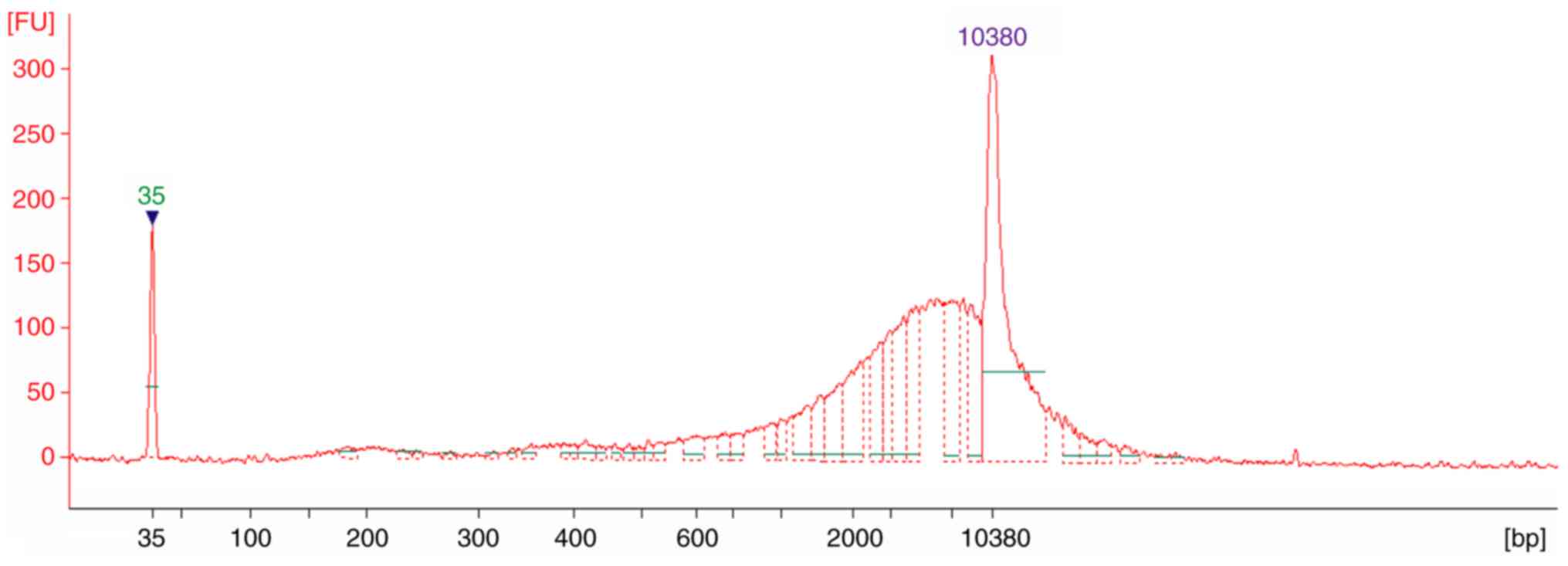
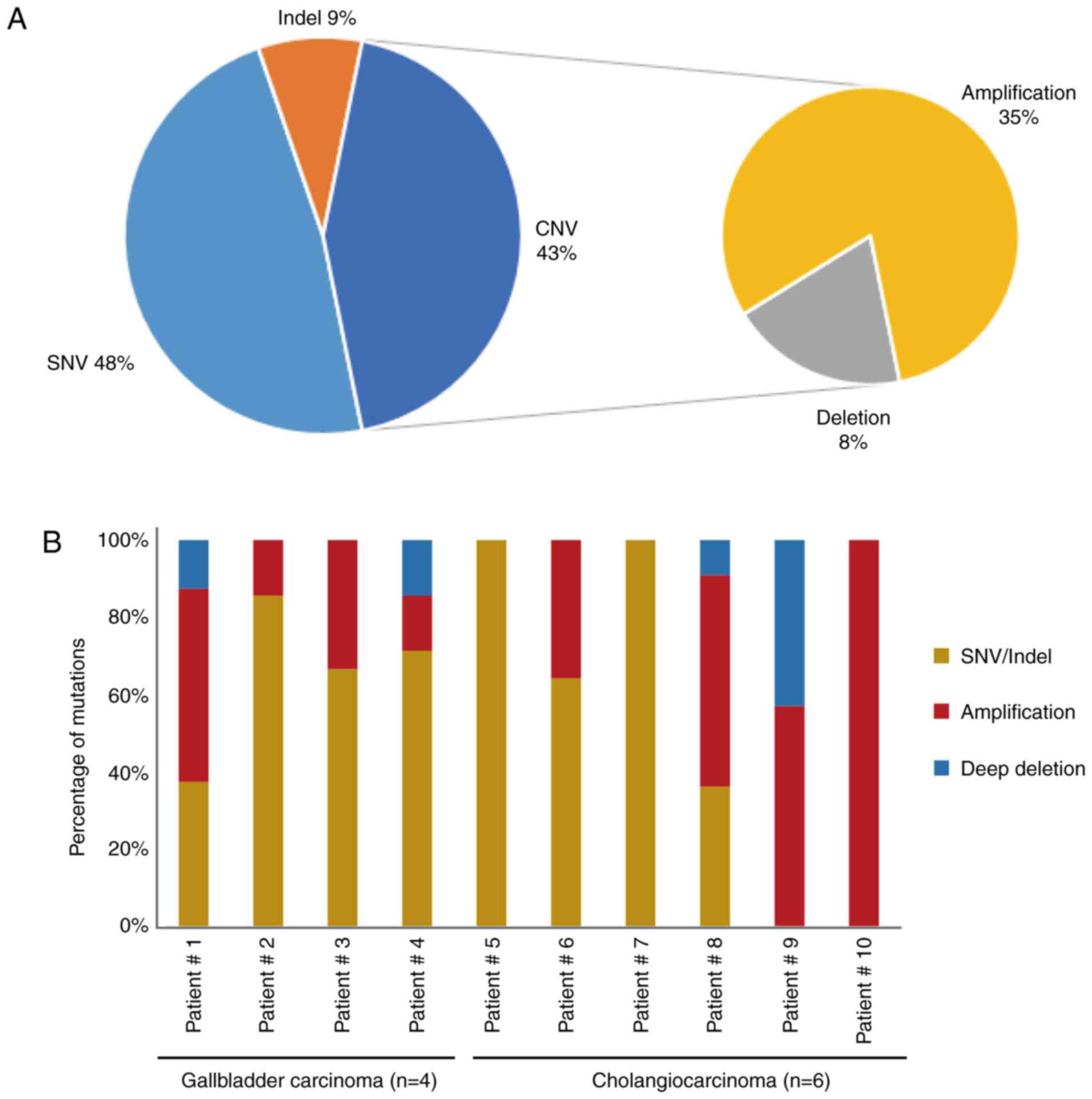
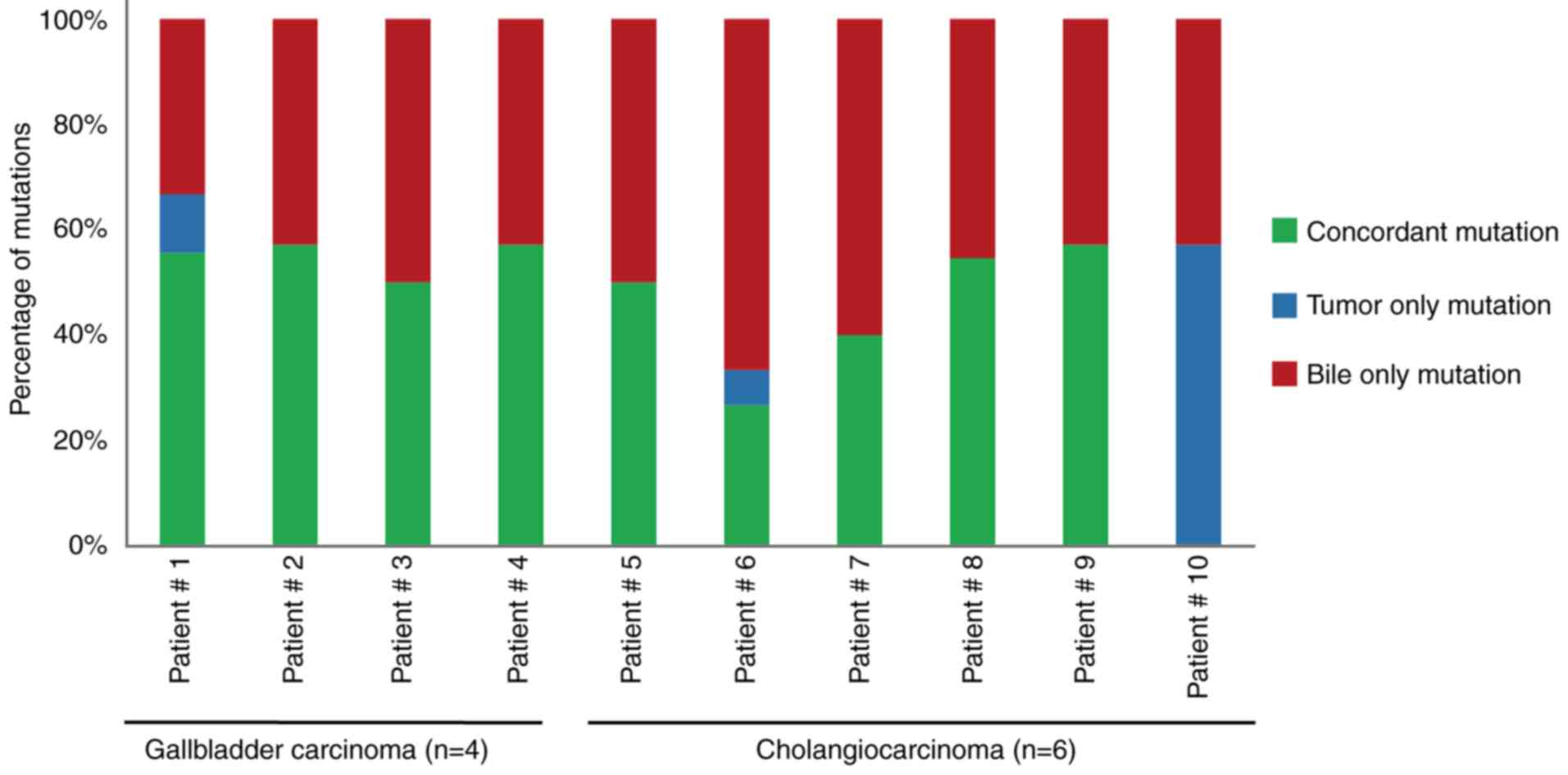
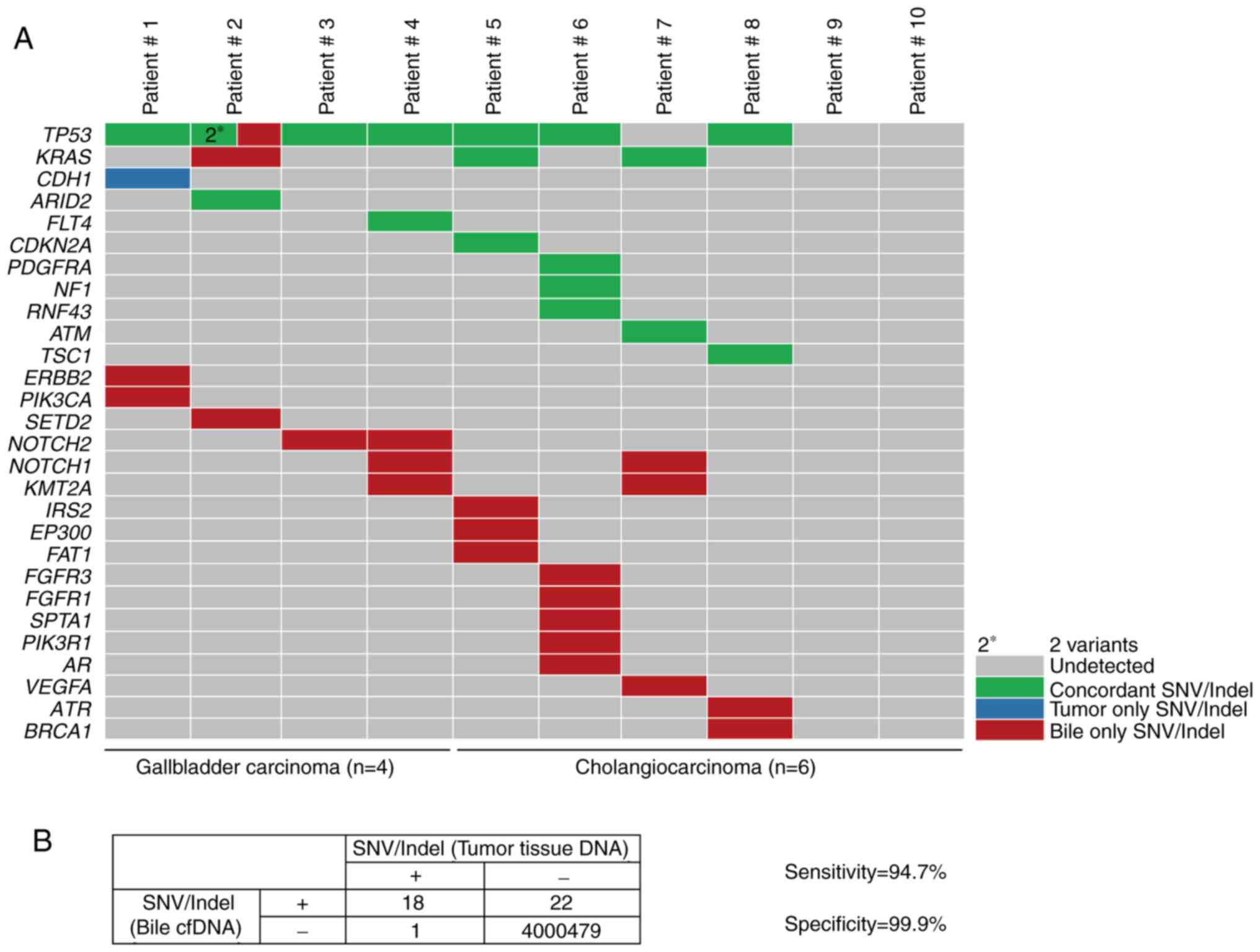
|
1
|
Augustine MM and Fong Y: Epidemiology and
risk factors of biliary tract and primary liver tumors. Surg Oncol
Clin N Am. 23:171–188. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marcano-Bonilla L, Mohamed EA, Mounajjed T
and Roberts LR: Biliary tract cancers: Epidemiology, molecular
pathogenesis and genetic risk associations. Chin Clin Oncol.
5:612016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Streeter OE Jr, Beron PJ and Iyer PN:
Precision medicine: Genomic profiles to individualize therapy.
Otolaryngol Clin North Am. 50:765–773. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran W Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nakamura H, Arai Y, Totoki Y, Shirota T,
Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T and Ohashi S:
Genomic spectra of biliary tract cancer. Nat Genet. 47:1003–1010.
2015. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nam AR, Kim JW, Cha Y, Ha H, Park JE, Bang
JH, Jin MH, Lee KH, Kim TY and Han SW: Therapeutic implication of
HER2 in advanced biliary tract cancer. Oncotarget. 7:58007–58021.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sakamoto Y, Yamagishi S, Tanizawa Y,
Tajimi M, Okusaka T and Ojima H: PI3K-mTOR pathway identified as a
potential therapeutic target in biliary tract cancer using a newly
established patient-derived cell panel assay. Jpn J Clin Oncol.
48:396–399. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rizvi S, Yamada D, Hirsova P, Bronk SF,
Werneburg NW, Krishnan A, Salim W, Zhang L, Trushina E, Truty MJ
and Gores GJ: A hippo and fibroblast growth factor receptor
autocrine pathway in cholangiocarcinoma. J Biol Chem.
291:8031–8047. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saha SK, Gordan JD, Kleinstiver BP, Vu P,
Najem MS, Yeo JC, Shi L, Kato Y, Levin RS, Webber JT, et al:
Isocitrate dehydrogenase mutations confer dasatinib
hypersensitivity and SRC dependence in intrahepatic
cholangiocarcinoma. Cancer Discov. 6:727–739. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wan JCM, Massie C, Garcia-Corbacho J,
Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R and Rosenfeld N:
Liquid biopsies come of age: Towards implementation of circulating
tumour DNA. Nat Rev Cancer. 17:223–238. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang N, Li Y, Liu Z, Qin H, Du D, Cao X,
Cao X, Li J, Li D, Jiang B, et al: The characteristics of ctDNA
reveal the high complexity in matching the corresponding tumor
tissues. BMC Cancer. 18:3192018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Siravegna G, Marsoni S, Siena S and
Bardelli A: Integrating liquid biopsies into the management of
cancer. Nat Rev Clin Oncol. 14:531–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang T, Ren S and Zhou C: Role of
circulating-tumor DNA analysis in non-small cell lung cancer. Lung
Cancer. 90:128–134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang T, Li X, Wang J, Su C, Han W, Zhao
C, Wu F, Gao G, Li W, Chen X, et al: Mutational landscape of cfDNA
identifies distinct molecular features associated with therapeutic
response to first-line platinum-based doublet chemotherapy in
patients with advanced NSCLC. Theranostics. 7:4753–4762. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Su D, Zhang D, Chen K, Lu J, Wu J, Cao X,
Ying L, Jin Q, Ye Y, Xie Z, et al: High performance of targeted
next generation sequencing on variance detection in clinical tumor
specimens in comparison with current conventional methods. J Exp
Clin Cancer Res. 36:1212017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Watanabe H, Okada M, Kaji Y, Satouchi M,
Sato Y, Yamabe Y, Onaya H, Endo M, Sone M and Arai Y: New response
evaluation criteria in solid tumours-revised RECIST guideline
(version 1.1). Gan To Kagaku Ryoho. 36:2495–2501. 2009.(In
Japanese). PubMed/NCBI
|
|
17
|
Ribiere SC, Léandri C, Guillaumot MA,
Brezault C and Coriat R: Therapeutic advances in the management of
biliary tract carcinoma. Presse Med. 47:419–422. 2018.(In French).
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zill OA, Greene C, Sebisanovic D, Siew LM,
Leng J, Vu M, Hendifar AE, Wang Z, Atreya CE, Kelley RK, et al:
Cell-free DNA next-generation sequencing in pancreatobiliary
carcinomas. Cancer Discov. 5:1040–1048. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kinugasa H, Nouso K, Ako S, Dohi C,
Matsushita H, Matsumoto K, Kato H and Okada H: Liquid biopsy of
bile for the molecular diagnosis of gallbladder cancer. Cancer Biol
Ther. 19:934–938. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Underhill HR, Kitzman JO, Hellwig S,
Welker NC, Daza R, Baker DN, Gligorich KM, Rostomily RC, Bronner MP
and Shendure J: Fragment length of circulating tumor DNA. PLoS
Genet. 12:e10061622016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xie D, Ren Z, Fan J and Gao Q: Genetic
profiling of intrahepatic cholangiocarcinoma and its clinical
implication in targeted therapy. Am J Cancer Res. 6:577–586.
2016.PubMed/NCBI
|
|
22
|
Li M, Zhang Z, Li X, Ye J, Wu X, Tan Z,
Liu C, Shen B, Wang XA, Wu W, et al: Whole-exome and targeted gene
sequencing of gallbladder carcinoma identifies recurrent mutations
in the ErbB pathway. Nat Genet. 46:872–876. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Oliveira DV, Zhang S, Chen X, Calvisi DF
and Andersen JB: Molecular profiling of intrahepatic
cholangiocarcinoma: The search for new therapeutic targets. Expert
Rev Gastroenterol Hepatol. 11:349–356. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee H and Ross JS: The potential role of
comprehensive genomic profiling to guide targeted therapy for
patients with biliary cancer. Therap Adv Gastroenterol. 10:507–520.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Javle M, Bekaii-Saab T, Jain A, Wang Y,
Kelley RK, Wang K, Kang HC, Catenacci D, Ali S, Krishnan S, et al:
Biliary cancer: Utility of next-generation sequencing for clinical
management. Cancer. 122:3838–3847. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zou S, Li J, Zhou H, Frech C, Jiang X, Chu
JS, Zhao X, Li Y, Li Q, Wang H, et al: Mutational landscape of
intrahepatic cholangiocarcinoma. Nat Commun. 5:56962014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chandra A, Gupta V, Rahul R, Kumar M and
Maurya A: Intraoperative ultrasonography of the biliary tract using
saline as a contrast agent: A fast and accurate technique to
identify complex biliary anatomy. Can J Surg. 60:316–322. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cao H, Liu J, Li T, Cao G, Xu G, Zhai S,
Xue J, Wang Z, Shi S and Bai W: Interventional therapy for the
treatment of severe hemobilia after percutaneous transhepatic
cholangial drainage: A case series. Int Surg. 98:223–228. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jin H, Pang Q, Liu H, Li Z, Wang Y, Lu Y,
Zhou L, Pan H and Huang W: Prognostic value of inflammation-based
markers in patients with recurrent malignant obstructive jaundice
treated by reimplantation of biliary metal stents: A retrospective
observational study. Medicine (Baltimore). 96:e58952017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li X, Yu J, Liang P, Yu X, Cheng Z, Han Z,
Duan S and Zheng J: Ultrasound-guided percutaneous microwave
ablation assisted by three-dimensional visualization operative
treatment planning system and percutaneous transhepatic cholangial
drainage with intraductal chilled saline perfusion for larger
hepatic hilum hepatocellular (D ≥ 3 cm): Preliminary results.
Oncotarget. 8:79742–79749. 2017.PubMed/NCBI
|
|
31
|
Njei B, McCarty TR, Varadarajulu S and
Navaneethan U: Systematic review with meta-analysis: endoscopic
retrograde cholangiopancreatography-based modalities for the
diagnosis of cholangiocarcinoma in primary sclerosing cholangitis.
Aliment Pharmacol Ther. 44:1139–1151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Newman AM, Bratman SV, To J, Wynne JF,
Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, et
al: An ultrasensitive method for quantitating circulating tumor DNA
with broad patient coverage. Nat Med. 20:548–554. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li J, Dittmar RL, Xia S, Zhang H, Du M,
Huang CC, Druliner BR, Boardman L and Wang L: Cell-free DNA copy
number variations in plasma from colorectal cancer patients. Mol
Oncol. 11:1099–1111. 2017. View Article : Google Scholar : PubMed/NCBI
|