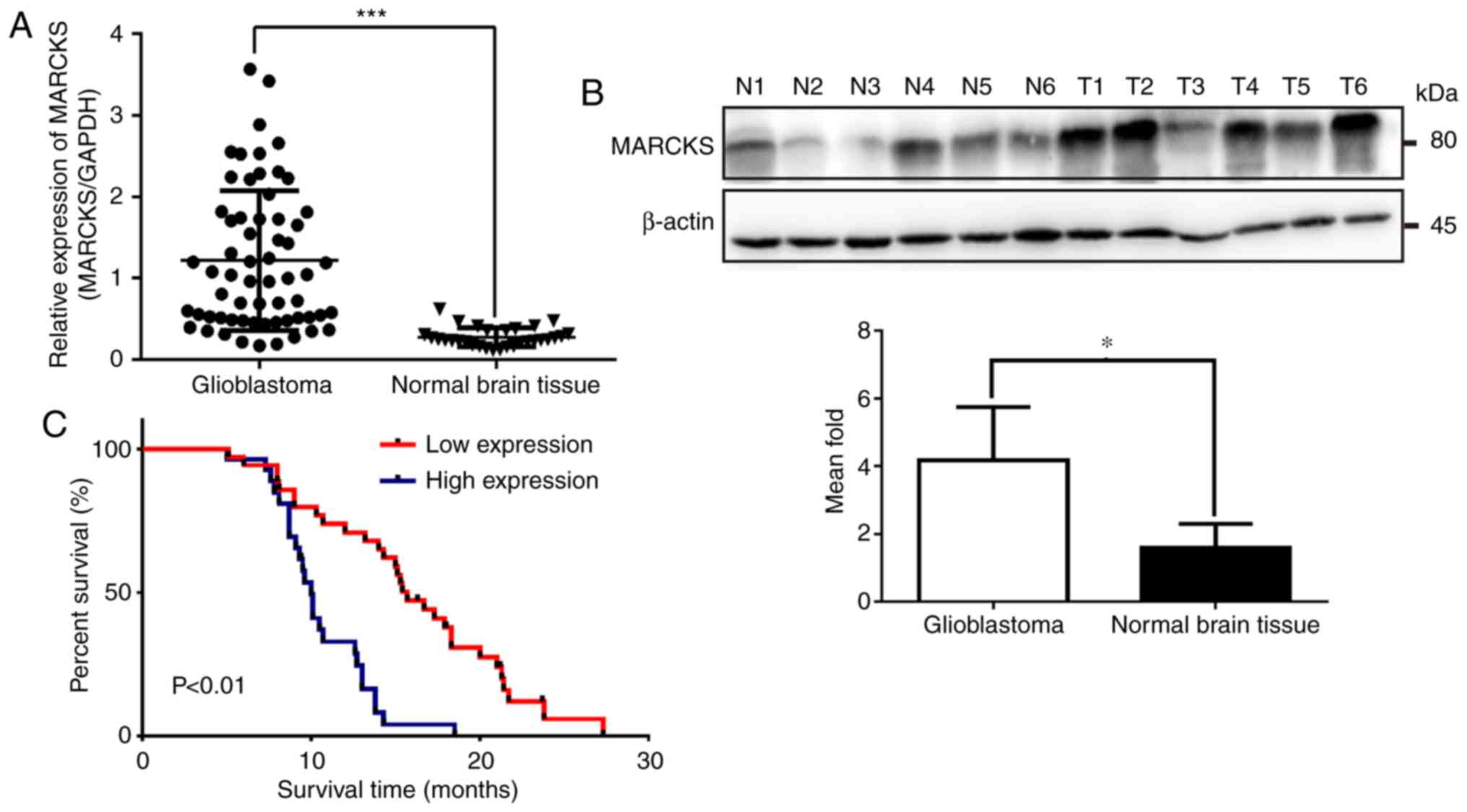
MARCKS is overexpressed in GBM tissues
and is associated with patient survival
To assess the role of MARCKS in GBM tumorigenesis,
RT-qPCR was performed to examine the expression of MARCKS mRNA in
62 GBM tissues and 30 normal brain tissues. The results showed that
MARCKS mRNA expression markedly increased in GBM tissues compared
with normal brain tissues (***P<0.001; Fig. 1A). Additionally, MARCKS protein
expression was upregulated in six GBM samples, when compared with
six normal brain tissues, as determined by western blot analysis
(*P<0.05; Fig. 1B). Furthermore,
to investigate the relationship between the expression level of
MARCKS and the outcome of patients with GBM, MARCKS expression in
all 62 GBM tissues was categorized as high or low expression
according to the mean value (Table
I). Kaplan-Meier analysis revealed that the patients with GBM
and high MARCKS expression had considerably worse outcomes than
those who had low MARCKS expression (P<0.01; Fig. 1C).
 | Table I.The level of expression of MARCKS
between glioblastoma and normal brain tissues. |
Table I.
The level of expression of MARCKS
between glioblastoma and normal brain tissues.
|
|
| mRNA expression |
|
|---|
|
|
|
|
|
|---|
| Group | Cases | High | Low | P-value |
|---|
| Glioblastoma | 62 | 40 | 22 | <0.001 |
| Normal brain
tissue | 30 | 9 | 21 |
|
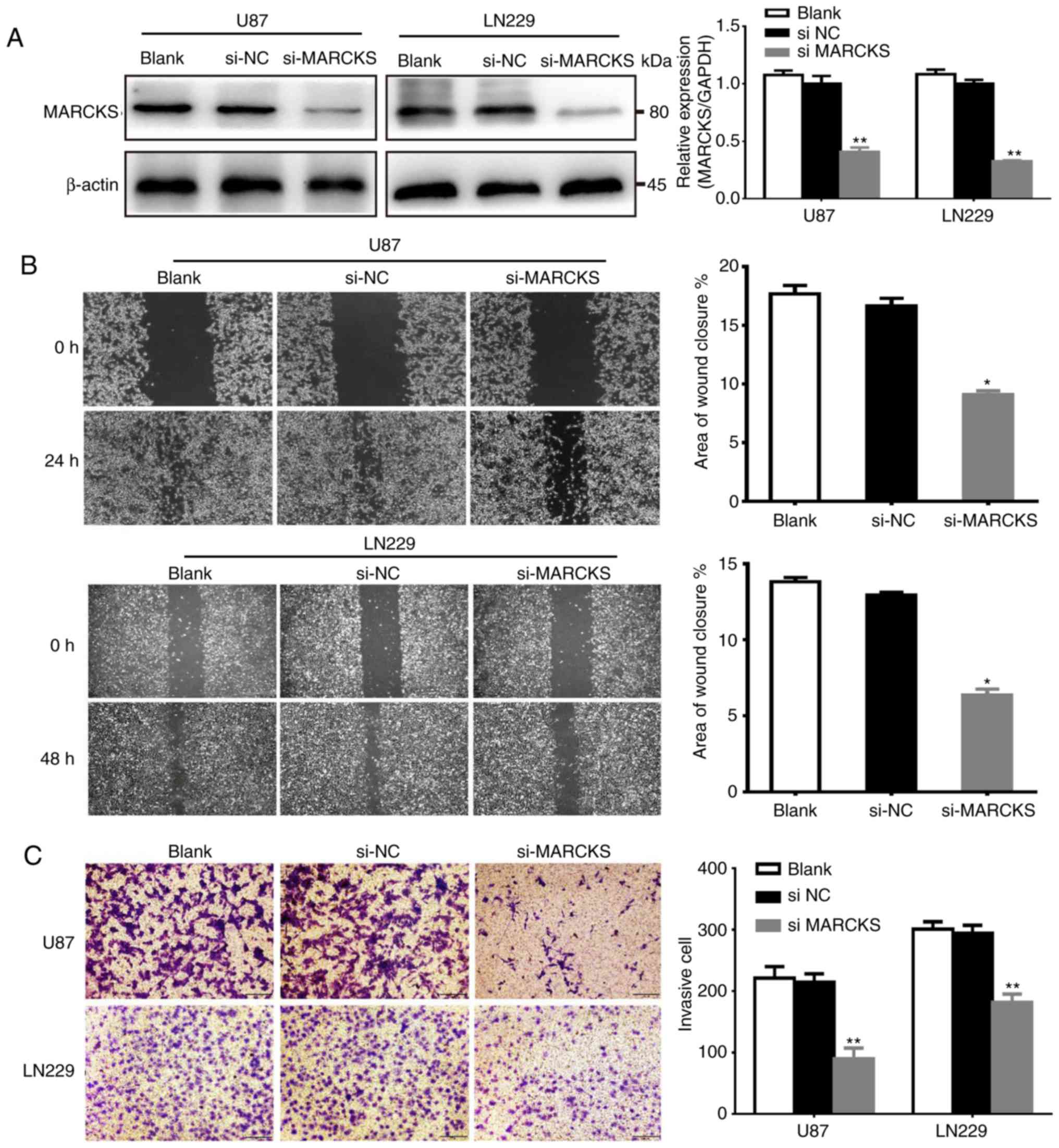
Downregulation of MARCKS suppresses
GBM cell migration and invasion in vitro
To determine the function of MARCKS in GBM, the
expression of MARCKS was knocked down in U87 and LN229 cell lines,
which were established from GBM via the transfection of siRNA. The
interference efficiency of siRNA was detected by qPCR and western
blot analysis. The data showed that the expression of MARCKS in the
si-MARCKS group was significantly downregulated in the U87 and
LN229 cell lines, compared with the si-NC and the untransfected
group (**P<0.01; Fig. 2A).
Subsequently, the effects of MARCKS downregulation
on the invasion and migration of both U87 and LN229 cells was
investigated in vitro with Matrigel invasion and scratch
assays. The data showed that downregulation of MARCKS markedly
decreased the invasive ability of GBM cells, compared with the
si-NC group. Furthermore, si-MARCKS also significantly inhibited
the migratory capacity of cells, compared with the
si-NC-transfected and Blank cells (*P<0.05, **P<0.01;
Fig. 2B and C).
MARCKS regulates the expression of
EMT-associated genes in GBM
To further investigate the mechanism of MARCKS in
GBM cell invasion and migration, the protein expression of
EMT-associated genes in U87 and LN229 cells with downregulated
MARCKS expression was examined by western blot analysis. Knocking
down MARCKS decreased the expression of β-catenin, vimentin and
N-cadherin, while increasing that of E-cadherin (*P<0.05,
**P<0.01; Fig. 3A).
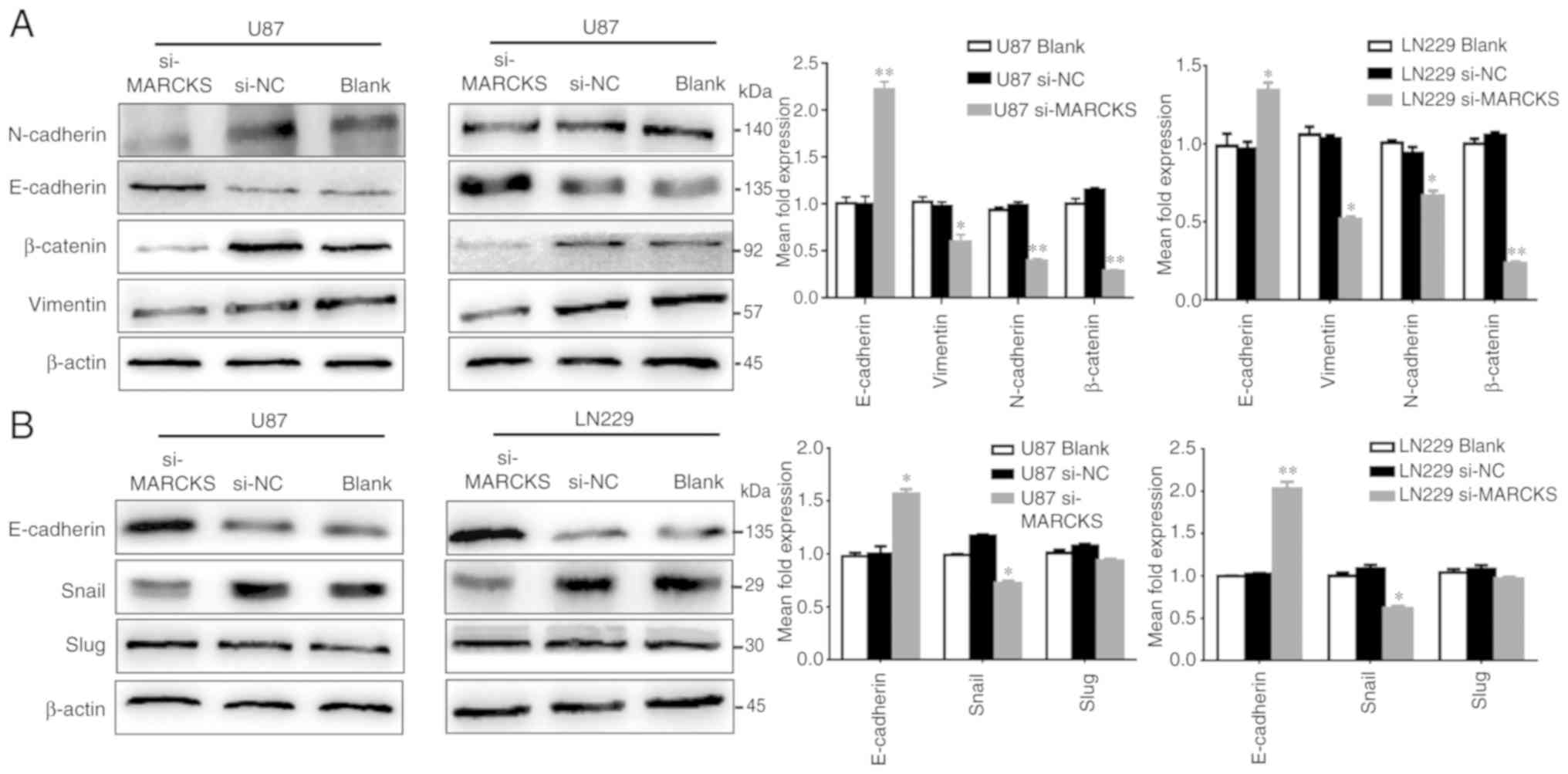 | Figure 3.MARCKS regulates the expression of
EMT-associated genes in GBM cells. (A) Knockdown of the expression
of MARCKS by si-MARCKS in U87 and LN229 cells enhanced E-cadherin
expression, but decreased the expression of EMT-marker genes
including β-catenin, N-cadherin and vimentin. *P<0.05,
**P<0.01 vs. si-NC. (B) In si-MARCKS-transfected U87 and LN229
cells, the protein expression of Snail was downregulated and
E-cadherin was upregulated. Slug expression was unchanged.
*P<0.05, **P<0.01 vs. si-NC. MARCKS, myristoylated
alanine-rich C kinase substrate; si, small interfering RNA; NC,
negative control; Snail, zinc finger protein SNAI1; Slug, zinc
finger protein SNAI2. |
MARCKS inhibits the expression of
E-cadherin by upregulating Snail expression in GBM cells
It has been reported that the expression of
E-cadherin may be inhibited by EMT-associated transcription
factors, such as Snail and Slug in cancer cells (17–20).
To study whether MARCKS modulates E-cadherin expression by
affecting the expression of these EMT-associated transcription
factors, the expression of MARCKS in U87 and LN229 cells was
knocked down and western blotting was used to examine the
alterations in E-cadherin, Snail and Slug protein expression. It
was found that the downregulation of MARCKS in U87 and LN229 cells
significantly inhibited Snail expression and increased E-cadherin
expression, whereas the expression of Slug remained unchanged
(*P<0.05; Fig. 3B).
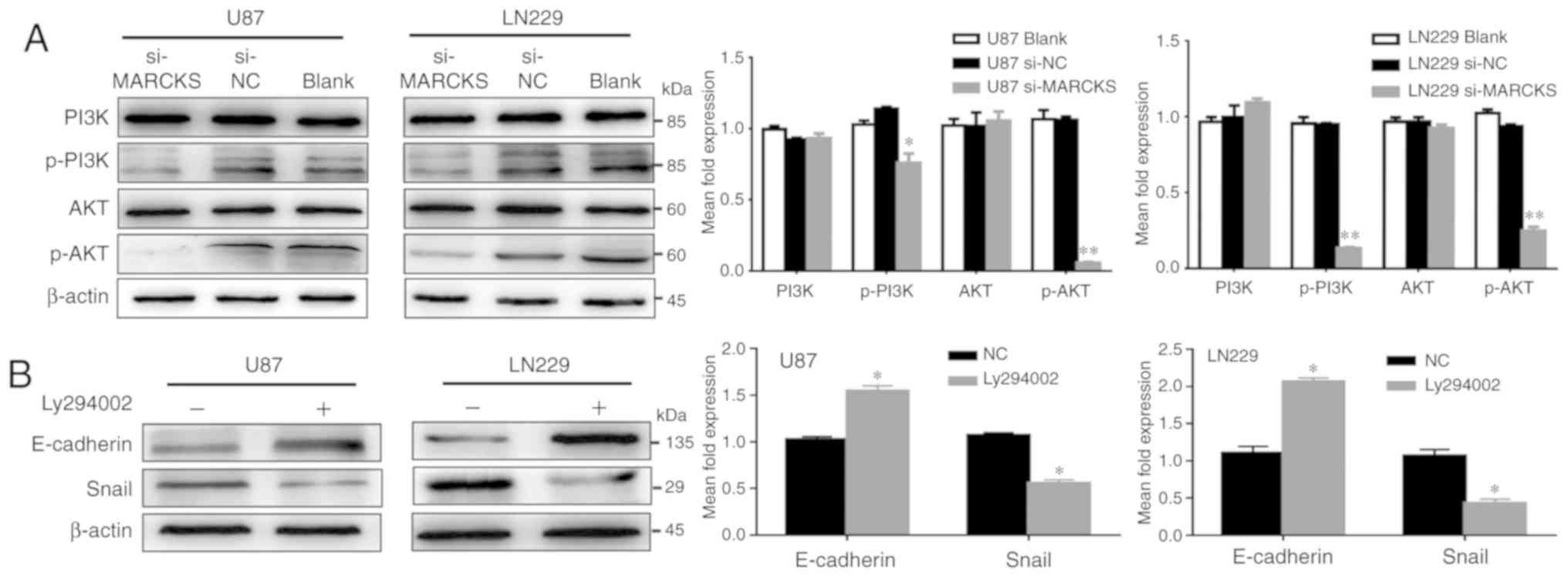
MARCKS modulates GBM cell invasion and
migration through the PI3K/Akt/Snail/E-cadherin pathways
A previous study reported that inhibition of the
PI3K/Akt pathway, which is known to be an upstream signaling
pathway involved regulating EMT signals, could downregulate the
expression of Snail (21,22). The present study investigated the
role of MARCKS in the PI3K/Akt pathway and found that
phosphorylated PI3K and Akt expression was significantly
downregulated in U87 and LN229 cells following the knockdown of
MARCKS, but total PI3K and Akt protein expression (*P<0.05,
**P<0.01; Fig. 4A). In addition,
the suppression of PI3K in U87 and LN229 GBM cells using LY294002,
also decreased Snail and increased E-cadherin expression, similar
to the effects of MARCKS downregulation (*P<0.05; Fig. 4B).
Discussion
Although the expression of MARCKS in glioma has been
reported, the biological effects, functions and underlying
molecular mechanisms of MARCKS in the invasion and migration of GBM
cells have not yet been well-characterized (12,13).
In the present study, using RT-qPCR and western blotting, it was
found that MARCKS was significantly upregulated in GBM specimens
compared with normal brain tissues. Furthermore, higher levels of
MARCKS were associated with worse outcomes in patients with GBM,
suggesting that MARCKS may be a prognostic factor. These results
were consistent with those of previous studies, which demonstrated
that MARCKS expression is upregulated in breast cancer,
osteosarcoma and hepatocellular carcinoma (23–25).
These data imply that MARCKS may play an oncogenic role in GBM
tumorigenesis.
Several studies have reported that MARCKS is
associated with the proliferation, invasion and migration of
several tumors, including lung cancer, prostate cancer and
hepatocellular carcinoma (6,25,26).
Furthermore, Micallef et al (13) showed that MARCKS serves as a
mediator of attachment and invasion in epidermal growth factor
receptor variant III-expressing GBM cells (13). However, the molecular mechanisms
underlying its effects on tumor cell invasion and migration remain
elusive. Therefore, in the current study, the role of MARCKS in GBM
cell invasion and migration was first determined. It was shown that
downregulated MARCKS expression inhibited GBM cell invasion and
migration in vitro.
Furthermore, EMT, which may be associated with
alterations in: Epithelial marker expression, such as E-cadherin;
mesenchymal marker expression, such as β-catenin, vimentin, and
N-cadherin; and the expression of several key transcription
repression factors, such as Snail and Slug. These proteins serve an
important role in cancer cell invasion and migration (27–31).
Thus, the underlying mechanisms involved in MARCKS-induced cell
invasion and migration were determined by examining the effects of
MARCKS on these EMT-associated proteins. Decreased Snail protein
expression and a concomitant increase in E-cadherin expression was
observed following the downregulation of MARCKS in GBM cell lines.
These data indicated that MARCKS may have modulated the expression
of E-cadherin via Snail, therefore resulting in EMT regulation.
Many studies have demonstrated that the PI3K/Akt
pathway is widely involved in human cancer migration, proliferation
and survival. Activation of the PI3K/Akt pathway increases Snail
and suppresses E-cadherin expression, thereby inducing EMT and
promoting invasion and migration (32–35).
The results of the present study found that the expression of
p-PI3K and p-Akt was downregulated following MARCKS knockdown, and
the treatment of U87 and LN229 cells with LY294002 had a similar
effect on E-cadherin and Snail expression, indicating that MARCKS
may be an upstream effector regulating the PI3K/Akt pathway in GBM.
Inactivation of the PI3K/Akt pathway was perhaps responsible for
the si-MARCKS-mediated inhibition of GBM cell invasion and
migration. Therefore, these results indicated that MARCKS may
conduce to GBM EMT via the PI3K/Akt pathway.
In summary, and to the best of our knowledge, the
present study is the first to demonstrate the function of MARCKS in
GBM, and to demonstrate that MARCKS promoted GBM cell invasion and
migration through activation of the PI3K/Akt pathway, which may
have inhibited EMT by increasing the expression of Snail and
reducing E-cadherin expression. Furthermore, it was shown that
MARCKS may be a poor prognostic marker in GBM. Therefore, it was
concluded that MARCKS may serve an important role in GBM
tumorigenesis and represents a potential therapeutic target for the
treatment of GBM.
Acknowledgements
We thank Dr Y.W. Liu of the Nanfang Hospital of
Southern Medical University (Guangzhou, China) for providing the
GBM U87 MG and LN-229 cell lines (American Type Culture Collection,
Manassas, VA, USA).
Funding
This study was funded by Medical Research Fund of Si
Chuan Medical Association (grant no. S16083), the Medical Research
Fund for Young Scholars of the Sichuan Medical Association (grant
no. Q16076), the Natural Science Foundation of Southwest Medical
University (grant no. 2016XNYD217) and Youth Fund of Southwest
Medical University (grant no. XNYD00030658).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JZ, WX, YM and LC conceived and designed the
experiments. JZ, WX, TP, LGC, YM, SL, KW, HW performed the
experiments. JZ, WX, TP, LC and YM analyzed the data. JZ, WX, TP,
LC provided materials and collected the clinical data. JZ, WX, TP,
YM and LC wrote the paper. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All procedures performed in studies involving human
participants were in accordance with the ethical standards of the
Ethics Committees of the Affiliated Hospital of Southwest Medical
University and with the 1964 Helsinki declaration and its later
amendments or comparable ethical standards. Informed consent was
obtained from all individual participants included in the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EMT
|
epithelial-mesenchymal transition
|
|
MARCKS
|
myristoylated alanine-rich C kinase
substrate
|
|
PI3K
|
phosphoinositide 3-kinase
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
siRNA
|
small interfering RNA
|
References
|
1
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Demuth T and Berens ME: Molecular
mechanisms of glioma cell migration and invasion. J Neurooncol.
70:217–28. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Albert KA, Nairn AC and Greengard P: The
87-kDa protein, a major specific substrate for protein kinase C:
Purification from bovine brain and characterization. Proc Natl Acad
Sci USA. 84:7046–7050. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arbuzova A, Schmitz AA and Vergères G:
Cross-talk unfolded: MARCKS proteins. Biochem J. 362:1–12. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Caroni P: New EMBO members' review: Actin
cytoskeleton regulation through modulation of PI(4,5)P(2) rafts.
EMBO J. 20:4332–4336. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hanada S, Kakehashi A, Nishiyama N, Wei M,
Yamano S, Chung K, Komatsu H, Inoue H, Suehiro S and Wanibuchi H:
Myristoylated alanine-rich C-kinase substrate as a prognostic
biomarker in human primary lung squamous cell carcinoma. Cancer
Biomark. 13:289–298. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rombouts K, Carloni V, Mello T, Omenetti
S, Galastri S, Madiai S, Galli A and Pinzani M: Myristoylated
Alanine-Rich protein Kinase C Substrate (MARCKS) expression
modulates the metastatic phenotype in human and murine colon
carcinoma in vitro and in vivo. Cancer Lett. 333:244–252. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Techasen A, Loilome W, Namwat N, Takahashi
E, Sugihara E, Puapairoj A, Miwa M, Saya H and Yongvanit P:
Myristoylated alanine-rich C kinase substrate phosphorylation
promotes cholangiocarcinoma cell migration and metastasis via the
protein kinase C-dependent pathway. Cancer Sci. 101:658–665. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang Y, Chen Y, Saha MN, Chen J, Evans K,
Qiu L, Reece D, Chen GA and Chang H: Targeting phospho-MARCKS
overcomes drug-resistance and induces antitumor activity in
preclinical models of multiple myeloma. Leukemia. 29:715–726. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang Z, Xu S, Jin P, Yang X, Li X, Wan D,
Zhang T, Long S, Wei X, Chen G, et al: MARCKS contributes to
stromal cancer-associated fibroblast activation and facilitates
ovarian cancer metastasis. Oncotarget. 7:37649–37663.
2016.PubMed/NCBI
|
|
12
|
Jarboe JS, Anderson JC, Duarte CW, Mehta
T, Nowsheen S, Hicks PH, Whitley AC, Rohrbach TD, McCubrey RO, Chiu
S, et al: MARCKS regulates growth and radiation sensitivity and is
a novel prognostic factor for glioma. Clin Cancer Res.
18:3030–3041. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Micallef J, Taccone M, Mukherjee J, Croul
S, Busby J, Moran MF and Guha A: Epidermal growth factor receptor
variant III-induced glioma invasion is mediated through
myristoylated alanine-rich protein kinase C substrate
overexpression. Cancer Res. 69:7548–7556. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xiang W, Qi ST, Liu YW, Li HZ, Zhou Q, Yi
GZ, Chen ZY and Yan L: RNA interference of PC4 and SFRS1
interacting protein 1 inhibits invasion and migration of U87 glioma
cells. Nan Fang Yi Ke Da Xue Xue Bao. 36:802–806. 2016.(In
Chinese). PubMed/NCBI
|
|
16
|
Yi GZ, Liu YW, Xiang W, Wang H, Chen ZY,
Xie SD and Qi ST: Akt and β-catenin contribute to TMZ resistance
and EMT of MGMT negative malignant glioma cell line. J Neurol Sci.
367:101–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bolós V, Peinado H, Pérez-Moreno MA, Fraga
MF, Esteller M and Cano A: The transcription factor Slug represses
E-cadherin expression and induces epithelial to mesenchymal
transitions: A comparison with Snail and E47 repressors. J Cell
Sci. 116:499–511. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moreno-Bueno G, Portillo F and Cano A:
Transcriptional regulation of cell polarity in EMT and cancer.
Oncogene. 27:6958–6969. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Peinado H, Ballestar E, Esteller M and
Cano A: Snail mediates E-cadherin repression by the recruitment of
the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell
Biol. 24:306–319. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang KJ, Wang DS, Zhang SY, Jiao XL, Li
CW, Wang XS, Yu QC and Cui HN: The E-cadherin repressor slug and
progression of human extrahepatic hilar cholangiocarcinoma. J Exp
Clin Cancer Res. 29:882010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lau MT and Leung PC: The PI3K/Akt/mTOR
signaling pathway mediates insulin-like growth factor 1-induced
E-cadherin down-regulation and cell proliferation in ovarian cancer
cells. Cancer Lett. 326:191–198. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jeon YK, Kim CK, Hwang KR, Park HY, Koh J,
Chung DH, Lee CW and Ha GH: Pellino-1 promotes lung carcinogenesis
via the stabilization of Slug and Snail through K63-mediated
polyubiquitination. Cell Death Differ. 24:469–480. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Manai M, Thomassin-Piana J, Gamoudi A,
Finetti P, Lopez M, Eghozzi R, Ayadi S, Lamine OB, Manai M, Rahal
K, et al: MARCKS protein overexpression in inflammatory breast
cancer. Oncotarget. 8:6246–6257. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu H, Su P, Zhi L and Zhao K: miR-34c-3p
acts as a tumor suppressor gene in osteosarcoma by targeting
MARCKS. Mol Med Red. 15:1204–1210. 2017. View Article : Google Scholar
|
|
25
|
Song J, Wang Q, Luo Y, Yuan P, Tang C, Hui
Y and Wang Z: miR-34c-3p inhibits cell proliferation, migration and
invasion of hepatocellular carcinoma by targeting MARCKS. Int J
Clin Exp Pathol. 8:12728–12737. 2015.PubMed/NCBI
|
|
26
|
Li T, Li D, Sha J, Sun P and Huang Y:
MicroRNA-21 directly targets MARCKS and promotes apoptosis
resistance and invasion in prostate cancer cells. Biochem Biophys
Res Commun. 383:280–285. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Grunert S, Jechlinger M and Beug H:
Diverse cellular and molecular mechanisms contribute to epithelial
plasticity and metastasis. Nat Rev Mol Cell Biol. 4:657–665. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Christofori G: New signals from the
invasive front. Nature. 441:444–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Batlle E, Sancho E, Francí C, Domínguez D,
Monfar M, Baulida J and García De Herreros A: The transcription
factor snail is a repressor of E-cadherin gene expression in
epithelial tumour cells. Nat Cell Biol. 2:84–89. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cano A, Pérez-Moreno MA, Rodrigo I,
Locascio A, Blanco MJ, del Barrio MG, Portillo F and Nieto MA: The
transcription factor snail controls epithelial-mesenchymal
transitions by repressing E-cadherin expression. Nat Cell Biol.
2:76–83. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Takkunen M, Grenman R, Hukkanen M,
Korhonen M, Garcia de Herreros A and Virtanen I: Snail-dependent
and -independent epithelial-mesenchymal transition in oral squamous
carcinoma cells. J Histochem Cytochem. 54:1263–1275. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bakin AV, Tomlinson AK, Bhowmick NA, Moses
HL and Arteaga CL: Phosphatidylinositol 3-kinase function is
required for transforming growth factor beta-mediated epithelial to
mesenchymal transition and cell migration. J Biol Chem.
275:36803–36810. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma NX, Sun W, Wu J, Liu SL, Weng L, Liu
YQ, Pu WX, Wu TT, Ding XL, Huang NG, et al: Compound wumei powder
inhibits the invasion and metastasis of gastric cancer via
Cox-2/PGE2-PI3K/AKT/GSK3β/β-catenin signaling pathway. Evid Based
Complement Alternat Med. 2017:30394502017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu WB, Xiao N and Liu XJ: Dietary
flavonoid tangeretin induces reprogramming of epithelial to
mesenchymal transition in prostate cancer cells by targeting the
PI3K/Akt/mTOR signaling pathway. Oncol Lett. 15:433–440.
2018.PubMed/NCBI
|
|
35
|
Li Z, Zhang TB, Jia DH, Sun WQ, Wang CL,
Gu AZ and Yang XM: Genipin inhibits the growth of human bladder
cancer cells via inactivation of PI3K/Akt signaling. Oncol Lett.
15:2619–2624. 2018.PubMed/NCBI
|