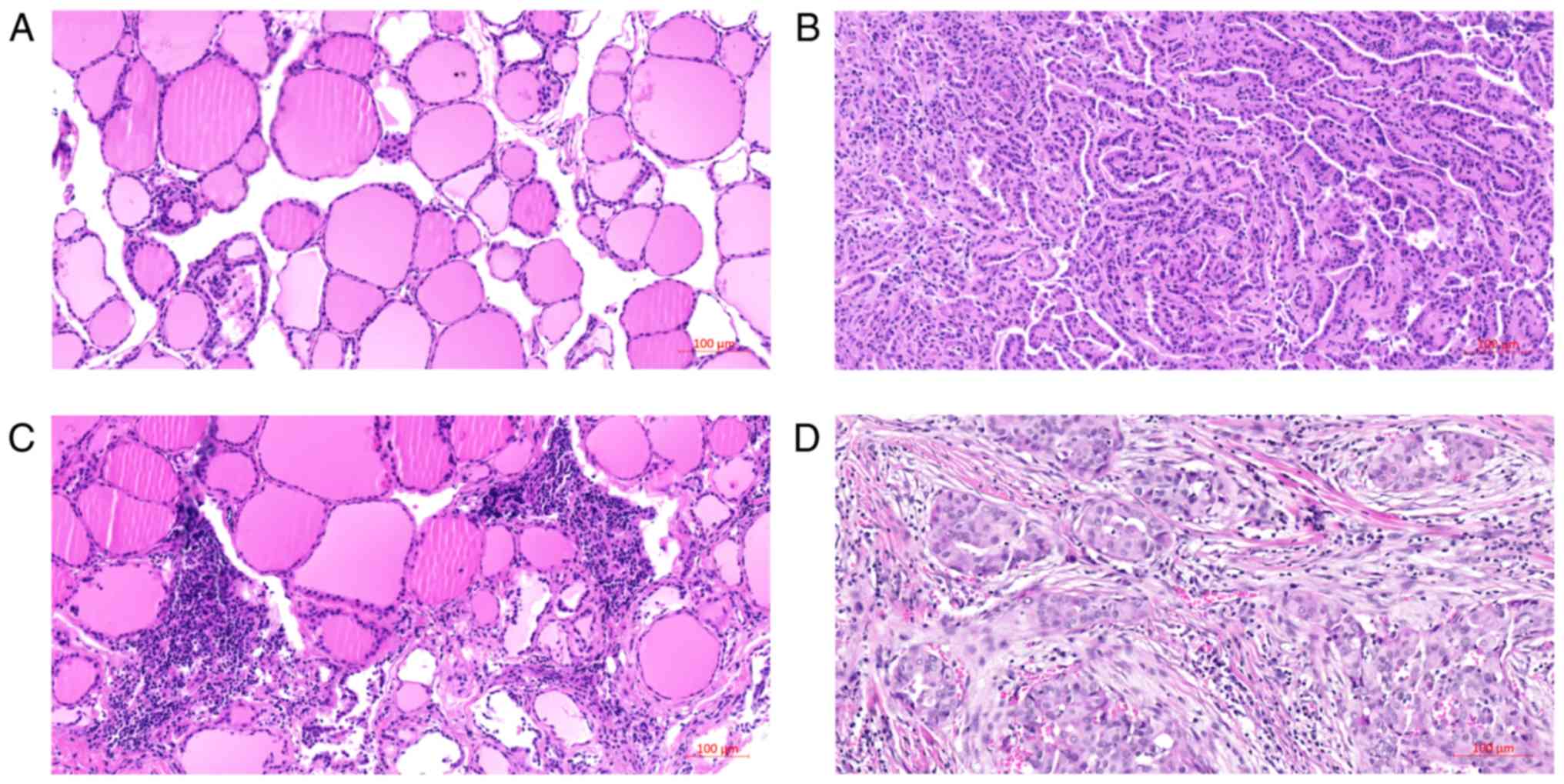
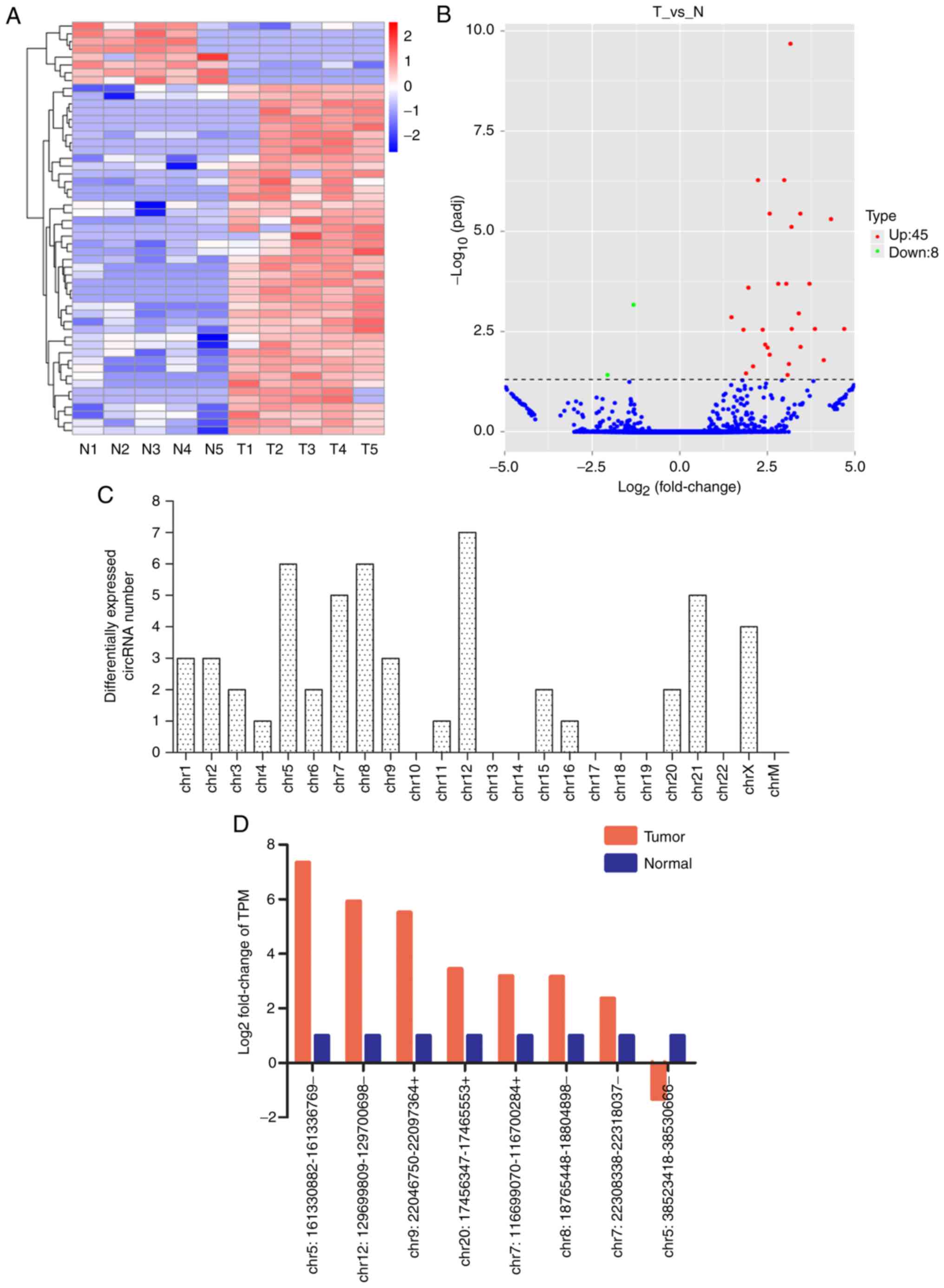
|
1
|
Lam AK, Lo CY and Lam KS: Papillary
carcinoma of thyroid: A 30-yr clinicopathological review of the
histological variants. Endocr Pathol. 16:323–330. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abdullah MI, Junit SM, Ng KL, Jayapalan
JJ, Karikalan B and Hashim OH: Papillary thyroid cancer: Genetic
alterations and molecular biomarker investigations. Int J Med Sci.
16:450–460. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ng SC, Kuo SF, Hua CC, Huang BY, Chiang
KC, Chu YY, Hsueh C and Lin JD: Differentiation of the follicular
variant of papillary thyroid carcinoma from classic papillary
thyroid carcinoma: An ultrasound analysis and complement to
fine-needle aspiration cytology. J Ultrasound Med. 37:667–674.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lin JD: Thyroglobulin and human thyroid
cancer. Clin Chim Acta. 388:15–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tuttle RM, Leboeuf R and Martorella AJ:
Papillary thyroid cancer: Monitoring and therapy. Endocrinol
Metabol Clin North Amer. 36:753–778. 2007. View Article : Google Scholar
|
|
6
|
Hurtado-Lopez LM, Fernandez-Ramirez F,
Martinez-Penafiel E, Carrillo Ruiz JD and Herrera Gonzalez NE:
Molecular analysis by gene expression of mitochondrial ATPase
subunits in papillary thyroid cancer: Is ATP5E transcript a
possible early tumor marker? Med Sci Monit. 21:1745–1751. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tang KT and Lee CH: BRAF mutation in
papillary thyroid carcinoma: Pathogenic role and clinical
implications. J Chin Med Assoc. 73:113–128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and Cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bachmayr-Heyda A, Reiner AT, Auer K,
Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW,
Zeillinger R and Pils D: Correlation of circular RNA abundance with
proliferation-exemplified with colorectal and ovarian cancer,
idiopathic lung fibrosis, and normal human tissues. Sci Rep.
5:80572015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z,
Yang J, Fan J, Liu L and Qin W: Hsa_circ_0001649: A circular RNA
and potential novel biomarker for hepatocellular carcinoma. Cancer
Biomarkers. 16:161–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ and Liang L: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang X, Zhang Y, Huang L, Zhang J, Pan F,
Li B, Yan Y, Jia B, Liu H, Li S and Zheng W: Decreased expression
of hsa_circ_001988 in colorectal cancer and its clinical
significances. Int J Clin Exp Pathol. 8:16020–16025.
2015.PubMed/NCBI
|
|
20
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tian M, Chen R, Li T and Xiao B: Reduced
expression of circRNA hsa_circ_0003159 in gastric cancer and its
clinical significance. J Clin Lab Anal. 32:e222812017. View Article : Google Scholar
|
|
22
|
Bi W, Huang J, Nie C, Liu B, He G, Han J,
Pang R, Ding Z, Xu J and Zhang J: CircRNA circRNA_102171 promotes
papillary thyroid cancer progression through modulating
CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin
Cancer Res. 37:2752018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wei H, Pan L, Tao D and Li R: Circular RNA
circZFR contributes to papillary thyroid cancer cell proliferation
and invasion by sponging miR-1261 and facilitating C8orf4
expression. Biochem Biophys Res Commun. 503:56–61. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lan X, Cao J, Xu J, Chen C, Zheng C, Wang
J, Zhu X, Zhu X and Ge M: Decreased expression of hsa_circ_0137287
predicts aggressive clinicopathologic characteristics in papillary
thyroid carcinoma. J Clin Lab Anal. 32:e225732018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang M, Chen B, Ru Z and Cong L: CircRNA
circ-ITCH suppresses papillary thyroid cancer progression through
miR-22-3p/CBL/β-catenin pathway. Biochem Biophys Res Commun.
504:283–288. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lan X, Xu J, Chen C, Zheng C, Wang J, Cao
J, Zhu X and Ge M: The landscape of circular RNA expression
profiles in papillary thyroid carcinoma based on RNA sequencing.
Cell Physiol Biochem. 47:1122–1132. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Doescher J, Veit JA and Hoffmann TK: The
8th edition of the AJCC Cancer Staging Manual. Updates in
otorhinolaryngology, head and neck surgery. HNO. 65:956–961.
2017.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
28
|
EI-Naggar AK, Chan JKC, Grandis JR, Takata
T and Slootweg PJ: WHO Classification of Head and Neck Tumours.
(Lyon). 2017.
|
|
29
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
D SJ: The positive false discovery rate: A
Bayesian interpretation and the q-value. Ann Statistics.
31:2013–2035. 2003. View Article : Google Scholar
|
|
31
|
Panda AC and Gorospe M: Detection and
analysis of circular RNAs by RT-PCR. Bio Protoc. 8(pii):
e27752018.PubMed/NCBI
|
|
32
|
Zhu M, Xu Y, Chen Y and Yan F: Circular
BANP, an upregulated circular RNA that modulates cell proliferation
in colorectal cancer. Biomed Pharmacother. 88:138–144. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sui W, Gan Q, Liu F, Chen H, Liu J and Dai
Y: The differentially expressed circular ribonucleic acids of
primary hepatic carcinoma following liver transplantation as new
diagnostic biomarkers for primary hepatic carcinoma. Tumour Biol.
40:10104283187669282018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shang X, Li G, Liu H, Li T, Liu J, Zhao Q
and Wang C: Comprehensive circular RNA profiling reveals that
hsa_circ_0005075, a new circular RNA biomarker, is involved in
hepatocellular crcinoma development. Medicine (Baltimore).
95:e38112016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ren H, Liu Z, Liu S, Zhou X, Wang H, Xu J,
Wang D and Yuan G: Profile and clinical implication of circular
RNAs in human papillary thyroid carcinoma. PeerJ. 6:e53632018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ouyang Q, Huang Q, Jiang Z, Zhao J, Shi GP
and Yang M: Using plasma circRNA_002453 as a novel biomarker in the
diagnosis of lupus nephritis. Mol Immunol. 101:531–538. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Peng Y, Song X, Zheng Y, Cheng H and Lai
W: circCOL3A1-859267 regulates type I collagen expression by
sponging miR-29c in human dermal fibroblasts. Eur J Dermatol.
28:613–620. 2018.PubMed/NCBI
|
|
38
|
Hu J, Li C, Liu C, Zhao S, Wang Y and Fu
Z: Expressions of miRNAs in papillary thyroid carcinoma and their
associations with the clinical characteristics of PTC. Cancer
Biomark. 18:87–94. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hoo ZH, Candlish J and Teare D: What is an
ROC curve? Emerg Med J. 34:357–359. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ebbesen KK, Hansen TB and Kjems J:
Insights into circular RNA biology. RNA Biol. 14:1035–1045. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dong Y, He D, Peng Z, Peng W, Shi W, Wang
J, Li B, Zhang C and Duan C: Circular RNAs in cancer: An emerging
key player. J Hematol Oncol. 10:22017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chang TH, Huang HY, Hsu BK, Weng SL, Horng
JT and Huang HD: An enhanced computational platform for
investigating the roles of regulatory RNA and for identifying
functional RNA motifs. BMC Bioinformatics. 14 (Suppl 2):S42013.
View Article : Google Scholar
|
|
43
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Backes C, Kehl T, Stöckel D, Fehlmann T,
Schneider L, Meese E, Lenhof HP and Keller A: miRPathDB: A new
dictionary on microRNAs and target pathways. Nucleic Acids Res.
45:D90–D96. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu R, Liu F, Li L, Sun M and Chen K:
MiR-498 regulated FOXO3 expression and inhibited the proliferation
of human ovarian cancer cells. Biomed Pharmacother. 72:52–57. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
You Y, Que K, Zhou Y, Zhang Z, Zhao X,
Gong J and Liu Z: MicroRNA-766-3p inhibits tumour progression by
targeting Wnt3a in hepatocellular carcinoma. Mol Cells. 41:830–841.
2018.PubMed/NCBI
|
|
48
|
Chen C, Xue S, Zhang J, Chen W, Gong D and
Zheng J, Ma J, Xue W, Chen Y, Zhai W and Zheng J:
DNA-methylation-mediated repression of miR-766-3p promotes cell
proliferation via targeting SF2 expression in renal cell carcinoma.
Int J Cancer. 141:1867–1878. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li Z, Liu YH, Diao HY, Ma J and Yao YL:
MiR-661 inhibits glioma cell proliferation, migration and invasion
by targeting hTERT. Biochem Biophys Res Commun. 468:870–876. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Reddy SD, Pakala SB, Ohshiro K, Rayala SK
and Kumar R: MicroRNA-661, a c/EBPalpha target, inhibits metastatic
tumor antigen 1 and regulates its functions. Cancer Res.
69:5639–5642. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Peng N, Shi L, Zhang Q, Hu Y, Wang N and
Ye H: Microarray profiling of circular RNAs in human papillary
thyroid carcinoma. PLoS One. 12:e01702872017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jin X, Wang Z, Pang W, Zhou J, Liang Y,
Yang J, Yang L and Zhang Q: Upregulated hsa_circ_0004458
Contributes to Progression of Papillary Thyroid Carcinoma by
Inhibition of miR-885-5p and Activation of RAC1. Med Sci Monit.
24:5488–5500. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hardee S, Prasad ML, Hui P, Dinauer CA and
Morotti RA: Pathologic characteristics, natural history, and
prognostic implications of BRAF(V600E) mutation in pediatric
papillary thyroid carcinoma. Pediatr Dev Pathol. 20:206–212. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Walts AE, Mirocha JM and Bose S:
Follicular variant of papillary thyroid carcinoma (FVPTC):
Histological features, BRAF V600E mutation, and lymph node status.
J Cancer Res Clin Oncol. 141:1749–1756. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kim H, Kim BH, Kim YK, Kim JM, Oh SY, Kim
EH, Lee MJ, Kim JH, Jeon YK, Kim SS, et al: Prevalence of
BRAFV600E mutation in follicular variant of
papillary thyroid carcinoma and non-invasive follicular tumor with
papillary-like nuclear features (NIFTP) in a
BRAFV600E prevalent area. J Korean Med Sci.
33:e752018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jin Y, Jin W, Zheng Z, Chen E, Wang Q,
Wang Y, Wang O and Zhang X: GABRB2 plays an important role in the
lymph node metastasis of papillary thyroid cancer. Biochem Biophys
Res Commun. 492:323–230. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Beltrami CM, Reis MBD, Barrosfilho MC,
Marchi FA, Kuasne H, Pinto CAL, Ambatipudi S, Herceg Z, Kowalski LP
and Rogatto SR: Integrated data analysis reveals potential drivers
and pathways disrupted by DNA methylation in papillary thyroid
carcinomas. Clin Epigenetics. 9:452017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Furlan JC, Bedard YC and Rosen IB:
Significance of tumor capsular invasion in well-differentiated
thyroid carcinomas. Am Surg. 73:484–491. 2007.PubMed/NCBI
|
|
59
|
Bolognamolina R, Gonzálezgonzález R,
Mosquedataylor A, Molinafrechero N, Damiánmatsumura P and
Dominguezmalagón H: Expression of syndecan-1 in papillary carcinoma
of the thyroid with extracapsular invasion. Arch Med Res. 41:33–37.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Meng X, Zhu P, Li N, Hu J, Wang S, Pang S
and Wang J: Expression of BMP-4 in papillary thyroid carcinoma and
its correlation with tumor invasion and progression. Pathol Res
Pract. 213:359–363. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Genpeng L, Jianyong L, Jiaying Y, Ke J,
Zhihui L, Rixiang G, Lihan Z and Jingqiang Z: Independent
predictors and lymph node metastasis characteristics of multifocal
papillary thyroid cancer. Medicine (Baltimore). 97:e96192018.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Erhardt A, Czibere L, Roeske D, Lucae S,
Unschuld PG, Ripke S, Specht M, Kohli MA, Kloiber S, Ising M, et
al: TMEM132D, a new candidate for anxiety phenotypes: Evidence from
human and mouse studies. Mol Psychiatr. 16:647–663. 2011.
View Article : Google Scholar
|
|
63
|
Karapetsas A, Giannakakis A, Dangaj D,
Lanitis E, Kynigopoulos S, Lambropoulou M, Tanyi JL, Galanis A,
Kakolyris S and Trypsianis G: Overexpression of GPC6 and TMEM132D
in early stage ovarian cancer correlates with CD8+ T-lymphocyte
infiltration and increased patient survival. Biomed Res Int.
2015:7124382015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Baek HJ, Kim DW and Ryu JH: Association
between TNM staging system and histopathological features in
patients with papillary thyroid carcinoma. Endocrine. 48:589–594.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chen D, Sun Y, Wei Y, Zhang P, Rezaeian
AH, Teruya-Feldstein J, Gupta S, Liang H, Lin HK, Hung MC and Ma L:
LIFR is a breast cancer metastasis suppressor upstream of the
Hippo-YAP pathway and a prognostic marker. Nat Med. 18:1511–1517.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Josson S, Gururajan M, Hu P, Shao C, Chu
GY, Zhau HE, Liu C, Lao K, Lu CL, Lu YT, et al: miR-409-3p/-5p
promotes tumorigenesis, epithelial-to-mesenchymal transition, and
bone metastasis of human prostate cancer. Clin Cancer Res.
20:4636–4646. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Josson S, Gururajan M, Sung SY, Hu P, Shao
C, Zhau HE, Liu C, Lichterman J, Duan P, Li Q, et al: Stromal
fibroblast-derived miR-409 promotes epithelial-to-mesenchymal
transition and prostate tumorigenesis. Oncogene. 34:2690–2699.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Yu CY and Kuo HC: The emerging roles and
functions of circular RNAs and their generation. J Biomed Sci.
26:292019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|