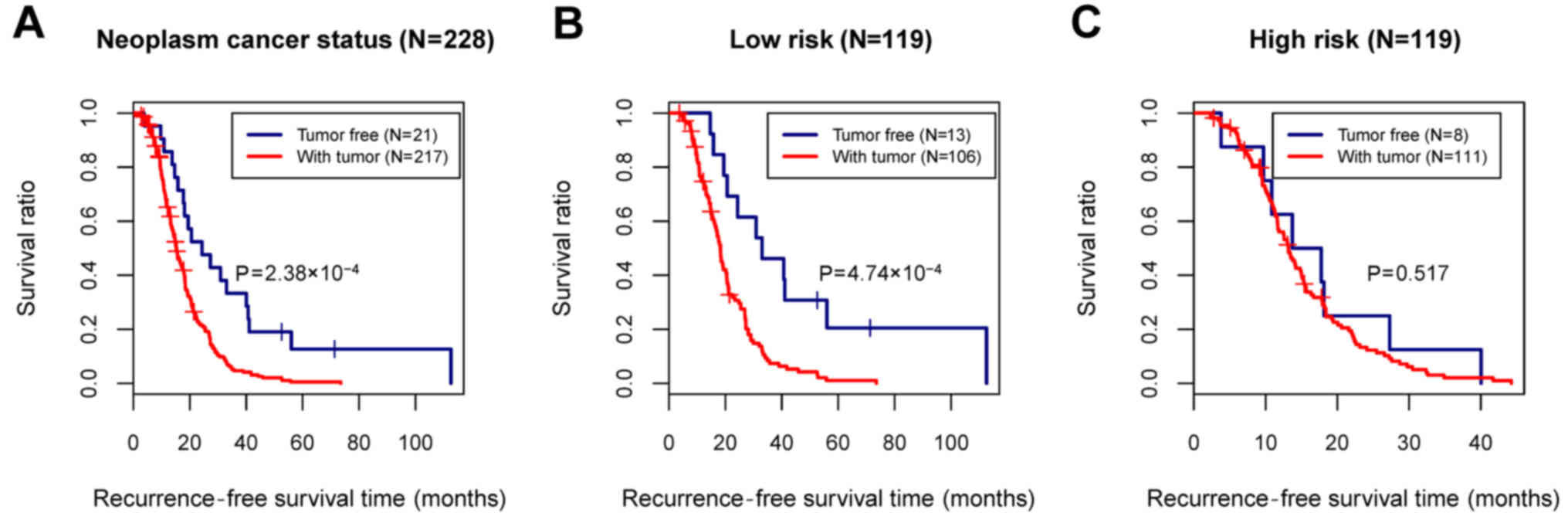
|
1
|
Moufarrij S, Dandapani M, Arthofer E,
Gomez S, Srivastava A, Lopez-Acevedo M, Villagra A and Chiappinelli
KB: Epigenetic therapy for ovarian cancer: Promise and progress.
Clin Epigenetics. 11:72019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mcguire S: World Cancer Report 2014.
Geneva, Switzerland: World Health Organization, International
Agency for research on cancer, WHO Press, 2015. Adv Nutr.
7:4182016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hanley GE, Mcalpine JN, Kwon JS and
Mitchell G: Opportunistic salpingectomy for ovarian cancer
prevention. Gynecol Oncol Res Pract. 2:52015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gibson SJ, Fleming GF, Temkin SM and Chase
DM: The application and outcome of standard of care treatment in
elderly women with ovarian cancer: A literature review over the
last 10 years. Front Oncol. 6:632016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gov E, Kori M and Arga KY: Multiomics
analysis of tumor microenvironment reveals Gata2 and miRNA-124-3p
as potential novel biomarkers in ovarian cancer. OMICS. 21:603–615.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Z, Ji G, Wu Q, Feng S, Zhao Y, Cao Z
and Tao C: Integrated microarray meta-analysis identifies miRNA-27a
as an oncogene in ovarian cancer by inhibiting FOXO1. Life Sci.
201:263–270. 2018. View Article : Google Scholar
|
|
7
|
Lv T, Song K, Zhang L, Li W, Chen Y, Diao
Y, Yao Q and Liu P: MiRNA-34a decreases ovarian cancer cell
proliferation and chemoresistance by targeting HDAC1. Biochem Cell
Biol. 96:663–671. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheng Y, Ban R, Liu W, Wang H, Li S, Yue
Z, Zhu G, Zhuan Y and Wang C: MiRNA-409-3p enhances
cisplatin-sensitivity of ovarian cancer cells by blocking the
autophagy mediated by Fip200. Oncol Res. Jan 2–2018.(Epub ahead of
print). doi: 10.3727/096504017X15138991620238. View Article : Google Scholar
|
|
9
|
Koutsaki M, Libra M, Spandidos DA and
Zaravinos A: The miR-200 family in ovarian cancer. Oncotarget.
8:66629–66640. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu X, Macdonald DM, Huettner PC, Feng Z,
El Naqa IM, Schwarz JK, Mutch DG, Grigsby PW, Powell SN and Wang X:
A miR-200 microRNA cluster as prognostic marker in advanced ovarian
cancer. Gynecol Oncol. 114:457–464. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hong F, Li Y, Xu Y and Zhu L: Prognostic
significance of serum microRNA-221 expression in human epithelial
ovarian cancer. J Int Med Res. 41:64–71. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao YC and Wu J: MicroRNA-200c and
microRNA-141 as potential diagnostic and prognostic biomarkers for
ovarian cancer. Tumor Biol. 36:4843–4850. 2015. View Article : Google Scholar
|
|
13
|
Wang S, Zhao X, Wang J, Wen Y, Zhang L,
Wang D, Chen H, Chen Q and Xiang W: Upregulation of microRNA-203 is
associated with advanced tumor progression and poor prognosis in
epithelial ovarian cancer. Med Oncol. 30:6812013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiaohong Z, Lichun F, Na X, Kejian Z,
Xiaolan X and Shaosheng W: MiR-203 promotes the growth and
migration of ovarian cancer cells by enhancing glycolytic pathway.
Tumor Biol. 37:14989–14997. 2016. View Article : Google Scholar
|
|
15
|
Xu YZ, Xi QH, Ge WL and Zhang XQ:
Identification of serum microRNA-21 as a biomarker for early
detection and prognosis in human epithelial ovarian cancer. Asian
Pac J Cancer Prev. 14:1057–1060. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wilczyński M, Żytko E, Danielska J,
Szymańska B, Dzieniecka M, Nowak M, Malinowski J, Owczarek D and
Wilczyński JR: Clinical significance of miRNA-21, −103, −129, −150
in serous ovarian cancer. Arch Gynecol Obstet. 297:741–748. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bagnoli M, De Cecco L, Granata A,
Nicoletti R, Marchesi E, Alberti P, Valeri B, Libra M, Barbareschi
M, Raspagliesi F, et al: Identification of a chrXq27.3 microRNA
cluster associated with early relapse in advanced stage ovarian
cancer patients. Oncotarget. 6:96432015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shih KK, Qin LX, Tanner EJ, Zhou Q,
Bisogna M, Dao F, Olvera N, Viale A, Barakat RR and Levine DA: A
microRNA survival signature (MiSS) for advanced ovarian cancer.
Gynecol Oncol. 121:444–450. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Therneau TM: Survival analysis [R package
survival version 2.41–3]. Technometrics. 46:111–112. 2015.
|
|
20
|
Stadler L, Welc A, Humer C and Jordan M:
Optimizing R language execution via aggressive speculation.
Symposium on Dynamic Languages. 84–95. 2016.
|
|
21
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng
Z, Zhu G, Qi J, Ma H, Nian H, et al: RNA-seq analyses of multiple
meristems of soybean: Novel and alternative transcripts,
evolutionary and functional implications. BMC Plant Biol.
14:1692014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang X, Zhang L, Wang B, Li F and Zhang
Z: Feature clustering based support vector machine recursive
feature elimination for gene selection. Appl Intelligence.
48:594–607. 2018. View Article : Google Scholar
|
|
24
|
Kumar A: Pre-processing and modelling
using caret package in R. Int J Comput Appl. 181:39–42. 2018.
|
|
25
|
Daliri MR: Feature selection using binary
particle swarm optimization and support vector machines for medical
diagnosis. Biomed Tech. 57:395–402. 2012. View Article : Google Scholar
|
|
26
|
Meyer D: Support vector machines the
interface to libsvm in package e1071. R News. 1:1–3. 2013.
|
|
27
|
Schröder MS, Culhane AC, Quackenbush J and
Haibe-Kains B: survcomp: An R/BioconductoR package for performance
assessment and comparison of survival models. Bioinformatics.
27:3206–3208. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12:772011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang DW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, et al: DAVID
Bioinformatics Resources: Expanded annotation database and novel
algorithms to better extract biology from large gene lists. Nucleic
Acids Res. 35:W169–W175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen K, Liu MX, Mak SL, Yung MM, Leung TH,
Xu D, Ngu SF, Chan KK, Yang H, Ngan HY, et al:
Methylation-associated silencing of miR-193a-3p promotes ovarian
cancer aggressiveness by targeting GRB7 and MAPK/ERK pathways.
Theranostics. 8:423–436. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang J, Qin J and Su Y: miR-193b-3p
possesses anti-tumor activity in ovarian carcinoma cells by
targeting p21-activated kinase 3. Biomed Pharmacother.
96:1275–1282. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mitra AK, Chiang CY, Tiwari P, Tomar S,
Watters KM, Peter ME and Lengyel E: Microenvironment-induced
downregulation of miR-193b drives ovarian cancer metastasis.
Oncogene. 34:5923–5932. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li H, Xu Y, Qiu W, Zhao D and Zhang Y:
Tissue miR-193b as a novel biomarker for patients with ovarian
cancer. Med Sci Monit. 21:3929–3934. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li N, Wang L, Tan G, Guo Z, Liu L, Yang M
and He J: MicroRNA-218 inhibits proliferation and invasion in
ovarian cancer by targeting Runx2. Oncotarget. 8:91530–91541.
2017.PubMed/NCBI
|
|
37
|
Ye Y, Gu B, Wang Y, Shen S and Huang W:
E2F1-mediated MNX1-AS1-miR-218-5p-SEC61A1 feedback loop contributes
to the progression of colon adenocarcinoma. J Cell Biochem.
120:6145–6153. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xia B, Yang S, Liu T and Lou G: miR-211
suppresses epithelial ovarian cancer proliferation and cell-cycle
progression by targeting Cyclin D1 and CDK6. Mol Cancer. 14:572015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tao F, Tian X, Ruan S, Shen M and Zhang Z:
miR-211 sponges lncRNA MALAT1 to suppress tumor growth and
progression through inhibiting PHF19 in ovarian carcinoma. FASEB J.
fj201800495RR2018.PubMed/NCBI
|
|
40
|
Zhao H, Ding Y, Tie B, Sun ZF, Jiang JY,
Zhao J, Lin X and Cui S: miRNA expression pattern associated with
prognosis in elderly patients with advanced OPSC and OCC. Int J
Oncol. 43:839–849. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chan CK, Pan Y, Nyberg K, Marra MA, Lim
EL, Jones SJ, Maar D, Gibb EA, Gunaratne PH, Robertson AG, et al:
Tumour-suppressor microRNAs regulate ovarian cancer cell physical
properties and invasive behaviour. Open Biol. 6:1602752016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hong L, Wang Y, Chen W and Yang S:
MicroRNA-508 suppresses epithelial-mesenchymal transition,
migration, and invasion of ovarian cancer cells through the
MAPK1/ERK signaling pathway. J Cell Biochem. 119:7431–7440. 2011.
View Article : Google Scholar
|
|
43
|
Xiao S, Zhang M, Liu C and Wang D: MiR-514
attenuates proliferation and increases chemoresistance by targeting
ATP binding cassette subfamily in ovarian cancer. Mol Genet
Genomics. May 11–2018.(Epub ahead of print). doi:
10.1007/s00438-018-1447-0. View Article : Google Scholar
|
|
44
|
Yin J, Lu K, Lin J, Wu L, Hildebrandt MA,
Chang DW, Meyer L, Wu X and Liang D: Genetic variants in TGF-β
pathway are associated with ovarian cancer Risk. PLoS One.
6:e255592011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Matsumura N, Huang Z, Mori S, Baba T,
Fujii S, Konishi I, Iversen ES, Berchuck A and Murphy SK:
Epigenetic suppression of the TGF-beta pathway revealed by
transcriptome profiling in ovarian cancer. Genome Res. 21:74–82.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lee SH and Dominguez R: Regulation of
actin cytoskeleton dynamics in cells. Mol Cells. 29:311–325. 2010.
View Article : Google Scholar : PubMed/NCBI
|