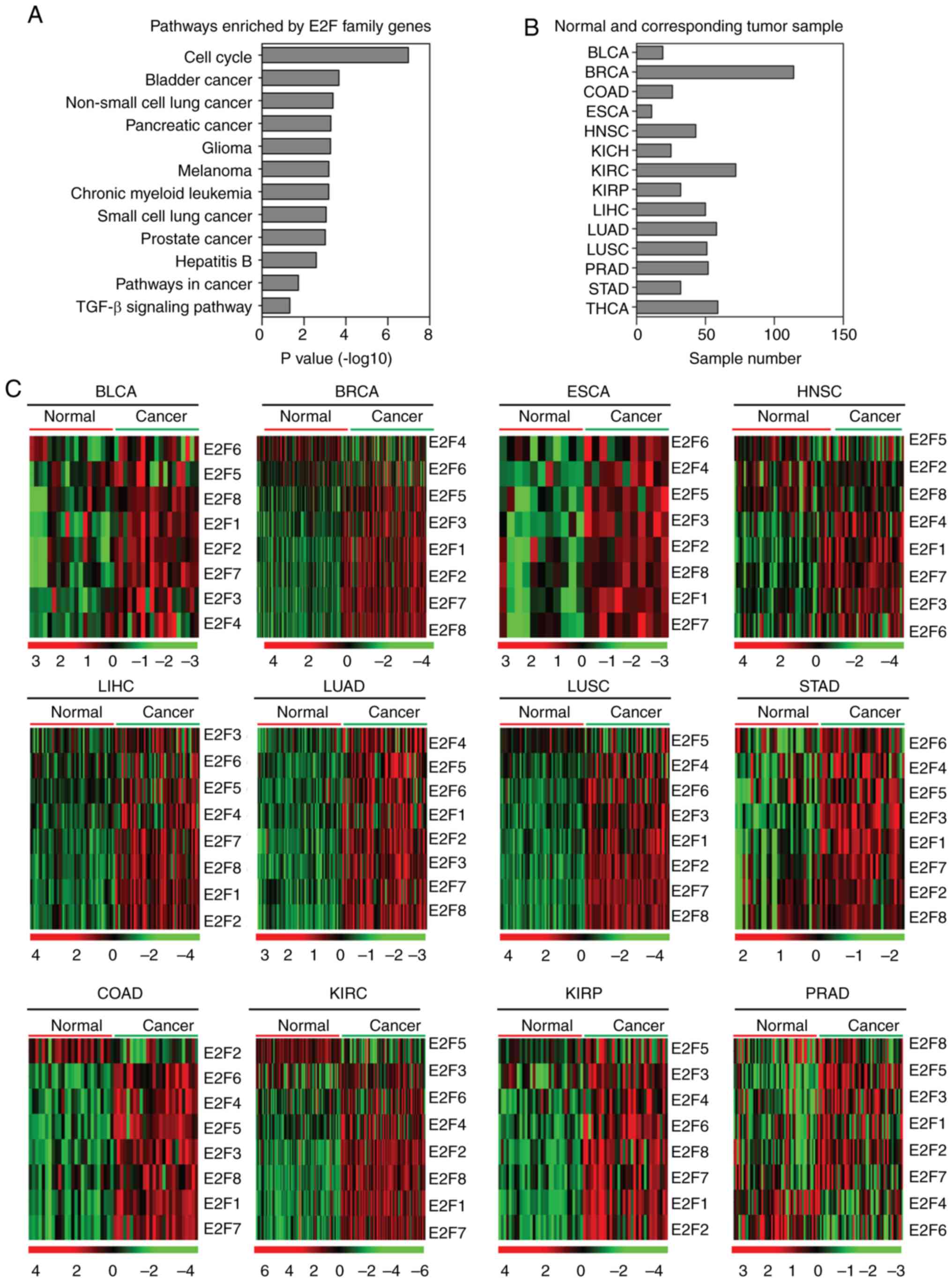
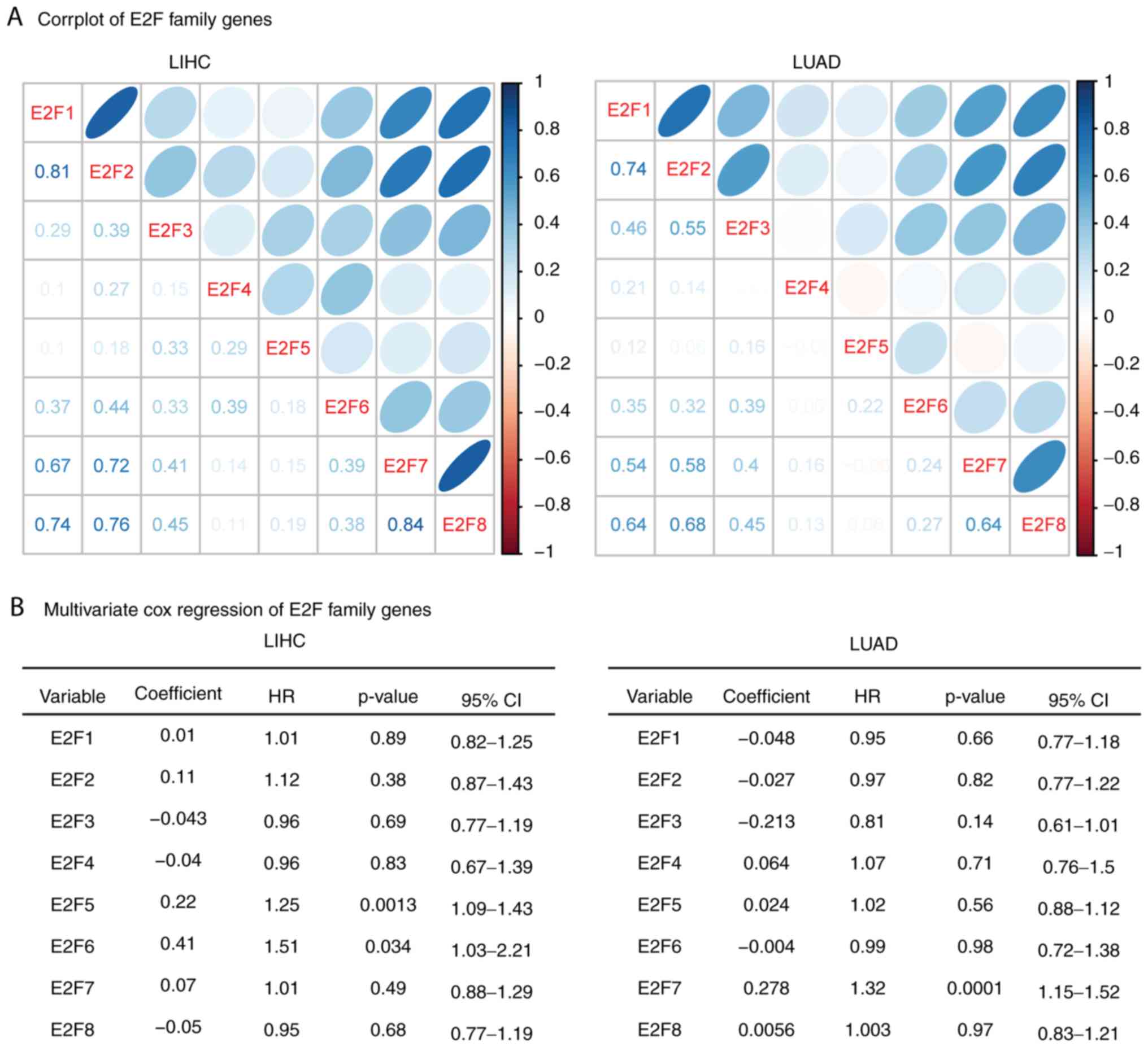
|
1
|
Kent LN and Leone G: The broken cycle: E2F
dysfunction in cancer. Nat Rev Cancer. 19:326–338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu H, Tang X, Srivastava A, Pécot T,
Daniel P, Hemmelgarn B, Reyes S, Fackler N, Bajwa A, Kladney R, et
al: Redeployment of Myc and E2f1-3 drives Rb-deficient cell cycles.
Nat Cell Biol. 17:1036–1048. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Morgunova E, Yin Y, Jolma A, Dave K,
Schmierer B, Popov A, Eremina N, Nilsson L and Taipale J:
Structural insights into the DNA-binding specificity of E2F family
transcription factors. Nat Commun. 6:100502015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen HZ, Tsai SY and Leone G: Emerging
roles of E2Fs in cancer: An exit from cell cycle control. Nat Rev
Cancer. 9:785–797. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang H, Martin V, Gomez-Manzano C,
Johnson DG, Alonso M, White E, Xu J, McDonnell TJ, Shinojima N and
Fueyo J: The RB-E2F1 pathway regulates autophagy. Cancer Res.
70:7882–7893. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Benevolenskaya EV and Frolov MV: Emerging
links between E2F control and mitochondrial function. Cancer Res.
75:619–623. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bertoli C, Herlihy AE, Pennycook BR,
Kriston-Vizi J and de Bruin RAM: Sustained E2F-dependent
transcription is a key mechanism to prevent
replication-stress-induced DNA damage. Cell Rep. 15:1412–1422.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kent LN, Bae S, Tsai SY, Tang X,
Srivastava A, Koivisto C, Martin CK, Ridolfi E, Miller GC, Zorko
SM, et al: Dosage-Dependent copy number gains in E2f1 and E2f3
drive hepatocellular carcinoma. J Clin Invest. 127:830–842. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tarangelo A, Lo N, Teng R, Kim E, Le L,
Watson D, Furth EE, Raman P, Ehmer U and Viatour P: Recruitment of
pontin/reptin by E2f1 amplifies E2f transcriptional response during
cancer progression. Nat Commun. 6:100282015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kent LN, Rakijas JB, Pandit SK, Westendorp
B, Chen HZ, Huntington JT, Tang X, Bae S, Srivastava A, Senapati S,
et al: E2f8 mediates tumor suppression in postnatal liver
development. J Clin Invest. 126:2955–2969. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Park SA, Platt J, Lee JW, Lopez-Giraldez
F, Herbst RS and Koo JS: E2F8 as a novel therapeutic target for
lung cancer. J Natl Cancer Inst. 18:1072015.
|
|
12
|
Fujiwara K, Yuwanita I, Hollern DP and
Andrechek ER: Prediction and genetic demonstration of a role for
activator E2Fs in myc-induced tumors. Cancer Res. 71:1924–1932.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lan W, Bian B, Xia Y, Dou S, Gayet O,
Bigonnet M, Santofimia-Castaño P, Cong M, Peng L, Dusetti N and
Iovanna J: E2F signature is predictive for the pancreatic
adenocarcinoma clinical outcome and sensitivity to E2F inhibitors,
but not for the response to cytotoxic-based treatments. Sci Rep.
8:83302018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Saenz-Ponce N, Pillay R, de Long LM,
Kashyap T, Argueta C, Landesman Y, Hazar-Rethinam M, Boros S,
Panizza B, Jacquemyn M, et al: Targeting the XPO1-dependent nuclear
export of E2F7 reverses anthracycline resistance in head and neck
squamous cell carcinomas. Sci Transl Med. 27:4472018.
|
|
15
|
Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao
SD, Zhang Y, Yang L, Shan W, He Q, et al: Comprehensive genomic
characterization of long non-coding RNAs across human cancers.
Cancer Cell. 28:529–540. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun W, Bunn P, Jin C, Little P,
Zhabotynsky V, Perou CM, Hayes DN, Chen M and Lin DY: The
association between copy number aberration, DNA methylation and
gene expression in tumor samples. Nucleic Acids Res. 46:3009–3018.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Weisenberger DJ: Characterizing DNA
methylation alterations from the cancer genome atlas. J Clin
Invest. 124:17–23. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Saghafinia S, Mina M, Riggi N, Hanahan D
and Ciriello G: Pan-Cancer landscape of aberrant DNA methylation
across human tumors. Cell Rep. 25:1066–1080. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Weinstein JN, Collisson EA, Mills GB, Shaw
KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C and Stuart JM;
Cancer Genome Atlas Research Network, : The cancer genome atlas
pan-cancer analysis project. Nat Genet. 45:1113–1120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hutter C and Zenklusen JC: The cancer
genome atlas: Creating lasting value beyond its data. Cell.
173:283–285. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Uhlen M, Zhang C, Lee S, Sjöstedt E,
Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et
al: A pathology atlas of the human cancer transcriptome. Science.
35:eaan2507. 7–2017.
|
|
22
|
Hoadley KA, Yau C, Hinoue T, Wolf DM,
Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, et
al: Cell-of-origin patterns dominate the molecular classification
of 10,000 tumors from 33 types of cancer. Cell. 173:291–304. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bailey MH, Tokheim C, Porta-Pardo E,
Sengupta S, Bertrand D, Weerasinghe A, Colaprico A, Wendl MC, Kim
J, Reardon B, et al: Comprehensive characterization of cancer
driver genes and mutations. Cell. 173:371–385. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cancer Genome Atlas Research Network.
Electronic address, . wheeler@bcm.edu and CancerGenome Atlas
Research Network: Comprehensive and integrative genomic
characterization of hepatocellular carcinoma. Cell. 169:1327–1341.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang HW, Hsieh TH, Huang SY, Chau GY, Tung
CY, Su CW and Wu JC: Forfeited hepatogenesis program and increased
embryonic stem cell traits in young hepatocellular carcinoma (HCC)
comparing to elderly HCC. BMC Genomics. 14:7362013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Melis M, Diaz G, Kleiner DE, Zamboni F,
Kabat J, Lai J, Mogavero G, Tice A, Engle RE, Becker S, et al:
Viral expression and molecular profiling in liver tissue versus
microdissected hepatocytes in hepatitis B virus-associated
hepatocellular carcinoma. J Transl Med. 12:2302014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sanchez-Palencia A, Gomez-Morales M,
Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R and Fárez-Vidal ME:
Gene expression profiling reveals novel biomarkers in nonsmall cell
lung cancer. Int J Cancer. 129:355–364. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hou J, Aerts J, den Hamer B, van Ijcken W,
den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens
JA, Hoogsteden HC, et al: Gene expression-based classification of
non-small cell lung carcinomas and survival prediction. PLoS One.
5:e103122010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fouret R, Laffaire J, Hofman P,
Beau-Faller M, Mazieres J, Validire P, Girard P, Camilleri-Bröet S,
Vaylet F, Leroy-Ladurie F, et al: A comparative and integrative
approach identifies ATPase family, AAA domain containing 2 as a
likely driver of cell proliferation in lung adenocarcinoma. Clin
Cancer Res. 18:5606–5616. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wei TY, Juan CC, Hisa JY, Su LJ, Lee YC,
Chou HY, Chen JM, Wu YC, Chiu SC, Hsu CP, et al: Protein arginine
methyltransferase 5 is a potential oncoprotein that upregulates G1
cyclins/cyclin-dependent kinases and the phosphoinositide
3-kinase/AKT signaling cascade. Cancer Sci. 103:1640–1650. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rousseaux S, Debernardi A, Jacquiau B,
Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY,
Lantuejoul S, Hainaut P, et al: Ectopic activation of germline and
placental genes identifies aggressive metastasis-prone lung
cancers. Sci Transl Med. 5:1862013. View Article : Google Scholar
|
|
33
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nagy A, Lanczky A, Menyhart O and Gyorffy
B: Validation of miRNA prognostic power in hepatocellular carcinoma
using expression data of independent datasets. Sci Rep. 8:92272018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ye QH, Qin LX, Forgues M, He P, Kim JW,
Peng AC, Simon R, Li Y, Robles AI, Chen Y, et al: Predicting
hepatitis B virus-positive metastatic hepatocellular carcinomas
using gene expression profiling and supervised machine learning.
Nat Med. 9:416–423. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hoshida Y, Villanueva A, Kobayashi M, Peix
J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J, et
al: Gene expression in fixed tissues and outcome in hepatocellular
carcinoma. N Engl J Med. 359:1995–2004. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim SM, Leem SH, Chu IS, Park YY, Kim SC,
Kim SB, Park ES, Lim JY, Heo J, Kim YJ, et al: Sixty-five
gene-based risk score classifier predicts overall survival in
hepatocellular carcinoma. Hepatology. 55:1443–1452. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kate RJ and Nadig R: Stage-Specific
predictive models for breast cancer survivability. Int J Med
Inform. 97:304–311. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jamal-Hanjani M, Wilson GA, McGranahan N,
Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R,
Rosenthal R, et al: Tracking the evolution of non-small-cell lung
cancer. N Engl J Med. 376:2109–2121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Peifer M, Fernandez-Cuesta L, Sos ML,
George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander
T, et al: Integrative genome analyses identify key somatic driver
mutations of small-cell lung cancer. Nat Genet. 44:1104–1110. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bertucci F, Ng CKY, Patsouris A, Droin N,
Piscuoglio S, Carbuccia N, Soria JC, Dien AT, Adnani Y and Kamal M:
Genomic characterization of metastatic breast cancers. Nature.
569:560–564. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Vousden KH and Lane DP: P53 in health and
disease. Nat Rev Mol Cell Biol. 8:275–283. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen Z, Trotman LC, Shaffer D, Lin HK,
Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al:
Crucial role of p53-dependent cellular senescence in suppression of
pten-deficient tumorigenesis. Nature. 436:725–730. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Meek DW: Tumour suppression by p53: A role
for the DNA damage response? Nat Rev Cancer. 9:714–723. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gyorffy B, Surowiak P, Budczies J and
Lanczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li Y, Huang J, Yang D, Xiang S, Sun J, Li
H and Ren G: Expression patterns of E2F transcription factors and
their potential prognostic roles in breast cancer. Oncol Lett.
15:9216–9230. 2018.PubMed/NCBI
|
|
49
|
Manicum T, Ni F, Ye Y, Fan X and Chen BC:
Prognostic values of E2F mRNA expression in human gastric cancer.
Biosci Rep. 21:382018.
|
|
50
|
Huang YL, Ning G, Chen LB, Lian YF, Gu YR,
Wang JL, Chen DM, Wei H and Huang YH: Promising diagnostic and
prognostic value of E2Fs in human hepatocellular carcinoma. Cancer
Manag Res. 11:1725–1740. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gao Z, Shi R, Yuan K and Wang Y:
Expression and prognostic value of E2F activators in NSCLC and
subtypes: A research based on bioinformatics analysis. Tumour Biol.
37:14979–14987. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Narita M, Nunez S, Heard E, Narita M, Lin
AW, Hearn SA, Spector DL, Hannon GJ and Lowe SW: Rb-Mediated
heterochromatin formation and silencing of E2F target genes during
cellular senescence. Cell. 113:703–716. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Aksoy O, Chicas A, Zeng T, Zhao Z,
McCurrach M, Wang X and Lowe SW: The atypical E2F family member
E2F7 couples the p53 and RB pathways during cellular senescence.
Genes Dev. 26:1546–1557. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ran LK, Chen Y, Zhang ZZ, Tao NN, Ren JH,
Zhou L, Tang H, Chen X, Chen K, Li WY, et al: SIRT6 overexpression
potentiates apoptosis evasion in hepatocellular carcinoma via
BCL2-associated X protein-dependent apoptotic pathway. Clin Cancer
Res. 22:3372–3382. 2016. View Article : Google Scholar : PubMed/NCBI
|