Introduction
Non-small cell lung cancer (NSCLC) is a common lung
cancer with a high incidence. In the past decades, although great
advances have been made in NSCLC diagnosis, surgical resection,
targeted therapies, radiotherapies, immunotherapies and
chemotherapies (1–5) the 5-year overall survival and recurrence
rate of NSCLC patients is still not satisfactory. The poor outcomes
may stem from the few biomarkers available for precise prognosis of
NSCLC and incompletely understanding the pathogenesis of NSCLC.
Therefore, identification of new prognostic markers and
understanding of the molecular mechanisms underlying the
pathogenesis of NSCLC would be of great significance in the
management of NSCLC patients.
Long non-coding RNAs (lncRNAs), a class of
non-coding RNAs with >200 nucleotides in length (6). are important regulators of many
biological activities, such as transcription, RNA protein
modification, and RNA interaction (7). lncRNAs demonstrate their own
characteristics: i) The sequence is less conserved, only ~12% of
lncRNAs can be found in other organisms other than humans,
indicating their specificity in humans; ii) some lncRNAs have a
unique subcellular location and may have a novel subcellular
composition; iii) lncRNAs can regulate gene expression from
chromatin remodeling, transcriptional regulation and
post-transcriptional processing; iv) lncRNAs can be recognized by
other complementary nucleic acids and direct the protein to
specific sequence sites, which makes lncRNAs more abundant in
cancers. Previous studies have revealed that altered expression of
lncRNAs is associated with the development and progression of human
malignant tumors, including lung and liver cancer, osteosarcoma,
glioblastoma and as well as other types of cancer (8–11).
Furthermore, lncRNAs can act as oncogenes or tumor suppressor
genes, modulating the progression of NSCLC and affecting the
overall survival (OS) of NSCLC patients (12,13).
Multiple RNA transcripts, such as mRNAs, lncRNAs,
pseudogenes, and circular RNAs, can act as competing endogenous
RNAs (ceRNAs) to mutually regulate their function by competitive
binding of miRNA response elements (MREs) (14). Abnormal expression of any transcript
may cause a failure to control a regulatory network, ultimately
leading to the development and progression of cancer (15). This type of regulatory network is
widely found in various cancer diseases, such as hepatocellular
carcinoma, pheochromocytoma, and ovarian cancer (16–18).
Previous studies have revealed that upregulated HOTAIR expression
was associated with metastasis and poor prognosis of NSCLC, and
HOTAIR could promote the proliferation, survival, invasion,
metastasis, and drug resistance of NSCLC cells (19) while MALAT1 altered the expression of
metastasis-associated genes in NSCLC (20). However, little is known on what type
of ceRNA network regulates the progression of NSCLC.
In the present study, the differentially expressed
lncRNAs, miRNAs and related mRNAs in NSCLC were extracted from The
Cancer Genome Atlas (TCGA) to construct a ceRNA network.
Subsequently, the expression of four selected lncRNAs, their
subcellular localization, and associated clinical parameters in
NSCLC tissues were analyzed. The present findings indicated that
these lncRNAs may be valuable biomarkers for prognosis and the
ceRNA network may provide new insights into the pathogenesis of
NSCLC.
Materials and methods
RNA sequencing data and analysis in
TCGA
Differentially expressed RNAs were screened from
RNA-seq data and miRNA-seq data in TCGA (cancergenome.nih.gov) separately, since lncRNAs and
mRNAs were included in the RNA-seq database and miRNAs were
included in the miRNA-seq database (21). Then, the data of RNA expression in
NSCLC were extracted from these two databases and normalized by
‘DESeq2’ and ‘edgeR’ package. The obtained P-values were corrected
by false discovery rate (FDR). Differentially expressed mRNAs,
lncRNAs and miRNAs were considered when the RNA transcript between
NSCLC and non-tumor tissues reached a P-value of <0.01, FDR
<0.01 and |logFC (fold change)|>2. Their expression levels
were expressed as Fragments Per Kilobase Million (FPKM).
Construction of a ceRNA network
A ceRNA network was established by three steps: i)
By miRcode matching, individual miRNAs that interacted with the
differentially expressed lncRNAs and overlapped with the
differentially expressed miRNAs were used for establishment of the
lncRNA/miRNA relationship; ii) the mRNAs targeted by miRNAs in the
lncRNA/miRNA network were predicted by miRDB (http://mirdb.org/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php),
and TargetScan databases (http://www.targetscan.org/vert_72/), and they were
overlapped with the previously selected differentially expressed
mRNAs to construct the miRNA-mRNA relationship network; iii)
finally, the ceRNA network was constructed, based on the
aforementioned two relationship networks using the ‘ggalluvial’ R
package.
Prognostic analysis of lncRNAs in the
network
Individual subjects in the TCGA database were
stratified, according to the levels of specific lncRNAs, and their
survival data were extracted. The survival difference of subjects
with varying levels of individual lncRNAs in the ceRNA network was
analyzed using ‘survminer’ R package and ‘surv_cutpoint’
function.
Bioinformatics
The potential functions and pathway enrichments of
the differentially expressed genes identified in the ceRNA network
were analyzed by gene ontology (GO) (http://geneontology.org/) (22) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) (https://www.kegg.jp/) (23) using the R language, based on a P-value
of <0.05. The diagnostic values of these differentially
expressed RNAs were analyzed by receiver operating characteristic
(ROC) curve.
Clinical specimens
Surgical NSCLC specimens were obtained from 48
individual patients (23 males and 25 females; age range, 45–81
years), who underwent NSCLC surgery at the Affiliated Shengjing
Hospital, China Medical University, Shenyang, China from May 2017
to July 2018. All samples were frozen at −80°C immediately after
excision during surgery. The NSCLC specimens were staged
pathologically by the TNM of the UICC (Union for International
Cancer Control) (24). The
experimental protocol was approved by the Shengjing Hospital Ethics
Committee (2018PS170K) and written informed consent was obtained
from all patients.
Cell culture
Human non-tumor lung epithelial BEAS-2B cells, 293
cells, NSCLC A549, NCI-H460, NCI-H1299 and NCI-H292 cells (<25
passages) were obtained from the Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China). BEAS-2B and 293
cells were cultured in Dulbecco's modified Eagle's medium
(DMEM)/F12 containing 10% heat-inactivated fetal bovine serum
(FBS), 100 IU/ml penicillin and 100 µg/ml streptomycin at 37°C in
5% CO2. NSCLC cells were maintained in 10% FBS RPMI-1640
medium.
RNA extraction and quantitative
RT-PCR
Total RNA was extracted from individual specimens
and cells using TRIzol reagent and reversely-transcribed into cDNA
using HisScript™ QRT SuperMix (Vazyme Biotech Co., Ltd., Nanjing,
China). The relative levels of RNA transcripts to the control were
determined by qRT-PCR on Agilent Technologies Stratagene Mx3000P
using ChamQ™ Universal SYBR qPCR Master Mix (Vazyme Biotech Co.,
Ltd.) and specific primers are presented in Table SI. The data were normalized and
analyzed by 2−ΔΔCq method (25).
Subcellular fractionation
The cells were harvested and after being washed, and
then they were treated with 500 µl of cell disruption buffer (PARIS
kit; cat. no. AM1921; Invitrogen; Thermo fisher Scientific, Inc.)
on ice for 10 min, followed by centrifugation at 500 × g. Their
supernatants were collected as the cytoplasmic samples. The pellets
were washed and solved in an equal volume of nuclear lysate buffer,
followed by centrifugation at 500 × g. The supernatants were used
as the nuclear samples. Subsequently, the RNAs in the nuclear and
cytoplasmic samples were extracted and subjected to qRT-PCR using
primers presented in Table SI.
Statistical analysis
Data are expressed as the mean ± SD. Difference
between two groups was analyzed by unpaired Student's t-test.
Difference of multiple comparisons was analyzed by ANOVA
(parametric) followed by Dunnett's post-test. The category data
were analyzed by Chi-square test. All statistical analyses were
performed using SPSS 22.0 software (IBM Corp.) and GraphPad Prism
version 7.0 (GraphPad Software, Inc.). A two-tailed P-value of
<0.05 was considered to indicate a statistically significant
difference.
Results
Screening of differentially expressed
RNAs of NSCLC in TCGA
To screen differentially expressed RNAs, RNA-seq
data of 535 NSCLC and 59 paracancerous lung samples and miRNA-seq
data of 521 NSCLC and 46 paracancerous lung samples from the TCGA
database were extracted. Based on statistical significance of
P<0.01, FDR<0.01 and |logFC|>2, these data were analyzed
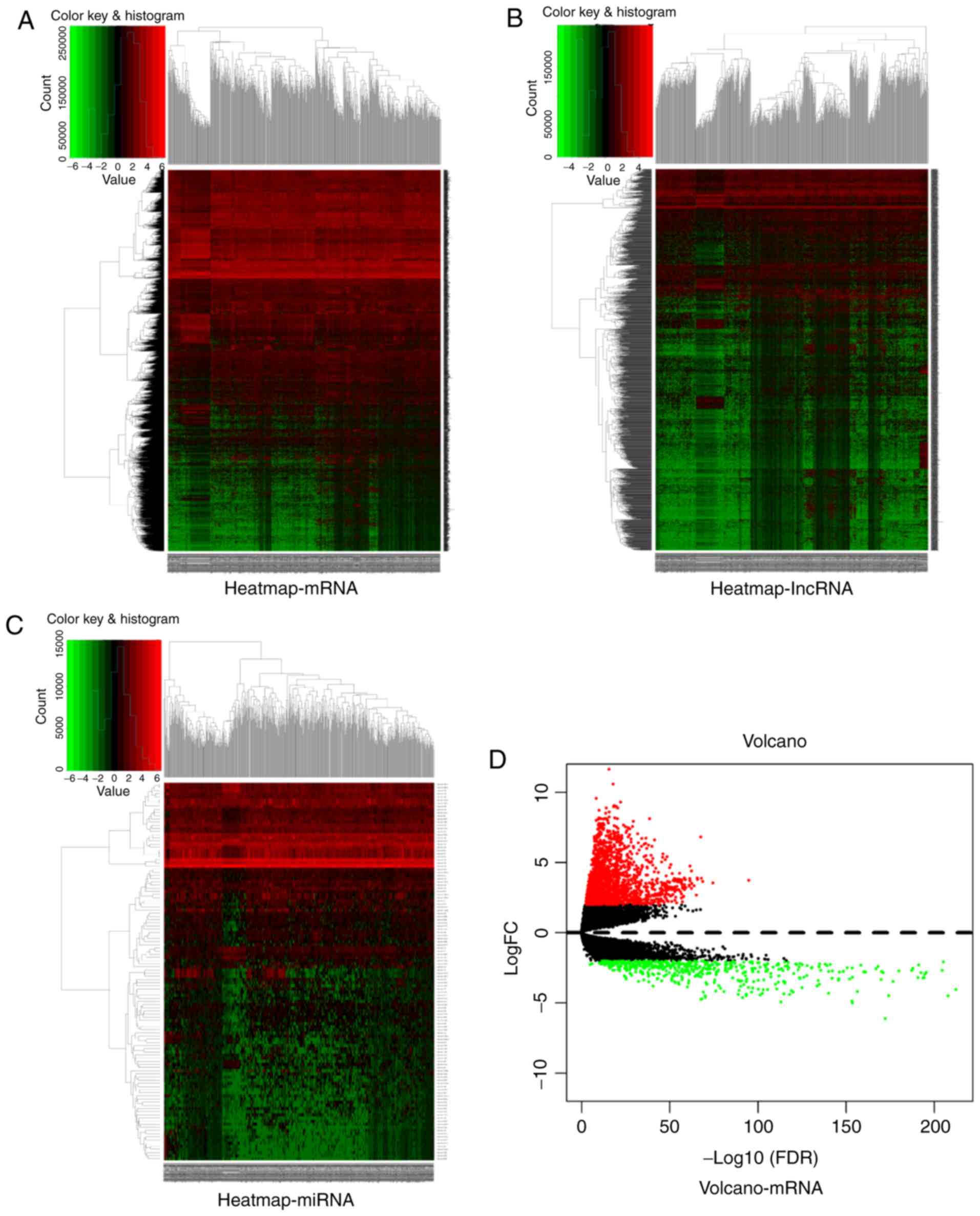
by heatmaps and volcanoes (Fig. 1).
There were 2,502 differentially expressed mRNAs (Fig. 1A), of which, 1,975 were upregulated
and 527 downregulated (Fig. 1D).
Furthermore, there were 1,685 differentially expressed lncRNAs
(Fig. 1B), of which, 1,486 were
upregulated and 199 downregulated (Fig.
1E). Moreover, there were 120 differentially expressed miRNAs
(Fig. 1C), of which, 104 were
upregulated and 16 downregulated (Fig.
1F).
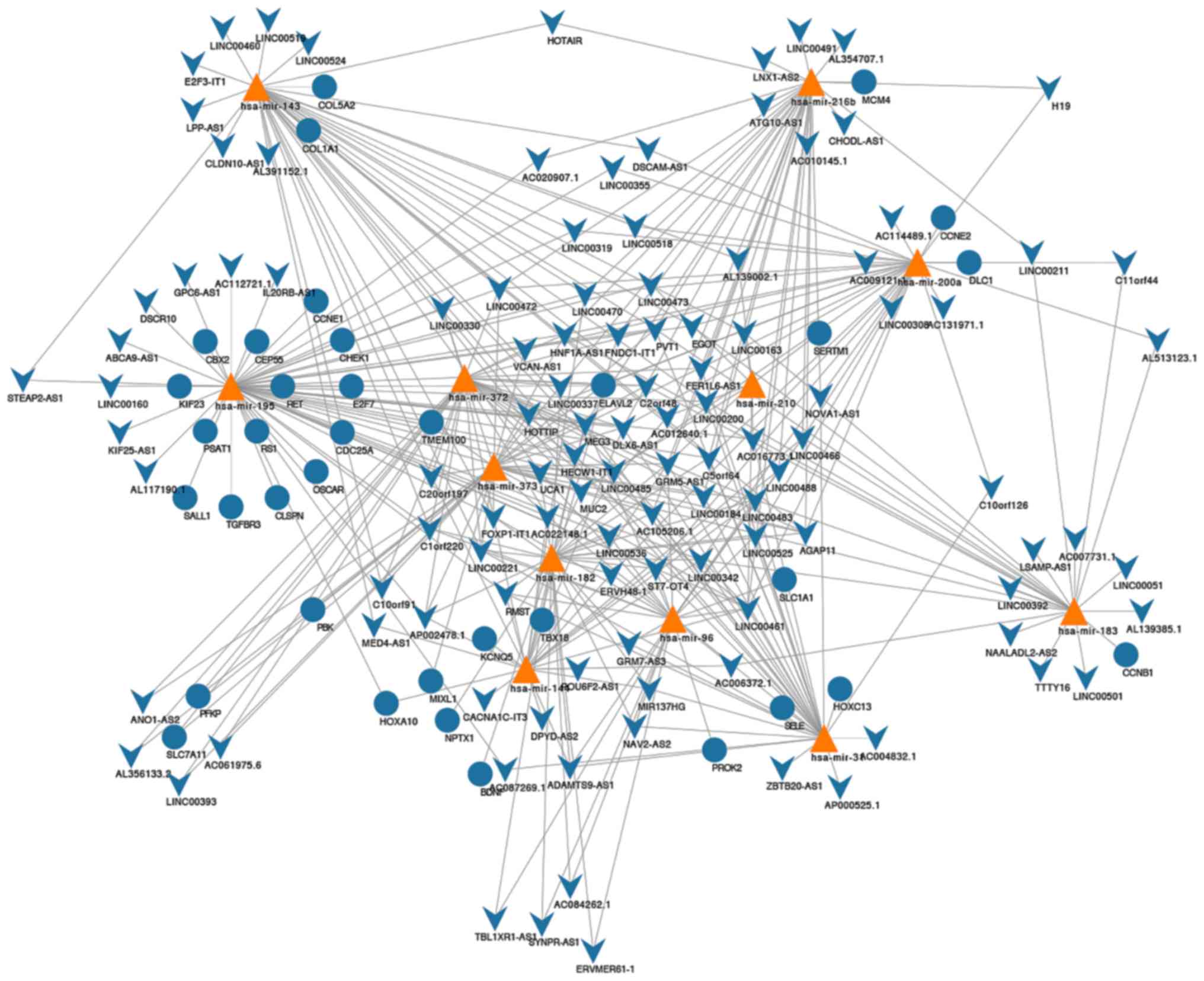
Construction of ceRNA network
The miRcode matching indicated that 144 lncRNAs and
22 miRNAs effectively generated 570 pairs of associations (Table SII). Furthermore, using the miRDB,
miRTarBase and TargetScan databases, we predicted that these 22
miNRAs targeted 722 mRNAs. After overlapping the 2,502
differentially expressed mRNAs with 722 mRNAs, 12 miRNAs formed 44
miRNA-mRNA association pairs (Table
SIII). Accordingly, a ceRNA network was constructed and
contained 113 DElncRNAs, 12 DEmiRNAs, and 36 DEmRNAs (Fig. 2). Further analysis of the network
revealed that it contained 161 nodes and 372 edges. The degrees of
the network were detailed in Table
SIV.
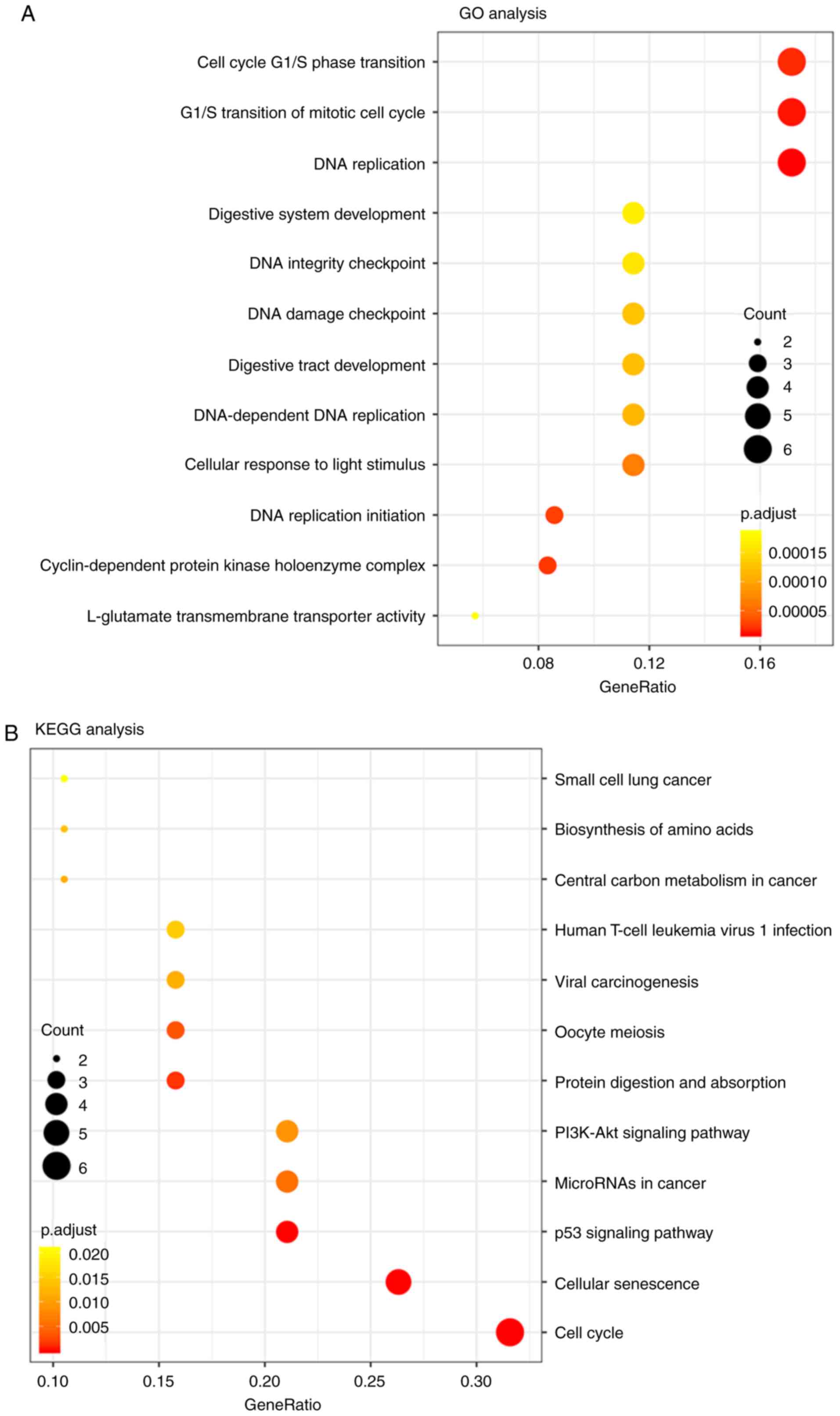
GO and KEGG pathway analyses
The potential biological functions of mRNA in ceRNA
networks were analyzed by GO and KEGG. The results revealed that
mRNAs in the ceRNA network were mainly involved in the cell cycle
G1/S phase transition and DNA replication (Fig. 3A) and participated in the processes of
the cell cycle, cellular senescence and p53 signaling (Fig. 3B).
Related lncRNAs in the ceRNA
network
After stratification of those subjects, based on the
differentially expressed lncRNAs, 103 out of 113 lncRNAs in the
network were significantly associated with prognosis in this
population (Table SIII). Since
LINC00525, MED4-AS1, STEAP2-AS1, SYNPR-AS1 were novel lncRNAs in
NSCLC and their differential fold and P-values were the most
significant (Table SV), their
functional association with survival was further analyzed (Fig. 4). The higher levels of LINC00525 or
STEAP2-AS1 were significantly associated with a shorter survival of
NSCLC patients while higher levels of MED4-AS1 or SYNPR-AS1 were
significantly associated with a longer survival of NSCLC patients.
Hence, these lncRNAs had opposite functions in regulating the
development and progression of NSCLC.
Expression and ROC curve analysis of
four lncRNAs in TCGA
To quantify lncRNA expression, four lncRNAs in tumor
and adjacent non-tumor tissues of individual subjects in TCGA were
quantified as FPKM values. The FPKM values of LINC00525, STEAP2-AS1
and SYNPR-AS1 in NSCLC tissues were significantly greater while the
FPKM values of MED4-AS1 in NSCLC tissues were significantly less
than that in the adjacent non-tumor tissues (Fig. 5A-D). Further ROC analysis revealed
that the area under the curve (AUC) values of LINC00525, MED4-AS1,
STEAP2-AS1, and SYNPR-AS1 were 0.841, 0.962, 0.816, and 0.749,
respectively.
Validation of four lncRNAs in NSCLC
samples and cells
To clarify the clinical significance of these four
lncRNAs, 48 pairs of NSCLC and adjacent tissue specimens were
collected. The relative expression levels of four lncRNAs in NSCLC
and adjacent tissues were determined by qRT-PCR. The relative
levels of four lncRNA transcripts in NSCLC tissues were
significantly higher than that in the adjacent tissues (Fig. 6A-D; P<0.05). Analyses of different
cell lines indicated various expression levels of these four
lncRNAs (Fig. 6E-H). In comparison
with that in non-tumor BEAS-2B cells, LINC00525, MED4-AS1 and
STEAP2-AS1 were significantly upregulated in A549 cells.
Significantly upregulated LINC00525 expression was also detected in
NCI-H460 cells. SYNPR-AS1 revealed no significant difference
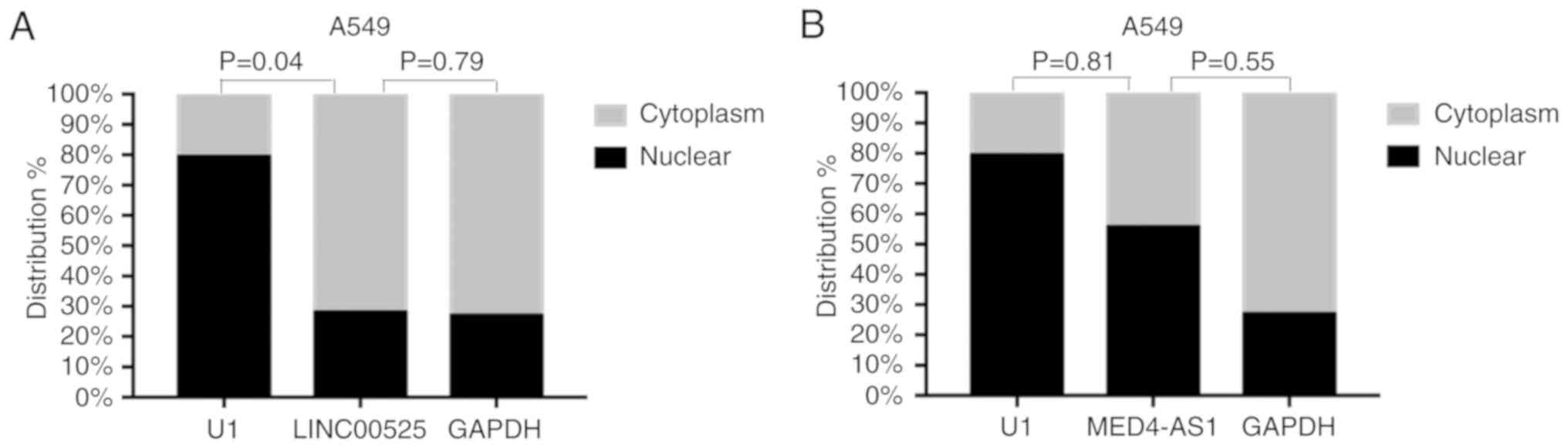
between groups. Further analysis revealed that LINC00525 was mainly
expressed in the cytoplasm while MED4-AS1 was detected in both the
nucleus and cytoplasm of A549 cells (Fig.
7A and B).
Higher expression levels of four
lncRNAs are associated with clinical characteristics
Finally, we analyzed the potential association
between higher expression levels of individual lncRNAs and clinical
parameters in 48 NSCLC patients. Notably, higher expression levels
of LINC00525 were significantly associated with smoker history
(P=0.036; Table I). Higher expression
levels of MED4-AS1 were significantly associated with females
(P=0.02), poor differentiation (P=0.046) and lymph node metastasis
(P=0.043) (Table II). Higher
expression levels of STEAP2-AS1 were significantly associated with
females (P=0.009; Table III) and
higher expression levels of SYNPR-AS1 were significantly associated
with females (P=0.043) and adenocarcinoma (P=0.035) (Table IV) in this population. After the
P-value was adjusted for sex, the expression of MED4-AS1 was
markedly associated with poor differentiation (adj.P=0.055) and
lymph node metastasis (adj.P=0.07). However, their expression
levels were not significantly associated with other parameters
assessed.
 | Table I.The association between LINC00525
expression and the clinical characteristics of NSCLC patients. |
Table I.
The association between LINC00525
expression and the clinical characteristics of NSCLC patients.
|
|
| Relative LINC00525
expression |
|---|
|
|
|
|
|---|
|
Characteristics | N | Low | High | P-value |
|---|
| Age (years) |
|
|
|
|
|
>65 | 9 | 5 | 4 | 0.712 |
|
≤65 | 39 | 19 | 20 |
|
| Sex |
|
|
|
|
|
Male | 23 | 13 | 10 | 0.386 |
|
Female | 25 | 11 | 14 |
|
|
Differentiation |
|
|
|
|
| Well,
moderate | 36 | 17 | 19 | 0.505 |
|
Poor | 12 | 7 | 5 |
|
| Tumor size (maximum
diameter) |
|
|
|
|
| >3
cm | 22 | 10 | 12 | 0.562 |
| ≤3
cm | 26 | 14 | 12 |
|
| Histological tumor
type |
|
|
|
|
|
Squamous cell carcinoma | 17 | 9 | 8 | 0.763 |
|
Adenocarcinoma | 31 | 15 | 16 |
|
| Smoking
history |
|
|
|
|
|
Smokers | 20 | 6 | 14 | 0.036a |
| Never
smoked | 28 | 17 | 11 |
|
| Lymph node
metastasis |
|
|
|
|
|
Positive | 21 | 9 | 12 | 0.383 |
|
Negative | 27 | 15 | 12 |
|
| TNM stage |
|
|
|
|
| Stage
I | 20 | 12 | 8 | 0.242 |
| Stages
II, III or IV | 28 | 12 | 16 |
|
 | Table II.The association between MED4-AS1
expression and the clinical characteristics of NSCLC patients. |
Table II.
The association between MED4-AS1
expression and the clinical characteristics of NSCLC patients.
|
|
| Relative MED4-AS1
expression |
|---|
|
|
|
|
|---|
|
Characteristics | N | Low | High | P-value | Adj.P-value |
|---|
| Age (years) |
|
|
|
|
|
|
>65 | 11 | 6 | 5 | 0.731 | 0.708 |
|
≤65 | 37 | 18 | 19 |
|
|
| Sex |
|
|
|
|
|
|
Male | 22 | 15 | 7 | 0.020a |
|
|
Female | 26 | 9 | 17 |
|
|
|
Differentiation |
|
|
|
|
|
| Well,
moderate | 36 | 21 | 15 | 0.046a | 0.055 |
|
Poor | 12 | 3 | 9 |
|
|
| Tumor size (maximum
diameter) |
|
|
|
|
|
| >3
cm | 22 | 8 | 14 | 0.141 | 0.109 |
| ≤3
cm | 26 | 15 | 11 |
|
|
| Histological tumor
type |
|
|
|
|
|
|
Squamous cell carcinoma | 16 | 8 | 8 | 0.999 | 0.097 |
|
Adenocarcinoma | 32 | 16 | 16 |
|
|
| Smoking
history |
|
|
|
|
|
|
Smokers | 19 | 12 | 7 | 0.14 | 0.477 |
| Never
smoked | 29 | 12 | 17 |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
Positive | 23 | 8 | 15 |
0.043a | 0.07 |
|
Negative | 25 | 16 | 9 |
|
|
| TNM stage |
|
|
|
|
|
| Stage
I | 19 | 12 | 7 | 0.14 | 0.596 |
| Stages
II, III or IV | 29 | 12 | 17 |
|
|
 | Table III.The association between STEAP2-AS1
expression and clinical characteristics of NSCLC patients. |
Table III.
The association between STEAP2-AS1
expression and clinical characteristics of NSCLC patients.
|
|
| Relative STEAP2-AS1
expression |
|---|
|
|
|
|
|---|
|
Characteristics | N | Low | High | P-value | Adj.P-value |
|---|
| Age (years) |
|
|
|
|
|
|
>65 | 10 | 7 | 3 | 0.155 | 0.151 |
|
≤65 | 38 | 17 | 21 |
|
|
| Sex |
|
|
|
|
|
|
Male | 23 | 16 | 7 | 0.009a |
|
|
Female | 25 | 8 | 17 |
|
|
|
Differentiation |
|
|
|
|
|
| Well,
moderate | 36 | 18 | 18 | 0.999 | 0.839 |
|
Poor | 12 | 6 | 6 |
|
|
| Tumor size (maximum
diameter) |
|
|
|
|
|
| >3
cm | 23 | 9 | 14 | 0.149 | 0.19 |
| ≤3
cm | 25 | 15 | 10 |
|
|
| Histological tumor
type |
|
|
|
|
|
|
Squamous cell carcinoma | 17 | 10 | 7 | 0.365 | 0.388 |
|
Adenocarcinoma | 31 | 14 | 17 |
|
|
| Smoking
history |
|
|
|
|
|
|
Smokers | 19 | 9 | 10 | 0.768 | 0.115 |
| Never
smoked | 29 | 15 | 14 |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
Positive | 21 | 9 | 12 | 0.383 | 0.489 |
|
Negative | 27 | 15 | 12 |
|
|
| TNM stage |
|
|
|
|
|
| 1
stage | 20 | 12 | 8 | 0.242 | 0.74 |
| 2.3.4
stage | 28 | 12 | 16 |
|
|
 | Table IV.The association between SYNPR-AS1
expression and the clinical characteristics of NSCLC patients. |
Table IV.
The association between SYNPR-AS1
expression and the clinical characteristics of NSCLC patients.
|
|
| Relative SYNPR-AS1
expression |
|---|
|
|
|
|
|---|
|
Characteristics | N | Low | High | P-value | Adj.P-value |
|---|
| Age (years) |
|
|
|
|
|
|
>65 | 10 | 5 | 5 | 0.999 | 0.964 |
|
≤65 | 38 | 19 | 19 |
|
|
| Sex |
|
|
|
|
|
|
Male | 23 | 15 | 8 |
0.043a |
|
|
Female | 25 | 9 | 16 |
|
|
|
Differentiation |
|
|
|
|
|
| Well,
moderate | 36 | 16 | 20 | 0.182 | 0.129 |
|
Poor | 12 | 8 | 4 |
|
|
| Tumor size (maximum
diameter) |
|
|
|
|
|
| >3
cm | 23 | 13 | 10 | 0.386 | 0.279 |
| ≤3
cm | 25 | 11 | 14 |
|
|
| Histological tumor
type |
|
|
|
|
|
|
Squamous cell carcinoma | 17 | 12 | 5 |
0.035a | 0.245 |
|
Adenocarcinoma | 31 | 12 | 19 |
|
|
| Smoking
history |
|
|
|
|
|
|
Smokers | 19 | 11 | 8 | 0.376 | 0.962 |
| Never
smoked | 29 | 13 | 16 |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
Positive | 23 | 10 | 13 | 0.386 | 0.578 |
|
Negative | 25 | 14 | 11 |
|
|
| TNM stage |
|
|
|
|
|
| Stage
I | 19 | 10 | 9 | 0.768 | 0.984 |
| Stages
I, II or III | 29 | 14 | 15 |
|
|
Discussion
NSCLC is a common lethal cancer in the world, and
the current 5-year survival rate of NSCLC remains low (26). In the present study, 2,502
differentially expressed mRNAs, 1,685 lncRNAs, and 120 miRNAs were
identified in NSCLC tissues from TCGA, respectively. Further GO and
KEGG analyses indicated that the differentially expressed mRNAs may
participate in cellular processes (27), molecular binding (28), signal transduction and survival
(29), which are important for the
progression of malignant tumors. Using these differentially
expressed RNAs, a ceRNA network was constructed, which reflected
the relationships among different RNA transcripts. Hence, such
findings may provide new insights into the pathogenesis of
NSCLC.
lncRNAs are abundant in different species of animals
and can regulate various biological processes and malignancies.
Previous studies have revealed that tumor-associated lncRNAs not
only bind to transcription factors and proteins, but also affect
tumor-associated mRNAs by competitive binding to miRNAs (30,31). In
fact, LINC00858 was revealed to act as a ceRNA to effectively
sponge miR-422a, modulating the expression of KLK4 to impair NSCLC
cell proliferation, migration and invasion (32). Similarly, ROR acted as a ceRNA of
miR-145 to promote the FSCN1-mediated metastasis of esophageal
squamous cell carcinoma (33). In
addition, DLG1-AS1, along with miR-107 and ZHX1, formed a ceRNA
network to regulate cervical cancer progression (34). MALAT1 may act as a sponger of miR-124
and attenuate the miR-124-downregulated FoxQ1 expression,
regulating the progression and metastasis of bladder transitional
cell carcinoma (35). In addition,
the MEG3/miR-181a/HOXA11 ceRNA network and the lncRNA/miRNA/mRNA
may reflect the regulatory communication among ceRNAs in multiple
myeloma and colorectal carcinoma (36,37). The
network construction visually demonstrated that ceRNAs are key
regulators in the communication among different RNA transcripts. It
may reveal the intrinsic regulatory network mechanism of NSCLC at
the transcriptional level. In addition, by analyzing and validating
the core factors in clinical specimen data and survival data within
this network, novel biomarkers may be identified to serve as
diagnostic and prognostic indicators for NSCLC. Thus, the
lncRNA/miRNA/mRNA ceRNA network we constructed in NSCLC extended
previous findings.
In the present study, 103 lncRNAs were identified in
the network that were significantly associated with prognosis,
including H19, HOTAIR, MEG3, UCA1 (38–41) and
other star molecules were associated with NSCLC cell proliferation,
invasion and apoptosis. Among them, LINC00525, MED4-AS1, STEAP2-AS1
and SYNPR-AS1 were new lncRNAs and their altered expression was
significantly associated with the survival of NSCLC patients.
Notably, these lncRNAs were associated with clinical parameters in
48 NSCLC patients. Accordingly, these lncRNAs may be valuable
biomarkers for prognosis of NSCLC patients. Notably, while the
expression of MED4-AS1 and SYNPR-AS1 was upregulated in NSCLC
tissues they were positively associated with OS of NSCLC patients.
Such data indicated that these lncRNAs may be the tumor-stimulated
compensatory lncRNAs that may, through a complex regulatory
network, inhibit the progression, recurrence and metastasis of
NSCLC (42). We are interested in
further investigating how these lncRNAs regulate the progression of
NSCLC.
Previous studies have revealed that BCAR4, XIST and
SNHG3 as well as other lncRNAs are significantly elevated in NSCLC
specimens (43–45) and they regulate the progression of
NSCLC. In the present study, it was revealed that LINC00525,
STEAP2-AS1 and SYNPR-AS1 transcripts were significantly increased
in NSCLC in the TCGA and 48 NSCLC tissues we collected. Notably,
the expression of MED4-AS1 was downregulated in NSCLC from the
TCGA, but upregulated in 48 NSCLC specimens we collected. The
discrepancy may stem from the different genetic backgrounds of
patients with varying stages of NSCLC. In addition, it was revealed
that the frequency of female NSCLC patients with increased
MED4-AS1, STEAP2-AS1 or SYNPR-AS1 expression was significantly
higher than male patients. We are interested in further examining
whether these upregulated lncRNAs can regulate the expression of
sex-determining region Y-box 2 (SOX2) transcription factor
(46). Moreover, it was revealed that
these four lncRNAs were variably expressed in different NSCLC
cells. While LINC00525 was mainly present in the cytoplasm,
MED4-AS1 was present in both the nucleus and cytoplasm of A549
cells. The inconsistent distribution may reflect different
functions of these lncRNAs in A549 cells, supporting the notion
that lncRNAs regulate the expression of many oncogenic or tumor
suppressive genes at transcriptional or post-transcriptional levels
(47).
Heavy smoking is a risk factor of NSCLC. A previous
study revealed that cigarette smoke exposure (CSE) increased
LINC00152 expression in airway epithelial cells in a dose-dependent
manner, and promoted cyclin D1 expression and G1/S transition,
leading to metastasis and proliferation of NSCLC (48). In the present study, it was revealed
that upregulated LINC00525 expression was positively associated
with a smoking history. In fact, it was revealed in a previous
study that >50% of NSCLC cases occured in non-smoking women, and
these cases were closely associated to EGFR and ALK mutations
(49). In addition, upregulated
MED4-AS1 expression was positively associated with poor
differentiation and lymphatic node metastasis, indicating that it
may promote invasion and metastasis of NSCLC. SYNPR-AS1 was
positively associated with adenocarcinoma. Notably, the present
study had limitations of small sample size and the lack of
functional studies of individual lncRNAs. This study mainly
predicted potential new biomarkers for NSCLC. Further
investigations are warranted in validating the diagnostic and
prognostic values of these lncRNAs and how these lncRNAs
synergistically regulate the progression of NSCLC by co-expression
of two or multiple lncRNAs.
In conclusion, the present data indicated that there
were numerous differentially expressed lncRNAs, and along with the
differentially expressed miRNAs and mRNAs, a ceRNA network was
constructed. These differentially expressed mRNAs were involved in
many biological processes. The expression of four selected lncRNAs
was significantly associated with OS of NSCLC patients and their
expression was validated in 48 NSCLC specimens we collected. These
lncRNAs were significantly associated with some clinical
parameters, and LINC00525 and MED4-AS1 were differentially present
in the nucleus and cytoplasm of A549 cells. Therefore, these four
lncRNAs may be valuable biomarkers for prognosis of NSCLC and may
provide new insights in the pathogenesis of NSCLC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
Shenyang Science and Technology Plan Project, Liaoning, China (Item
no. 17-231-1-51). The funder had no role in the study design, the
data collection and analysis, the decision to publish, or the
preparation of the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available upon reasonable request from the corresponding
author.
Authors' contributions
XWW and QQG wrote the study, conducted the
bioinformatics analysis and performed the experiments. YW, KMR and
HYZ performed lobectomy to provide lung cancer specimens. TJ and
FSZ performed the statistical analysis of the data. JGZ designed
the project, supervised the experiments and corrected the draft.
All authors read and approved the final manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The experimental protocol was approved by the
Shengjing Hospital Ethics Committee and written informed consent
was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
ceRNA
|
competing endogenous RNA
|
|
MREs
|
miRNA response elements
|
|
lncRNA
|
long non-coding RNA
|
|
miRNA
|
microRNA
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
TCGA
|
The Cancer Genome Atlas
|
References
|
1
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kris MG, Gaspar LE, Chaft JE, Kennedy EB,
Azzoli CG, Ellis PM, Lin SH, Pass HI, Seth R, Shepherd FA, et al:
Adjuvant systemic therapy and adjuvant radiation therapy for stage
I to IIIA completely resected non-small-cell lung cancers: American
society of clinical oncology/cancer care ontario clinical practice
guideline update. J Clin Oncol. 35:2960–2974. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moghissi K and Dixon K: Image-guided
surgery and therapy for lung cancer: A critical review. Future
Oncol. 13:2383–2394. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nagasaka M and Gadgeel SM: Role of
chemotherapy and targeted therapy in early-stage non-small cell
lung cancer. Expert Rev Anticancer Ther. 18:63–70. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oak CH, Wilson D, Lee HJ, Lim HJ and Park
EK: Potential molecular approaches for the early diagnosis of lung
cancer (review). Mol Med Rep. 6:931–936. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery, and classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun W, Shi Y, Wang Z, Zhang J, Cai H,
Zhang J and Huang D: Interaction of long-chain non-coding RNAs and
important signaling pathways on human cancers (Review). Int J
Oncol. 53:2343–2355. 2018.PubMed/NCBI
|
|
8
|
Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu
J, Miao N, Shen J and Peng T: lncRNA DANCR promotes tumor
progression and cancer stemness features in osteosarcoma by
upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 405:46–55.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Osielska MA and Jagodzinski PP: Long
non-coding RNA as potential biomarkers in non-small-cell lung
cancer: What do we know so far? Biomed Pharmacother. 101:322–333.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qiu L, Tang Q, Li G and Chen K: Long
non-coding RNAs as biomarkers and therapeutic targets: Recent
insights into hepatocellular carcinoma. Life Sci. 191:273–282.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vecera M, Sana J, Lipina R, Smrcka M and
Slaby O: Long non-coding RNAs in gliomas: From molecular pathology
to diagnostic biomarkers and therapeutic Targets. Int J Mol Sci.
19(pii): E27542018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu YH, Tu JR, Zhao TT, Xie SG and Tang SB:
Overexpression of lncRNA EGFRAS1 is associated with a poor
prognosis and promotes chemotherapy resistance in nonsmall cell
lung cancer. Int J Oncol. 54:295–305. 2019.PubMed/NCBI
|
|
13
|
Chang L, Xu W, Zhang Y and Gong F: Long
non-coding RNA-NEF targets glucose transportation to inhibit the
proliferation of non-small-cell lung cancer cells. Oncol Lett.
17:2795–2801. 2019.PubMed/NCBI
|
|
14
|
Chen ZP, Wei JC, Wang Q, Yang P, Li WL, He
F, Chen HC, Hu H, Zhong JB and Cao J: Long noncoding RNA 00152
functions as a competing endogenous RNA to regulate NRP1 expression
by sponging with miRNA206 in colorectal cancer. Int J Oncol.
53:1227–1236. 2018.PubMed/NCBI
|
|
15
|
Chan JJ and Tay Y: Noncoding RNA:RNA
regulatory networks in cancer. Int J Mol Sci. 19(pii): E13102018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liang YC, Wu YP, Chen DN, Chen SH, Li XD,
Sun XL, Wei Y, Ning X and Xue XY: Building a competing endogenous
RNA network to find potential long non-coding RNA biomarkers for
pheochromocytoma. Cell Physiol Biochem. 51:2916–2924. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang X, Han L, Zhou L, Wang L and Zhang
LM: Prediction of candidate RNA signatures for recurrent ovarian
cancer prognosis by the construction of an integrated competing
endogenous RNA network. Oncol Rep. 40:2659–2673. 2018.PubMed/NCBI
|
|
18
|
Yan Y, Yu J, Liu H, Guo S, Zhang Y, Ye Y,
Xu L and Ming L: Construction of a long non-coding RNA-associated
ceRNA network reveals potential prognostic lncRNA biomarkers in
hepatocellular carcinoma. Pathol Res Pract. 214:2031–2038. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Loewen G, Jayawickramarajah J, Zhuo Y and
Shan B: Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol.
7:902014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gutschner T, Hammerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lou X, Li J, Yu D, Wei YQ, Feng S and Sun
JJ: Comprehensive analysis of five long noncoding RNAs expression
as competing endogenous RNAs in regulating hepatoma carcinoma.
Cancer Med. 8:5735–5749. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gene Ontology Consortium: The gene
ontology (GO) project in 2006. Nucleic Acids Res. 34((Database
Issue)): D322–D326. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Detterbeck FC, Boffa DJ, Kim AW and Tanoue
LT: The eighth edition lung cancer stage classification. Chest.
151:193–203. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KG and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zeng H, Zheng R, Guo Y, Zhang S, Zou X,
Wang N, Zhang L, Tang J, Chen J, Wei K, et al: Cancer survival in
China, 2003–2005: A population-based study. Int J Cancer.
136:1921–1930. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kimmelman AC and White E: Autophagy and
tumor metabolism. Cell Metab. 25:1037–1043. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Domschke P, Trucu D, Gerisch A and
Chaplain MAJ: Structured models of cell migration incorporating
molecular binding processes. J Math Biol. 75:1517–1561. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Franci G, Manfroni G, Cannalire R,
Felicetti T, Tabarrini O, Salvato A, Barreca ML, Altucci L and
Cecchetti V: Tumour cell population growth inhibition and cell
death induction of functionalized 6-aminoquinolone derivatives.
Cell Prolif. 48:705–717. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li C, Miao R, Zhang J, Qu K and Liu C:
Long non-coding RNA KCNQ1OT1 mediates the growth of hepatocellular
carcinoma by functioning as a competing endogenous RNA of miR-504.
Int J Oncol. Mar 12–2018.(Epub ahead of print).
|
|
31
|
Mou K, Liu B, Ding M, Mu X, Han D, Zhou Y
and Wang LJ: lncRNA-ATB functions as a competing endogenous RNA to
promote YAP1 by sponging miR-590-5p in malignant melanoma. Int J
Oncol. 53:1094–1104. 2018.PubMed/NCBI
|
|
32
|
Zhu SP, Wang JY, Wang XG and Zhao JP: Long
intergenic non-protein coding RNA 00858 functions as a competing
endogenous RNA for miR-422a to facilitate the cell growth in
non-small cell lung cancer. Aging (Albany NY). 9:475–486. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shang M, Wang X, Zhang Y, Gao Z, Wang T
and Liu R: LincRNA-ROR promotes metastasis and invasion of
esophageal squamous cell carcinoma by regulating miR-145/FSCN1.
Onco Targets Ther. 11:639–649. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rui X, Xu Y, Huang Y, Ji L and Jiang X:
lncRNA DLG1-AS1 promotes cell proliferation by competitively
binding with miR-107 and up-regulating ZHX1 expression in cervical
cancer. Cell Physiol Biochem. 49:1792–1803. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jiao D, Li Z, Zhu M, Wang Y, Wu G and Han
X: LncRNA MALAT1 promotes tumor growth and metastasis by targeting
miR-124/foxq1 in bladder transitional cell carcinoma (BTCC). Am J
Cancer Res. 8:748–760. 2018.PubMed/NCBI
|
|
36
|
Shen X, Bai H, Zhu H, Yan Q, Yang Y, Yu W,
Shi Q, Wang J, Li J and Chen L: Long non-coding RNA MEG3 functions
as a competing endogenous RNA to regulate HOXA11 expression by
sponging miR-181a in multiple myeloma. Cell Physiol Biochem.
49:87–100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu J, Li H, Zheng B, Sun L, Yuan Y and
Xing C: Competitive endogenous RNA (ceRNA) regulation network of
lncRNA-miRNA-mRNA in colorectal carcinogenesis. Dig Dis Sci.
64:1868–1877. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huang Z, Lei W, Hu HB, Zhang H and Zhu Y:
H19 promotes non-small-cell lung cancer (NSCLC) development through
STAT3 signaling via sponging miR-17. J Cell Physiol. 233:6768–6776.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jiang C, Yang Y, Yang Y, Guo L, Huang J,
Liu X, Wu C and Zou J: Long noncoding RNA (lncRNA) hOTAIR affects
tumorigenesis and metastasis of non-small cell lung cancer by
upregulating miR-613. Oncol Res. 26:725–734. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu JL, Meng FM and Li HJ: High expression
of lncRNA MEG3 participates in non-small cell lung cancer by
regulating microRNA-7-5p. Eur Rev Med Pharmacol Sci. 22:5938–5945.
2018.PubMed/NCBI
|
|
41
|
Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao
X, Chen WS and Li B: LncRNA-UCA1 exerts oncogenic functions in
non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett.
371:99–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chang N, Ahn SH, Kong DS, Lee HW and Nam
DH: The role of STAT3 in glioblastoma progression through dual
influences on tumor cells and the immune microenvironment. Mol Cell
Endocrinol. 451:53–65. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang H, Yan L, Sun K, Sun X, Zhang X, Cai
K and Song T: lncRNA BCAR4 increases viability, invasion, and
migration of non-small cell lung cancer cells by targeting
glioma-associated oncogene 2 (GLI2). Oncol Res. 27:359–369. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li C, Wan L, Liu Z, Xu G, Wang S, Su Z,
Zhang Y, Zhang C, Liu X, Lei Z and Zhang HT: Long non-coding RNA
XIST promotes TGF-β-induced epithelial-mesenchymal transition by
regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer.
Cancer Lett. 418:185–195. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu L, Ni J and He X: Upregulation of the
long noncoding RNA SNHG3 promotes lung adenocarcinoma
proliferation. Dis Markers. 2018:57367162018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zeng H, Wang J, Chen T, Zhang K, Chen J,
Wang L, Li H, Tuluhong D, Li J and Wang S: Downregulation of long
non-coding RNA Opa interacting protein 5-antisense RNA 1 inhibits
breast cancer progression by targeting sex-determining region Y-box
2 by microRNA-129-5p upregulation. Cancer Sci. 110:289–302.
2019.PubMed/NCBI
|
|
47
|
Renganathan A and Felley-Bosco E: Long
noncoding RNAs in cancer and therapeutic potential. Adv Exp Med
Biol. 1008:199–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu Z, Liu A, Nan A, Cheng Y, Yang T, Dai
X, Chen L, Li X, Jia Y, Zhang N and Jiang Y: The linc00152 controls
cell cycle progression by regulating CCND1 in 16HBE cells
malignantly transformed by cigarette smoke extract. Toxicol Sci.
167:496–508. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Saito S, Espinoza-Mercado F, Liu H, Sata
N, Cui X and Soukiasian HJ: Current status of research and
treatment for non-small cell lung cancer in never-smoking females.
Cancer Biol Ther. 18:359–368. 2017. View Article : Google Scholar : PubMed/NCBI
|