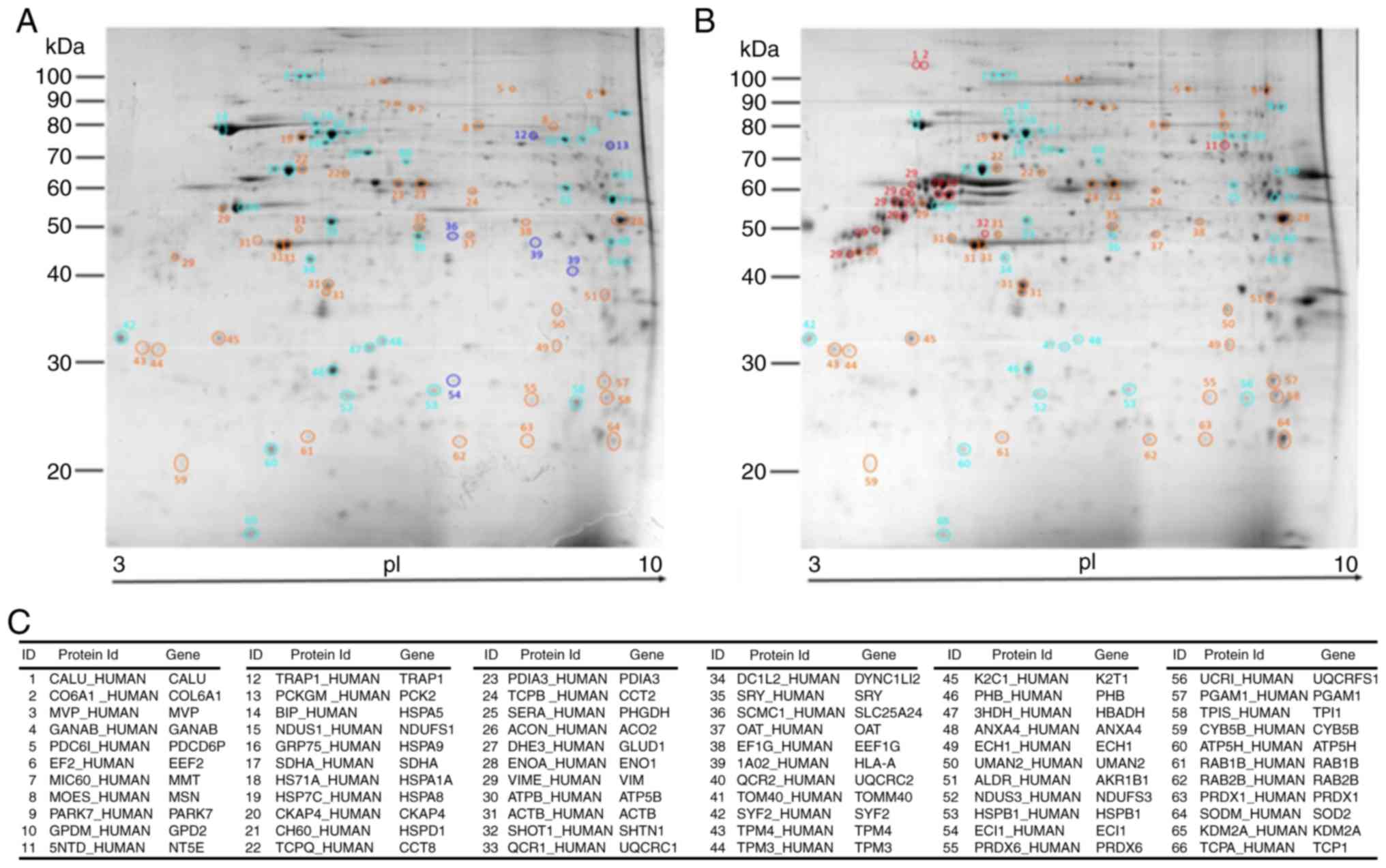
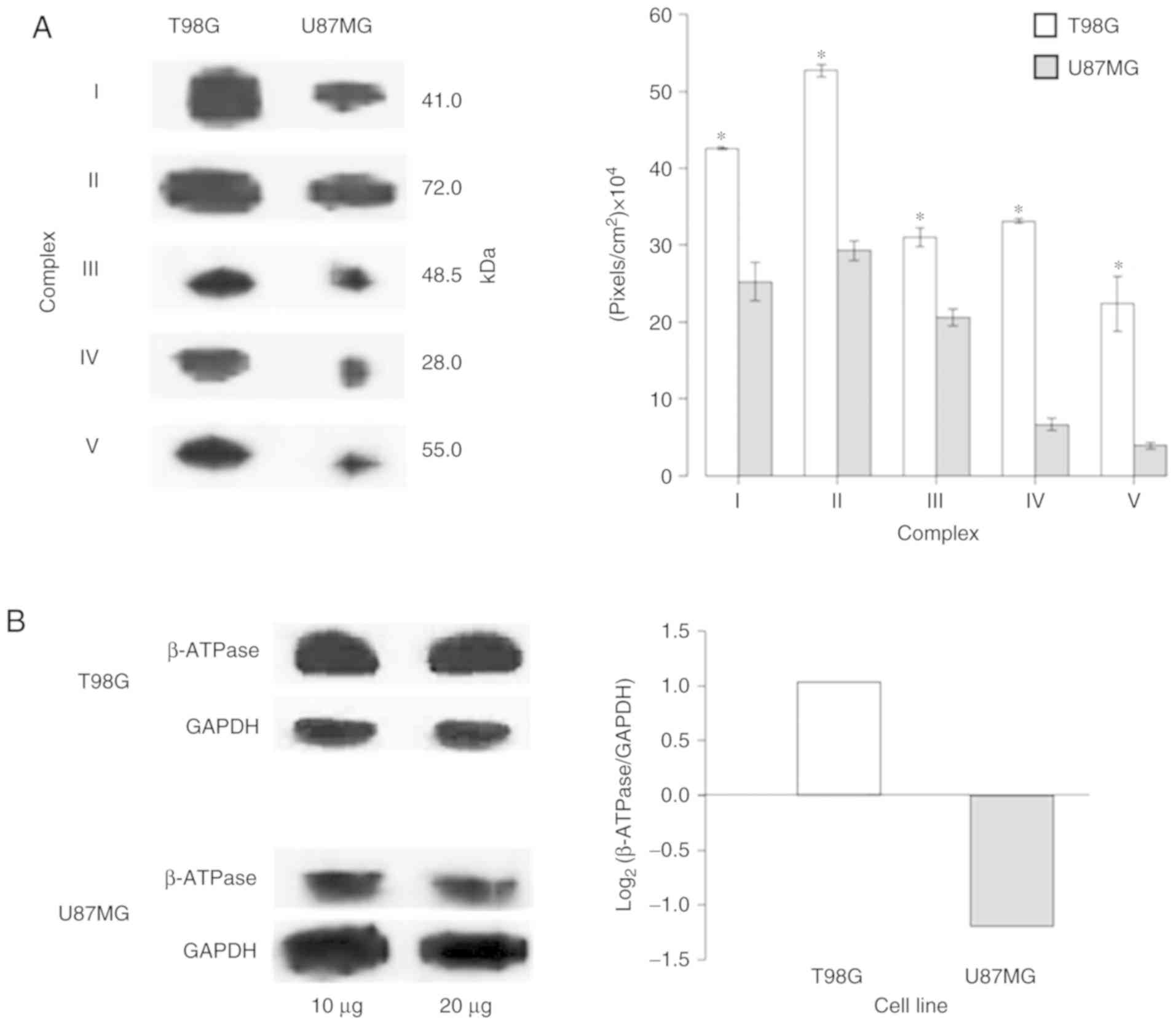
|
1
|
Luna B, Bhatia S, Yoo C, Felty Q, Sandberg
DI, Duchowny M, Khatib Z, Miller I, Ragheb J, Prasanna J and Roy D:
Proteomic and mitochondrial genomic analyses of pediatric brain
tumors. Mol Neurobiol. 52:1341–1363. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pooladi M, Rezaei-Tavirani M, Hashemi M,
Hesami-Tackallou S, Khaghani-Razi-Abad S, Moradi A, Zali AR,
Mousavi M, Firozi-Dalvand L, Rakhshan A and Zamanian Azodi M:
Cluster and Principal Component Analysis of Human Glioblastoma
Multiforme (GBM) Tumor Proteome. Iran J cancer Prev. 7:87–95.
2014.PubMed/NCBI
|
|
3
|
Fangusaro J: Pediatric high grade glioma:
A review and update on tumor clinical characteristics and biology.
Front Oncol. 2:1052012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Batash R, Asna N, Schaffer P, Francis N
and Schaffer M: Glioblastoma multiforme, diagnosis and treatment;
recent literature review. Curr Med Chem. 24:3002–3009. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kafka A, Tomas D, Lechpammer M, Gabud T,
Pažanin L and Pećina-Šlaus N: Expression Levels and Localizations
of DVL3 and sFRP3 in Glioblastoma. Dis Markers. 2017:9253495. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hagberg H, Mallard C, Rousset CI and
Thornton C: Mitochondria: Hub of injury responses in the developing
brain. Lancet Neurol. 13:217–232. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Raefsky SM and Mattson MP: Adaptive
responses of neuronal mitochondria to bioenergetic challenges:
Roles in neuroplasticity and disease resistance. Free Radic Biol.
102:203–216. 2017. View Article : Google Scholar
|
|
8
|
Son G and Han J: Roles of mitochondria in
neuronal development. BMB Rep. 51:549–556. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hanahan D and Weinberg RA: Hallmarks of
cancer. The next generation. 144:646–674. 2011.
|
|
10
|
Floor SL, Dumont JE, Maenhaut C and Raspe
E: Hallmarks of cancer: Of all cancer cells, all the time? Trends
Mol Med. 18:509–515. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Czarnecka AM, Czarnecki JS, Kukwa W,
Cappello F, Ścińska A and Kukwa A: Molecular oncology focus-is
carcinogenesis a ‘mitochondriopathy’? J Biomed Sci. 17:312010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kroemer G and Pouyssegur J: Tumor cell
metabolism: Cancer's Achilles' heel. Cancer Cell. 13:472–482. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Galluzzi L, Morselli E, Kepp O, Vitale I,
Rigoni A, Vacchelli E, Michaud M, Zischka H, Castedo M and Kroemer
G: Mitochondrial gateways to cancer. Mol Aspects Med. 31:1–20.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vyas S, Zaganjor E and Haigis MC:
Mitochondria and Cancer. Cell. 166:555–566. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Petrak J, Ivanek R, Toman O, Cmejla R,
Cmejlova J, Vyoral D, Zivny J and Vulpe CD: Déjà vu in proteomics.
A hit parade of repeatedly identified differentially expressed
proteins. Proteomics. 8:1744–1749. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Deighton RF, McGregor R, Kemp J, McCulloch
J and Whittle IR: Glioma pathophysiology: Insights emerging from
proteomics. Brain Pathol. 20:691–703. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Valledor L and Jorrín J: Back to the
basics: Maximizing the information obtained by quantitative two
dimensional gel electrophoresis analyses by an appropriate
experimental design and statistical analyses. J Proteomics.
74:1–18. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meehan MC: General system theory:
Foundations, development, applications. JAMA. 208((5)):
87019691969.
|
|
19
|
Obre E and Rossignol R: Emerging concepts
in bioenergetics and cancer research: Metabolic flexibility,
coupling, symbiosis, switch, oxidative tumors, metabolic
remodeling, signaling and bioenergetic therapy. Int J Biochem Cell
Biol. 59:167–181. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tyanova S, Temu T and Cox J: The MaxQuant
computational platform for mass spectrometry-based shotgun
proteomics. Nat Protoc. 11:2301–2319. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stekhoven DJ and Buhlmann P:
MissForest-non-parametric missing value imputation for mixed-type
data. Bioinformatics. 28:112–118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lê S, Josse J and Husson F: FactoMineR: An
R package for multivariate analysis. J Stat Softw. 1–18.
2008.doi.org/10.18637/jss.v025.i01 PMID: 18676020.
|
|
23
|
Abdi H and Williams LJ: Principal
component analysis. Wiley Interdiscip Rev Comput Stat. 2:433–459.
2010. View Article : Google Scholar
|
|
24
|
Hurkman WJ and Tanaka CK: Solubilization
of plant membrane proteins for analysis by two-dimensional gel
electrophoresis. Plant Physiol. 81:802–806. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Salazar E, Díaz-Mejía JJ, Moreno-Hagelsieb
G, Martínez- Batallar G, Mora Y, Mora J and Encarnación S:
Characterization of the Nif A-RpoN regulon in rhizobium etli in
free life and in symbiosis with phaseolus vulgaris. Appl Environ
Microbiol. 76:4510–4520. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mead R, Curnow RN and Hasted AM:
Statistical Methods in Agriculture and Experimental Biology. (3rd).
Chapman and Hall/CRC Press. (New York, NY). 4722003.
|
|
27
|
Cochran WG: Sampling Techniques. (3rd).
Wiley. (New Jersey). 4281977.
|
|
28
|
Perkins DN, Pappin DJ, Creasy DM and
Cottrell JS: Probability-based protein identification by searching
sequence databases using mass spectrometry data. Electrophoresis.
20:3551–3567. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
The UniProt Consortium: UniProt: The
universal protein knowledge base. Nucleic Acids Res. 45:D158–D169.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Montojo J, Zuberi K, Rodriguez H, Kazi F,
Wright G, Donaldson SL, Morris Q and Bader GD: GeneMANIA Cytoscape
plugin: Fast gene function predictions on the desktop.
Bioinformatics. 26:2927–2928. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ashkenazi M, Bader GD, Kuchinsky A,
Moshelion M and States DJ: Cytoscape ESP: Simple search of complex
biological networks. Bioinformatics. 24:1465–1466. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mi H, Huang X, Muruganujan A, Tang H,
Mills C, Kang D and Thomas PD: PANTHER version 11: Expanded
annotation data from Gene Ontology and Reactome pathways, and data
analysis tool enhancements. Nucleic Acids Res. 45:D183–D189. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Supek F, Bošnjak M, Škunca N and Šmuc T:
Revigo summarizes and visualizes long lists of gene ontology terms.
PLoS One. 6:e218002011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
R Core Team, . R: A Language and
Environment for Statistical Computing. (Vienna, Austria).
2017.
|
|
36
|
Laemmli UK: Cleavage of structural
proteins during the assembly of the head of bacteriophage T4.
Nature. 227:680–685. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Towbin H, Staehelin T and Gordon J:
Electrophoretic transfer of proteins from polyacrylamide gels to
nitrocellulose sheets: Procedure and some applications. Proc Natl
Acad Sci USA. 76:4350–4354. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lowry OH, Rosebrough NJ, Farr AL and
Randall RJ: Protein measurement with the folin phenol reagent. J
Biol Chem. 193:265–275. 1951.PubMed/NCBI
|
|
39
|
Cuezva JM, Krajewska M, de Heredia ML,
Krajewski S, Santamaría G, Kim H, Zapata JM, Marusawa H, Chamorro M
and Reed JC: The Bioenergetic Signature of Cancer: A Marker of
Tumor Progression. Cancer Res. 62:6674–6681. 2002.PubMed/NCBI
|
|
40
|
Hair JF, Anderson RE, Tatham RL and Black
WC: Multivariate Data Analysis. Int J Pharmaceutics. 816:21335075.
1998.
|
|
41
|
Everitt B and Hothorn T: An Introduction
to Applied Multivariate Analysis with R. Springer-Verlag; New York,
NY: 2011, View Article : Google Scholar
|
|
42
|
Heinisch O: An introduction to sampling
theory with application to agriculture. Biom Z. 5:2122017.
View Article : Google Scholar
|
|
43
|
Cuezva JM, Ortega AD, Willers I,
Sánchez-Cenizo L, Aldea M and Sánchez-Aragó M: The tumor suppressor
function of mitochondria: Translation into the clinics. Biochim
Biophys Acta. 1792:1145–58. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Siegelin MD, Plescia J, Raskett CM,
Gilbert CA, Ross AH and Altieri DC: Global targeting of subcellular
heat shock protein-90 networks for therapy of glioblastoma. Mol
Cancer Ther. 9:1638–1646. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wolf DA: Is reliance on mitochondrial
respiration a ‘chink in the armor’ of therapy-resistant cancer?
Cancer Cell. 26:788–795. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jose C, Bellance N and Rossignol R:
Choosing between glycolysis and oxidative phosphorylation: A
tumor's dilemma? Biochim Biophys Acta. 1807:552–561. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pooladi M, Abad SK and Hashemi M:
Proteomics analysis of human brain glial cell proteome by 2D gel.
Indian J Cancer. 51:159–162. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ramão A, Gimenez M, Laure HJ, Izumi C,
Vida RC, Oba-Shinjo S, Marie SK and Rosa JC: Changes in the
expression of proteins associated with aerobic glycolysis and cell
migration are involved in tumorigenic ability of two glioma cell
lines. Proteome Sci. 10:532012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Banerjee HN, Mahaffey K, Riddick E,
Banerjee A, Bhowmik N and Patra M: Search for a
diagnostic/prognostic biomarker for the brain cancer glioblastoma
multiforme by 2D-DIGE-MS technique. Mol Cell Biochem. 367:59–63.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Karpel-Massler G, Ishida CT, Bianchetti E,
Shu C, Perez-Lorenzo R, Horst B, Banu M, Roth KA, Bruce JN, Canoll
P, Altieri DC and Siegelin MD: Inhibition of mitochondrial matrix
chaperones and antiapoptotic bcl-2 family proteins empower
antitumor therapeutic responses. Cancer Res. 77:3513–3526. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chae JI, Jeon YJ and Shim JH:
Downregulation of Sp1 is involved in honokiol-induced cell cycle
arrest and apoptosis in human malignant pleural mesothelioma cells.
Oncol Rep. 29:2318–2324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Porporato PE, Filigheddu N, Pedro JMB,
Kroemer G and Galluzzi L: Mitochondrial metabolism and cancer. Cell
Res. 28:265–280. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
White E: Deconvoluting the
context-dependent role for autophagy in cancer. Nat Rev Cancer.
12:401–410. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Giatromanolaki A, Sivridis E, Mitrakas A,
Kalamida D, Zois CE, Haider S, Piperidou C, Pappa A, Gatter KC,
Harris AL and Koukourakis MI: Autophagy and lysosomal related
protein expression patterns in human glioblastoma. Cancer Biol
Ther. 15:1468–1478. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sica V, Galluzzi L, Bravo-San Pedro JM,
Izzo V, Maiuri MC and Kroemer G: Organelle-Specific Initiation of
Autophagy. Mol Cell. 59:522–539. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Gautam P, Nair SC, Gupta MK, Sharma R,
Polisetty RV, Uppin MS, Sundaram C, Puligopu AK, Ankathi P, Purohit
AK, et al: Proteins with altered levels in plasma from glioblastoma
patients as revealed by iTRAQ-based quantitative proteomic
analysis. PLoS One. 7:e461532012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Locasale JW, Melman T, Song S, Yang X,
Swanson KD, Cantley LC, Wong ET and Asara JM: Metabolomics of human
cerebrospinal fluid identifies signatures of malignant glioma. Mol
Cell Proteomics. 11:M111.014688. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Iwadate Y, Sakaida T, Hiwasa T, Nagai Y,
Ishikura H, Takiguchi M and Yamaura A: Molecular classification and
survival prediction in human gliomas based on proteome analysis.
Cancer Res. 64:2496–2501. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ordys BB, Launay S, Deighton RF, McCulloch
J and Whittle IR: The role of mitochondria in glioma
pathophysiology. Mol Neurobiol. 42:64–75. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Collet B, Guitton N, Saïkali S, Avril T,
Pineau C, Hamlat A, Mosser J and Quillien V: Differential analysis
of glioblastoma multiforme proteome by a 2D-DIGE approach. Proteome
Sci. 9:162011. View Article : Google Scholar : PubMed/NCBI
|