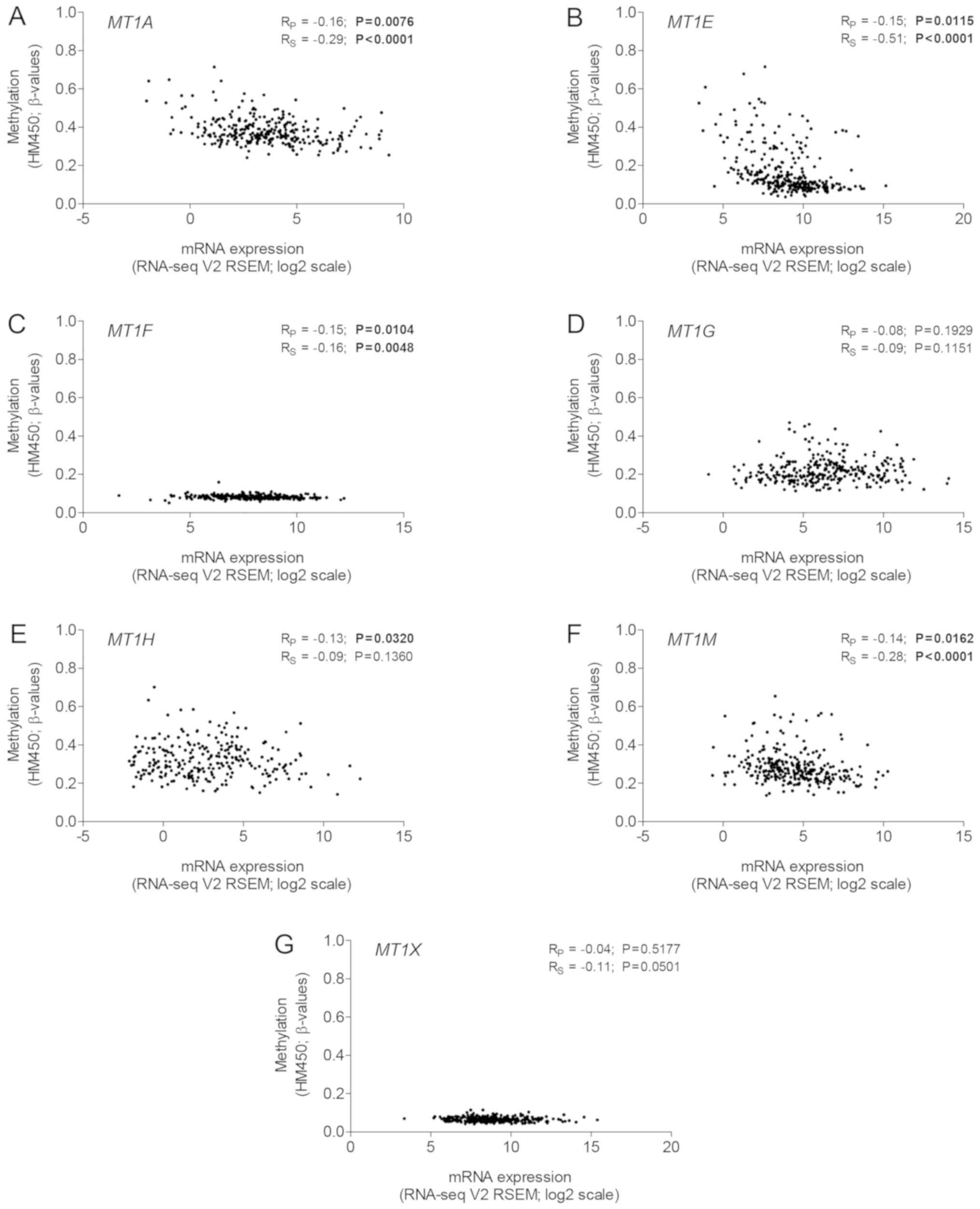
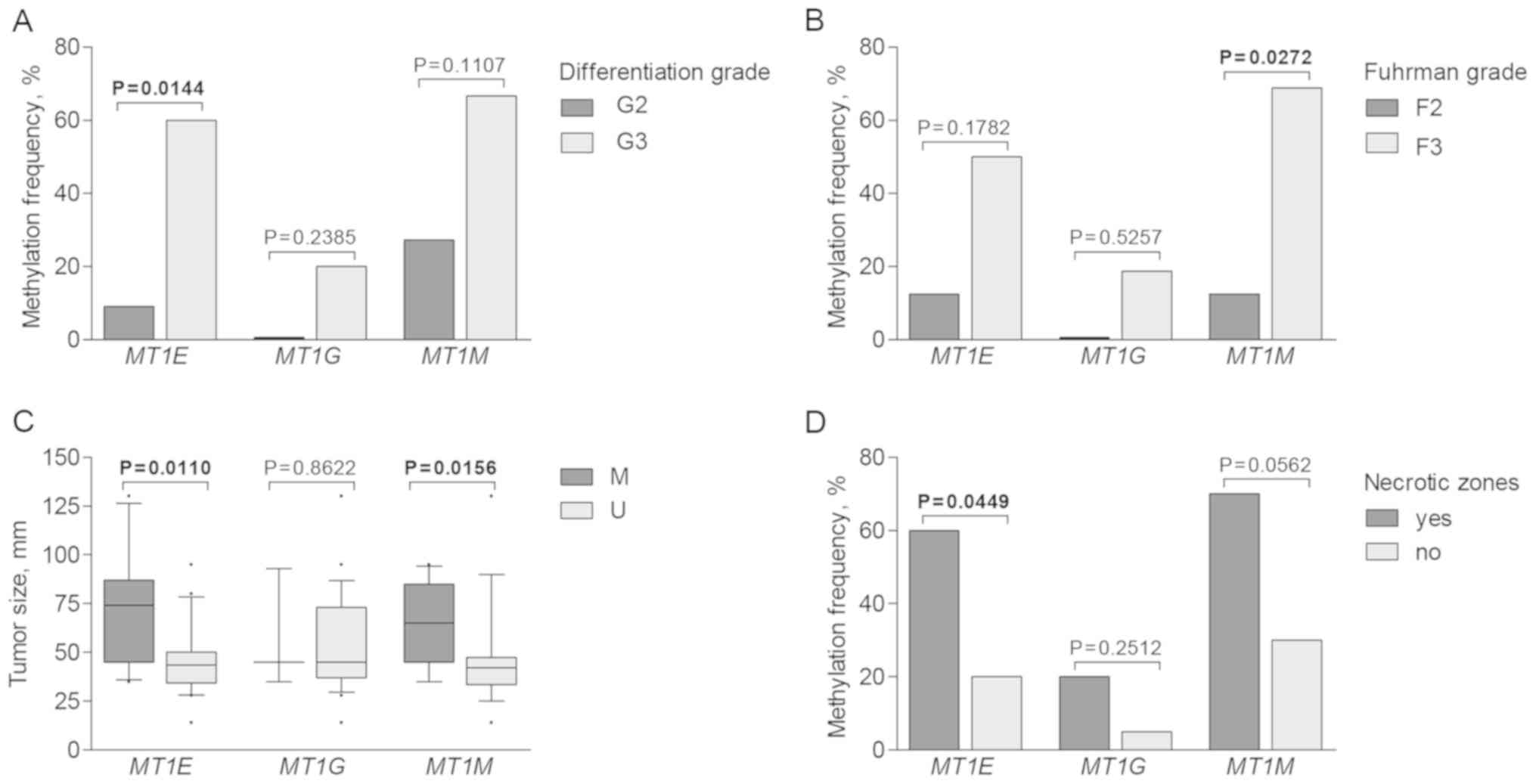
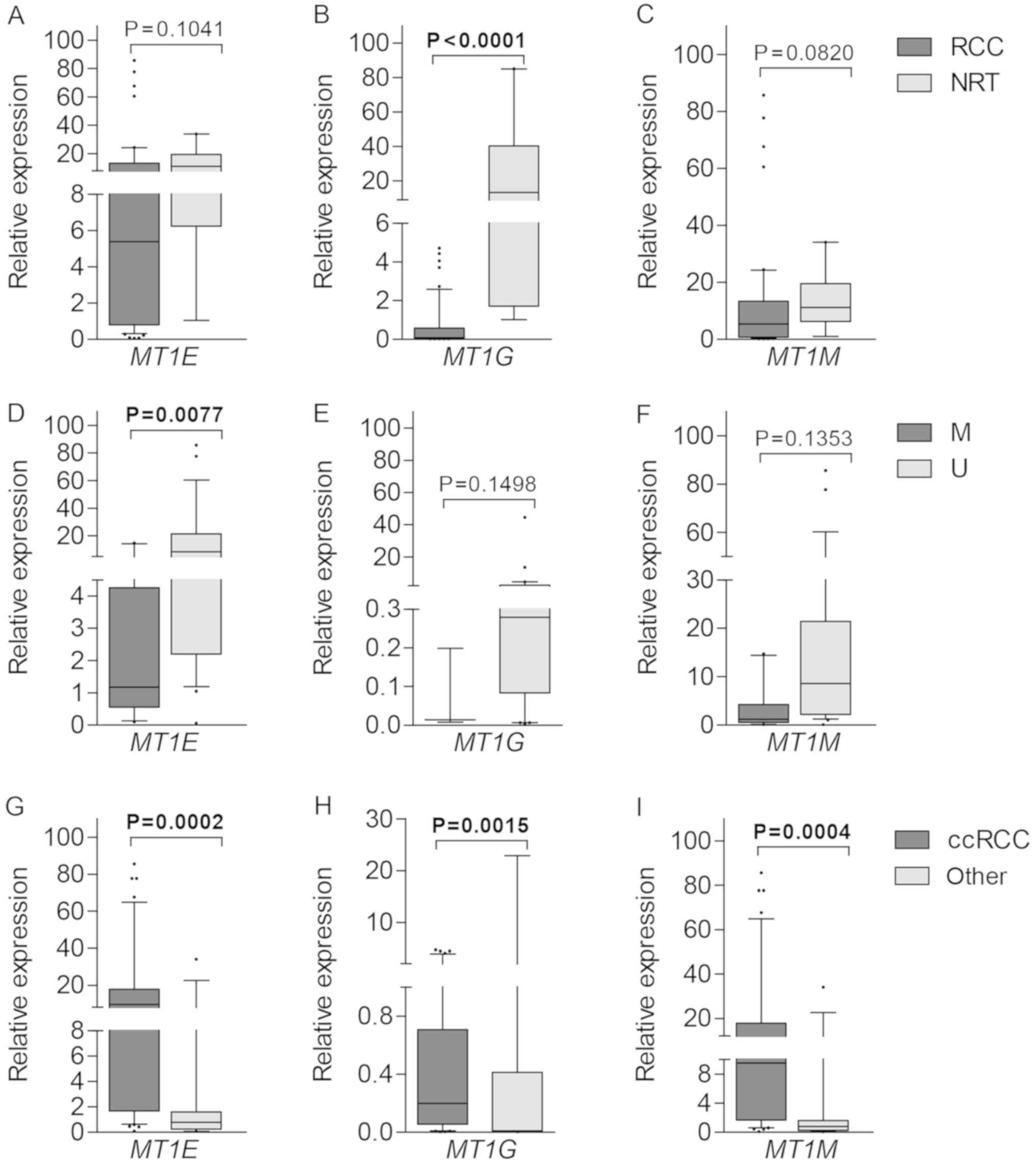
|
1
|
Krężel A and Maret W: The functions of
metamorphic metallothioneins in zinc and copper metabolism. Int J
Mol Sci. 18:E12372017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Klaassen CD, Liu J and Diwan BA:
Metallothionein protection of cadmium toxicity. Toxicol Appl
Pharmacol. 238:215–220. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Agrawal S, Flora G, Bhatnagar P and Flora
SJ: Comparative oxidative stress, metallothionein induction and
organ toxicity following chronic exposure to arsenic, lead and
mercury in rats. Cell Mol Biol. 60:13–21. 2014.PubMed/NCBI
|
|
4
|
Dutsch-Wicherek M, Sikora J and
Tomaszewska R: The possible biological role of metallothionein in
apoptosis. Front Biosci. 13:4029–4038. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Si M and Lang J: The roles of
metallothioneins in carcinogenesis. J Hematol Oncol. 11:1072018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun X, Niu X, Chen R, He W, Chen D, Kang R
and Tang D: Metallothionein-1G facilitates sorafenib resistance
through inhibition of ferroptosis. Hepatology. 64:488–500. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gansukh T, Donizy P, Halon A, Lage H and
Surowiak P: In vitro analysis of the relationships between
metallothionein expression and cisplatin sensitivity of non-small
cellular lung cancer cells. Anticancer Res. 33:5255–5260.
2013.PubMed/NCBI
|
|
8
|
Smith DJ, Jaggi M, Zhang W, Galich A, Du
C, Sterrett SP, Smith LM and Balaji KC: Metallothioneins and
resistance to cisplatin and radiation in prostate cancer. Urology.
67:1341–1347. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jing L, Yang M, Li Y, Yu Y, Liang B, Cao
L, Zhou X, Peng S and Sun Z: Metallothionein prevents doxorubicin
cardiac toxicity by indirectly regulating the uncoupling proteins
2. Food Chem Toxicol. 110:204–213. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Park Y and Yu E: Expression of
metallothionein-1 and metallothionein-2 as a prognostic marker in
hepatocellular carcinoma. J Gastroenterol Hepatol. 28:1565–1572.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Theocharis S, Karkantaris C, Philipides T,
Agapitos E, Gika A, Margeli A, Kittas C and Koutselinis A:
Expression of metallothionein in lung carcinoma: Correlation with
histological type and grade. Histopathology. 40:143–151. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Demidenko R, Daniunaite K, Bakavicius A,
Sabaliauskaite R, Skeberdyte A, Petroska D, Laurinavicius A,
Jankevicius F, Lazutka JR and Jarmalaite S: Decreased expression of
MT1E is a potential biomarker of prostate cancer
progression. Oncotarget. 8:61709–61718. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Weinlich G, Eisendle K, Hassler E, Baltaci
M, Fritsch PO and Zelger B: Metallothionein-overexpression as a
highly significant prognostic factor in melanoma: A prospective
study on 1,270 patients. Br J Cancer. 94:835–841. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hengstler JG, Pilch H, Schmidt M,
Dahlenburg H, Sagemüller J, Schiffer I, Oesch F, Knapstein PG,
Kaina B and Tanner B: Metallothionein expression in ovarian cancer
in relation to histopathological parameters and molecular markers
of prognosis. Int J Cancer. 95:121–127. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tai SK, Tan OJ, Chow VT, Jin R, Jones JL,
Tan PH, Jayasurya A and Bay BH: Differential expression of
metallothionein 1 and 2 isoforms in breast cancer lines with
different invasive potential: Identification of a novel nonsilent
metallothionein-1H mutant variant. Am J Pathol. 163:2009–2019.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wong MCS, Goggins WB, Yip BHK, Fung FDH,
Leung C, Fang Y, Wong SYS and Ng CF: Incidence and mortality of
kidney cancer: Temporal patterns and global trends in 39 countries.
Sci Rep. 7:156982017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rabjerg M, Mikkelsen MN, Walter S and
Marcussen N: Incidental renal neoplasms: Is there a need for
routine screening? A Danish single-center epidemiological study.
APMIS. 122:708–714. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Linehan WM, Bratslavsky G, Pinto PA,
Schmidt LS, Neckers L, Bottaro DP and Srinivasan R: Molecular
diagnosis and therapy of kidney cancer. Annu Rev Med. 61:329–343.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ricketts CJ, De Cubas AA, Fan H, Smith CC,
Lang M, Reznik E, Bowlby R, Gibb EA, Akbani R, Beroukhim R, et al:
The cancer genome atlas comprehensive molecular characterization of
renal cell carcinoma. Cell Rep. 23:313–326. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang WY, Hsu SD, Huang HY, Sun YM, Chou
CH, Weng SL and Huang HD: MethHC: A database of DNA methylation and
gene expression in human cancer. Nucleic Acids Res. 43:D856–D861.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tse KY, Liu VW, Chan DW, Chiu PM, Tam KF,
Chan KK, Liao XY, Cheung AN and Ngan HY: Epigenetic alteration of
the metallothionein 1E gene in human endometrial carcinomas. Tumour
Biol. 30:93–99. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Faller WJ, Rafferty M, Hegarty S, Gremel
G, Ryan D, Fraga MF, Esteller M, Dervan PA and Gallagher WM:
Metallothionein 1E is methylated in malignant melanoma and
increases sensitivity to cisplatin-induced apoptosis. Melanoma Res.
20:392–400. 2010.PubMed/NCBI
|
|
24
|
Chai H and Brown RE: Field effect in
cancer-an update. Ann Clin Lab Sci. 39:331–337. 2009.PubMed/NCBI
|
|
25
|
Ding J and Lu SC: Low metallothionein 1M
expression association with poor hepatocellular carcinoma prognosis
after curative resection. Genet Mol Res. 15:2016. View Article : Google Scholar :
|
|
26
|
Oka D, Yamashita S, Tomioka T, Nakanishi
Y, Kato H, Kaminishi M and Ushijima T: The presence of aberrant DNA
methylation in noncancerous esophageal mucosae in association with
smoking history: A target for risk diagnosis and prevention of
esophageal cancers. Cancer. 115:3412–3426. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jadhav RR, Ye Z, Huang RL, Liu J, Hsu PY,
Huang YW, Rangel LB, Lai HC, Roa JC, Kirma NB, et al: Genome-wide
DNA methylation analysis reveals estrogen-mediated epigenetic
repression of metallothionein-1 gene cluster in breast cancer. Clin
Epigenetics. 7:132015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fu CL, Pan B, Pan JH and Gan MF:
Metallothionein 1M suppresses tumorigenesis in hepatocellular
carcinoma. Oncotarget. 8:33037–33046. 2017.PubMed/NCBI
|
|
29
|
Dalgin GS, Drever M, Williams T, King T,
DeLisi C and Liou LS: Identification of novel epigenetic markers
for clear cell renal cell carcinoma. J Urol. 180:1126–1130. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo R, Wu G, Li H, Qian P, Han J, Pan F,
Li W, Li J and Ji F: Promoter methylation profiles between human
lung adenocarcinoma multidrug resistant A549/cisplatin (A549/DDP)
cells and its progenitor A549 cells. Biol Pharm Bull. 36:1310–1316.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nguyen A, Jing Z, Mahoney PS, Davis R,
Sikka SC, Agrawal KC and Abdel-Mageed AB: In vivo gene expression
profile analysis of metallothionein in renal cell carcinoma. Cancer
Lett. 160:133–140. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chan KY, Lai PB, Squire JA, Beheshti B,
Wong NL, Sy SM and Wong N: Positional expression profiling
indicates candidate genes in deletion hotspots of hepatocellular
carcinoma. Mod Pathol. 19:1546–1554. 2006. View Article : Google Scholar : PubMed/NCBI
|