Introduction
Currently, breast cancer remains a leading health
problem and constitutes one of the most severe burdensome diseases
in females around the world despite understanding of underlying
molecular mechanisms (1). Tumor
metastasis is diagnosed in approximately 30% of breast cancer
patients and is the major cause of cancer-related deaths (2). The prognosis for most patients with
metastatic breast cancer is unfavorable with a median overall
survival range from 2 to 3 years (3). Generally, breast cancer can be
categorized into four subtypes [luminal A, luminal B, human
epidermal growth factor receptor 2 (HER2) positive and triple
negative], which are defined using immunohistochemical breast tumor
markers (4). These four subtypes
have the potential risk of distant metastasis but with differential
site-specific metastatic patterns (5). Currently, breast cancer metastasis
from primary tumor to distant organs occurs through a sequential
molecular cascade including local angiogenesis for tumor growth,
invasion of the surrounding tissue, intravasation of the carcinoma
cells into the blood or lymphatic vessels, dissemination and
proliferation at secondary neoplastic foci (6). These carcinoma cells obtain
mesenchymal features and suppress their epithelial features through
the epithelial-to-mesenchymal transition (EMT) process to promote
an invasive and metastatic phenotype (7). Multiple transcription factors
coordinate EMT programs. Among them, zinc-finger E-box-binding
(ZEB) transcription factors, ZEB1 and ZEB2, are two EMT regulators
that either repress or activate transcription in various types of
cancer (8). Furthermore, ZEB2 was
reported to negatively correlate with the epithelial marker
E-cadherin in breast cancer cells involved in breast cancer
progression (9).
Long non-coding RNAs (lncRNAs) are a class of
transcripts containing more than 200 nucleotides in length with
limited protein-coding capacity (10). Recent findings have shown that
dysregulation of lncRNAs is involved in cell proliferation, tumor
progression and metastasis in cancers (11). Functionally, lncRNAs interact with
proteins and other RNAs to regulate their activities and cellular
location. Furthermore, lncRNAs act as molecular sponges for miRNAs
that block the binding activity for target transcripts (12). In breast cancer, several lncRNAs
have been identified as either oncogenic or tumor suppressive
factors, such as X-inactive-specific-transcript (XIST), HOX
antisense intergenic RNA (HOTAIR), growth arrest specific 5 (GAS5)
and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1)
(13–16). A systematic analysis of the
correlation has been carried out between these dysregulated lncRNAs
and breast cancer clinicopathology and survival suggesting a
pivotal role in cancer development (17). Increasing lncRNAs have been shown to
participate in specific cancer types but more often exert general
function in a broad spectrum of cancer.
OPA-interacting protein 5 antisense transcript 1
(OIP5-AS1) is an evolutionarily conserved long non-coding RNA that
is transcribed from opposite direction to the OIP5 gene. It
was first shown to be expressed in the nervous system and was
essential for neurogenesis during embryonic development (18). The functions of OIP5-AS1 in multiple
human cancers have been reported to be associated with oncogenesis
(19,20). In breast cancer, OIP5-AS1 levels are
upregulated in breast tumor tissue and correlated with tumor size,
metastatic status of lymph nodes, pathological grading and TNM
stage (21).
In the present study, we investigated the role of
OIP5-AS1 in breast cancer metastasis using the in vitro and
in vivo models showing that OIP5-AS1 regulates ZEB2
expression by acting as ceRNA for miR-340-5p.
Materials and methods
Cell culture and transfection
Breast cancer cell lines MCF-7, MDA-MB-231, ZR-75,
MDA-MB-468, SKBR3 and normal human epithelial cell line MCF-10A
were purchased from the American Type Culture Collection (ATCC).
MCF-10A cells were cultured in MEBM (Lonza) and supplemented with
100 ng/ml cholera toxin. ZR-75 and SKBR3 cell lines were maintained
in RPMI-1640 medium (Sigma) containing 10% FBS, 2 mM L-glutamine
and 2% penicillin and streptomycin. MCF-7, MDA-MB-231, and
MDA-MB-468 cell lines were maintained in DMEM supplemented with 10%
FBS plus 2% penicillin and streptomycin. Cells were cultured in a
humidified incubator at 37°C with 5% CO2.
The pre-designed siOIP5-AS1 and siZEB2 were
purchased from ThermoFisher (no. 4390771, no. AM16708) and
transfected into cells using Lipofectamine RNAiMAX reagent
(ThermoFisher) following the manufacturer's instructions.
miR-340-5p mimics, negative control mimics, miR-340-5p inhibitors
and negative control inhibitors were purchased from GenePharma and
transfected into cells using Lipofectamine 2000 reagent
(ThermoFisher), according to the manufacturers' instructions.
miR-340-5p mimic: 5′-UUAUAAAGCAAUGAGACUGAUU-3′ and miR-340-5p
inhibitor: 5′-AAUCAGUCUCAUUGCUUUAUAA-3′. Cells were used for
further experiments at 48 h after transfection.
Wound healing assay
MCF-7 and MDA-MB-231 cells were seeded in 12-well
plates and transfected with either siNC or siOIP5-AS1. A linear
wound was scratched across the center of the well using a sterile
pipette tip. The images of wound closure were captured after 24 h
using Olympus microscope (×10).
Transwell invasion assay
The invasion of MCF-7 and MDA-MB-231 cells was
detected using matrigel-coated or non-coated chambers with a pore
size of 0.8 µM. The transfected cells were seeded into the upper
chamber in DMEM with 1% FBS and the lower chamber was filled with
10% FBS as a chemoattractant. After 24-h incubation in the
humidified incubator at 37°C with 5% CO2, cells in the
upper chamber were removed and cells in the lower side were fixed
with 4% PFA and stained by 1% crystal violet. Stained cells were
then visualized and imaged at a ×20 magnification by a light
microscope.
Western blot analysis
The cellular proteins were extracted in RIPA buffer
supplemented with protease inhibitor. The protein concentration was
quantified using BCA protein assay and 20 µg of each protein sample
was loaded and analyzed by 10% sodium dodecyl sulfate
(SDS)-polyacrylamide gel electrophoresis (PAGE) system. Then,
proteins were transferred to a PVDF membrane. The membrane was
blocked in 5% BSA and probed with primary antibodies: Anti-ZEB2
(1:1,000; no. ab138222, Abcam), anti-E-cadherin (1:1,000; no.
14472, Cell signaling), anti-N-cadherin (1:1,000; no. ab18203,
Abcam), anti-vimentin (1:1,000; no. ab92547, Abcam), anti-ZEB1
(1:1,000; no. 70512, Cell signaling), anti-Snail (1:1,000; no.
IMG-6639A, Novus Biologicals), anti-Slug (1:1,000; no. 9585, Cell
signaling), anti-Twist (1:1,000; no. 69366, Cell signaling) and
anti-GAPDH (1:2,000; no. ab8245, Abcam). Then, the membrane was
incubated with peroxidase-conjugated anti-mouse or anti-rabbit
secondary antibody (1:2,000, nos. NEF822001EA; NEF812001EA,
PerkinElmer). Immunoreactivity bands were detected by
chemiluminescence and the intensity of the bands was quantified
using Image Lab Software (Bio Rad, China).
RNA fluorescence in situ hybridization
(FISH)
A Cy3-labeled set of probes recognizing OIP5-AS1 was
designed and synthesized by Biosearch Technologies. The MCF-7 cells
were cultured on coverslips for 24 h and then fixed in 4% PFA.
After permeabilization with 70% ethanol at 4°C for 1 h, cells were
hybridized with the OIP5-AS1 probes dissolved in hybridization
buffer (no. SMF-HB1-10, Biosearch Technologies) at 37°C in the dark
for 16 h. The nucleus was stained with DAPI. Images were captured
using a confocal microscope (Olympus).
Quantitative real-time PCR
(RT-PCR)
Total RNA was extracted from cells (MCF-7,
MDA-MB-231, ZR-75, MDA-MB-468, SKBR3, and MCF-10A) using
TRIzol® reagent (Thermo Fisher). Total RNA (1 µg) was
reverse transcribed into cDNA and a SYBR-Green quantitative
real-time PCR Master Mix kit was used to detect qPCR signals. The
targeted gene expression was normalized with GAPDH and
calculated using 2−ΔΔCq method (22). The primer sequences used were:
OIP5-AS1: 5′-TGCAACCCAAGGTGGATACT-3′ and
5′-GAGAGACTGCAGTGAGCAGA-3′; ZEB2: 5′-CAGCTCTTCCACCTCAAAGC-3′ and
5′-TCCTTGTTTCCGCTGGTACT-3′; GAPDH: 5′-GTCGGAGTCAACGGATTTGG-3′ and
5′-TGACGGTGCCATGGAATTTG-3′. For the detection of miR-340-5p,
stem-loop qRT-PCR was performed using miScript SYBR-Green PCR Kit
with U6 small nuclear RNA as an internal control (Qiagen). The
following thermocycling conditions were used in the experiments:
PCR initial activation at 95°C for 15 min, followed by 40 cycles of
denaturation at 94°C for 15 sec, annealing at 55°C for 30 sec and
an extension at 70°C for 30 sec.
RNA immunoprecipitation assay
Magna RIP kit (Millipore) was used for RNA
immunoprecipitation experiments. The procedure was performed
following the manufacturer's protocol. Briefly, after miR-340-5p
mimics or NC mimics transfection, the cells were lysed in RIP lysis
buffer. The cell lysate was incubated with either Ago2 antibody or
control IgG together with protein A/G magnetic beads. Then the
beads were washed and incubated with Proteinase K at 55°C for 30
min to digest proteins. The purified RNA was obtained and analyzed
by RT-qPCR.
Dual-luciferase reporter assay
In this study, the OIP5-AS1/miRNA interactions were
predicted using Starbase (http://starbase.sysu.edu.cn/) and DIANA-LncBase
database (http://www.microrna.gr/LncBase). For OIP5-AS1 and
miR-340-5p binding activity, OIP5-AS1 fragment containing the
binding sites of miR-340-5p, as well as those of the wild-type and
mutant sequences were cloned into a pmirGLO Dual-luciferase Vector
designated as OIP5-AS1 WT or OIP5-AS1 MUT. For ZEB2 and miR-340-5p
binding activity, fragment of 3′UTR ZEB2 containing the binding
sites of miR-340-5p, as well as the wild-type and mutant sequences
were cloned into a pmirGLO Dual-luciferase Vector designated as
ZEB2 WT or ZEB2 MUT. These vectors were co-transfected with either
NC mimics or miR-340-5p mimics using Lipofectamine 2000 reagent. At
48 h after transfection, the relative luciferase activities were
recorded by dual- luciferase reporter assay system (Promega) and
the values were normalized to the Renilla luciferase
activity.
Immunohistochemistry staining and
hematoxylin and eosin (H&E) staining
The lung of nude mice was dissected and fixed in 10%
formaldehyde at room temperature overnight. The embedded samples in
paraffin were sectioned into 5 µm slices and mounted on glass
slides. For immunohistochemical staining, the slides of interest
were probed with anti-Ki-67 antibodies (1:500; Abcam) and then the
secondary streptavidin-horseradish peroxidase-conjugated antibody
staining. Immunoreactivity was visualized by DAB and lightly
counterstained with 5% hematoxylin. For H&E staining, slides
were deparaffinized and rehydrated in graded ethanol solutions,
then in distilled water. After H&E staining, slides were
mounted and examined under a light microscope.
Lentivirus production and in vivo
metastasis assay
Full-length cDNA of human OIP5-AS1 was amplified
from the mRNA of MCF-7 cells and subcloned into pcDNA3.1 (AddGene).
The lentiviral and packaging vectors (AddGene) were co-transfected
into HEK293FT cells using Lipofectamine 2000 reagent (ThermoFisher)
according to the manufacturer's instructions. Virus was collected
and concentrated at 48 h after transfection.
Twenty healthy 6- to 8-week-old female BALB/c nude
mice (The Animal Institute, Jilin University) were used in this
study and randomly divided into two groups. The mice were housed in
a specific pathogen-free (SPF) facility and exposed to a 12-h
light/dark cycle. Water and food were offered ad libitum.
After 1 week of acclimatization, MCF-7 cells (1×106)
infected with LV-OIP5-AS1 or LV-NC were intravenously injected
through the tail vein of BALB/c nude mice under isoflurane
anaesthesia. After 8 weeks of inoculation, the mice were euthanized
and the number of lung metastatic tumors per lung were counted
under a dissecting microscope and confirmed by H&E staining.
The experimental protocols were approved by the Animal Care
Committee of China-Japan Union Hospital Affiliated to Jilin
University.
Statistical analysis
Data are expressed as mean ± standard error mean
(SEM). The Student's t test was employed to compare two groups and
one-way ANOVA with post hoc test was used to analyze differences
among multiple groups. A value of P<0.05 was considered as
statistically significant.
Results
Interference of OIP5-AS1 represses
epithelial-to-mesenchymal transition (EMT) in breast cancer cells
by regulating ZEB family proteins
The dysregulation of long non-coding RNA OIP5-AS1
was involved in multiple cancer types associating with overall
survival, TNM stage and prognosis (21,23–25).
In breast cancer, studies reported that OIP5-AS1 is upregulated in
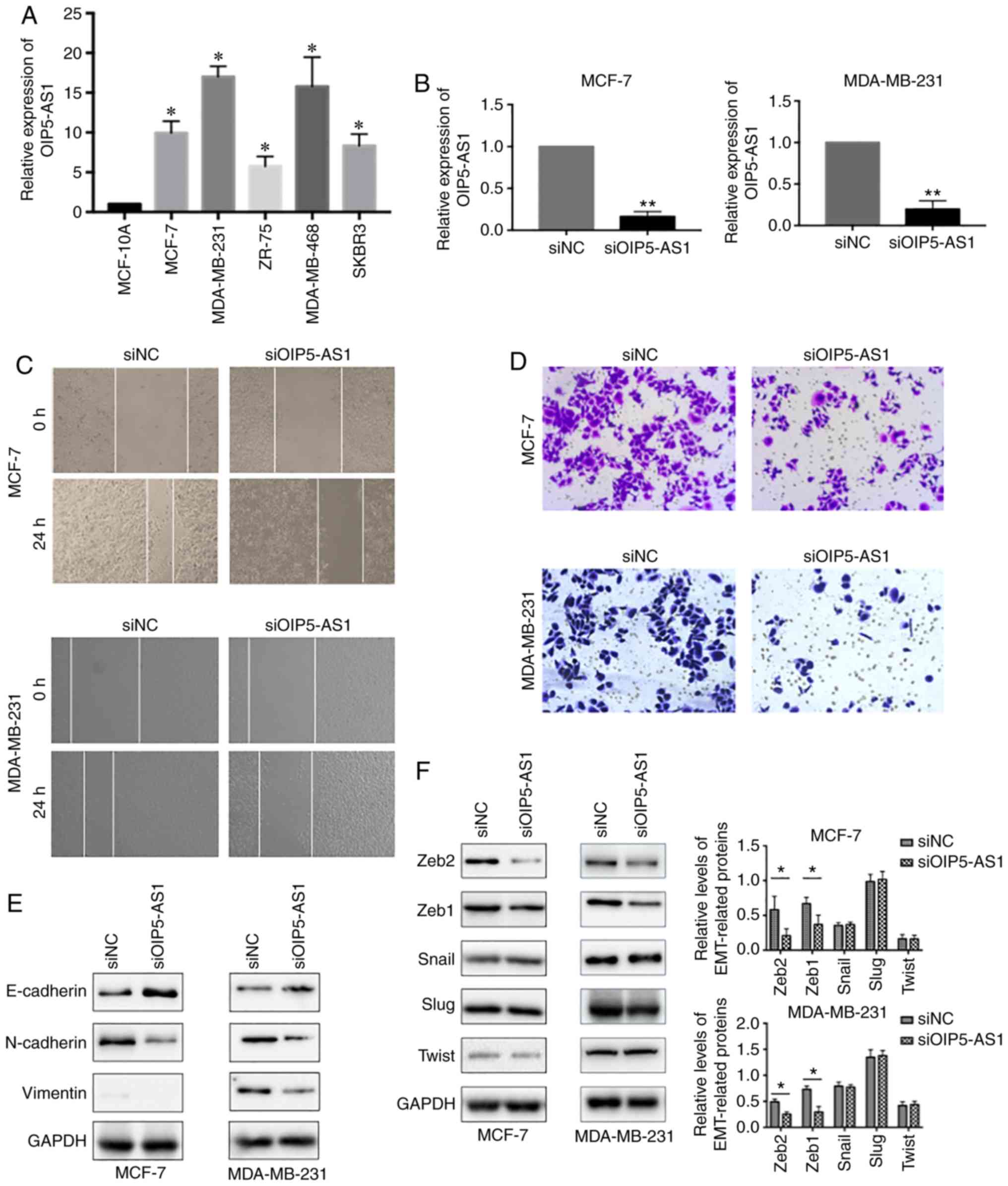
both tumor samples and cell lines (21). We first evaluated the expression
levels of OIP5-AS1 in five breast cancer cell lines. The results
showed that the relative expression levels of OIP5-AS1 were much
higher in the five breast cancer cell lines than in the normal
epithelial cell line MCF-10A (Fig.
1A). Then, we chose luminal-type breast cancer cell line MCF-7
and basal B TNBC cell line MDA-MB-231 for further functional
studies. To investigate the role of OIP5-AS1 in breast cancer
metastasis, we efficiently knocked down OIP5-AS1 with siRNAs in
MCF-7 and MDA-MB-231 cell lines (Fig.
1B) and analyzed the cell migration and invasion properties. In
the wound healing assay, siOIP5-AS1 groups showed a slower
migration rate than the siNC group (Fig. 1C). Furthermore, knockdown of
OIP5-AS1 in the two cell lines significantly inhibited cell
invasion (Fig. 1D). Next, we
assessed the effects of the downregulation of OIP5-AS1 on the
expression of epithelial-to-mesenchymal transition (EMT) markers.
The protein analysis results indicated that the epithelial marker
E-cadherin was increased whereas the mesenchymal markers N-cadherin
and Vimentin were decreased (Fig.
1E). These results suggested that downregulation of OIP5-AS1
repressed epithelial-to-mesenchymal transition (EMT). Considering
the importance of transcription factors in EMT co-ordination, we
further tested the expression of EMT-related transcription factors
(ZEB1, ZEB2, Snail, Slug and Twist). In the siOIP5-AS1 group, ZEB1
and ZEB2 were significantly downregulated whereas the expression of
Snail, Slug and Twist were not affected (Fig. 1F). Thus, we speculated that OIP5-AS1
may exert functions in EMT through regulating ZEB family
proteins.
OIP5-AS1 directly targeted
miR-340-5p
LncRNAs exert function in various aspects of
cellular function and biological process in either nucleus or
cytoplasm. In nucleus, lncRNAs may take part in chromatin
remodeling and modification or gene expression prior to
transcription, whereas lncRNAs in cytoplasm mainly participate in
post-transcriptional regulation and post-translational modification
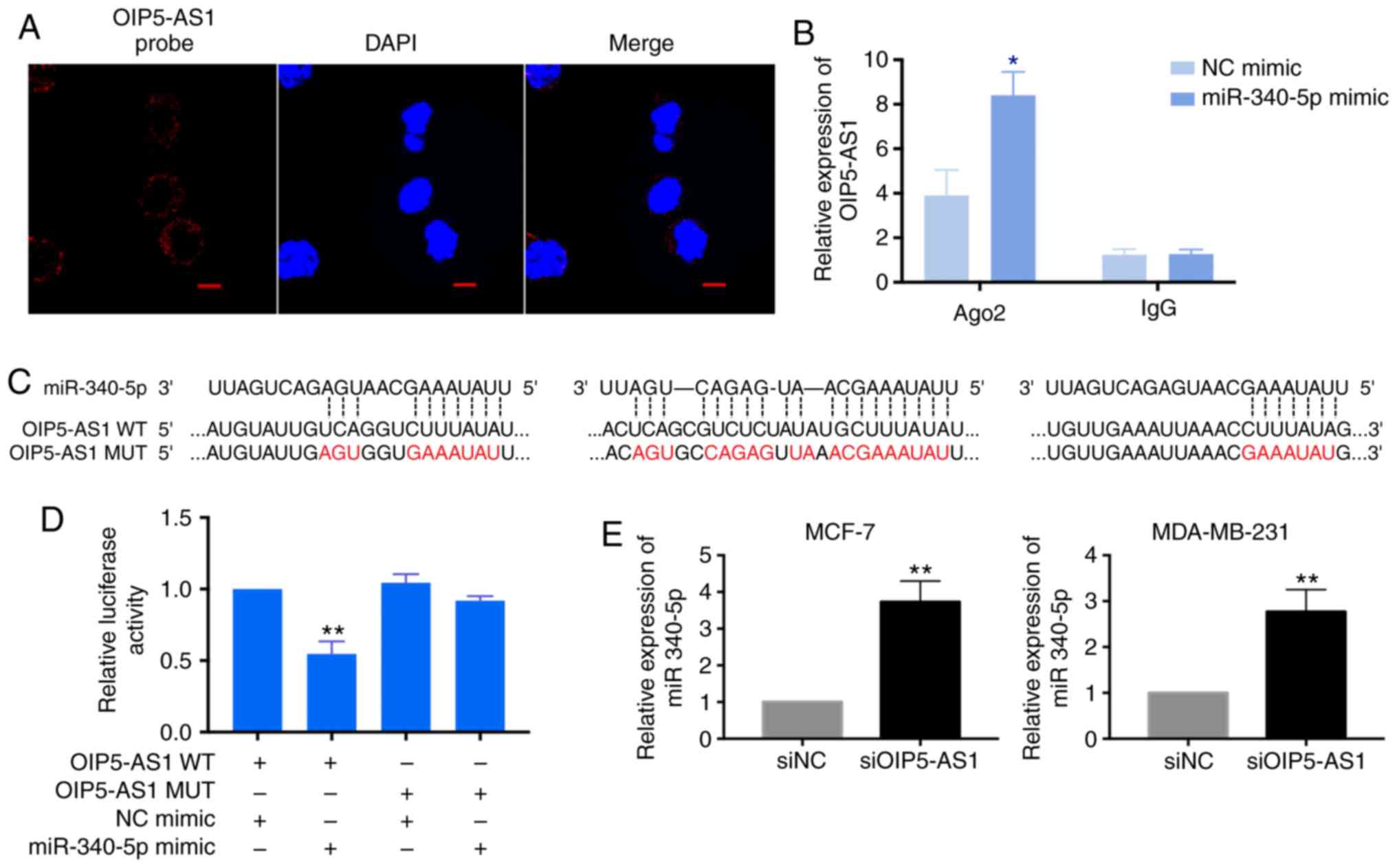
(26,27). Thus, we assessed subcellular
location of OIP5-AS1 by fluorescence in situ hybridization
(FISH). The detected OIP5-AS1 was mainly localized in the cytoplasm
in MCF-7 cells which indicated OIP5-AS1 may serve as a ceRNA in
breast cancer cells (Fig. 2A).
In this study, the OIP5-AS1/miRNA interactions were
predicted using Starbase and DIANA tools. Among the predicted
miRNAs, miR-340-5p possesses three target sites on OIP5-AS1. In
order to confirm that miR-340-5p is the target gene of OIP5-AS1, we
performed anti-Ago2 RIP assay and dual luciferase reporter assay.
In anti-Ago2 RIP assay, the endogenous OIP5-AS1 was specifically
enriched in miR-340-5p mimics-transfected cells when compared with
NC mimics group (Fig. 2B). We
constructed the OIP5-AS1 wild-type and mutant reporter plasmids
according to the binding sequences of miR-340-5p (Fig. 2C). The dual luciferase reporter
assay showed that the reduced luciferase activity was only found in
the miR-340-5p mimics and OIP5-AS1 wild-type co-transfection groups
but not in the OIP5-AS1 mutant co-transfection group (Fig. 2D). Moreover, we tested the
expression of miR-340-5p with OIP5-AS1 knockdown in breast cancer
cells. After two days with siOIP5-AS1 transfection, the level of
miR-340-5p was increased in MCF-7 and MDA-MB-231 cells (Fig. 2E). Taken together, these results
confirmed the direct binding activity between OIP5-AS1 and
miR-340-5p in breast cancer cells.
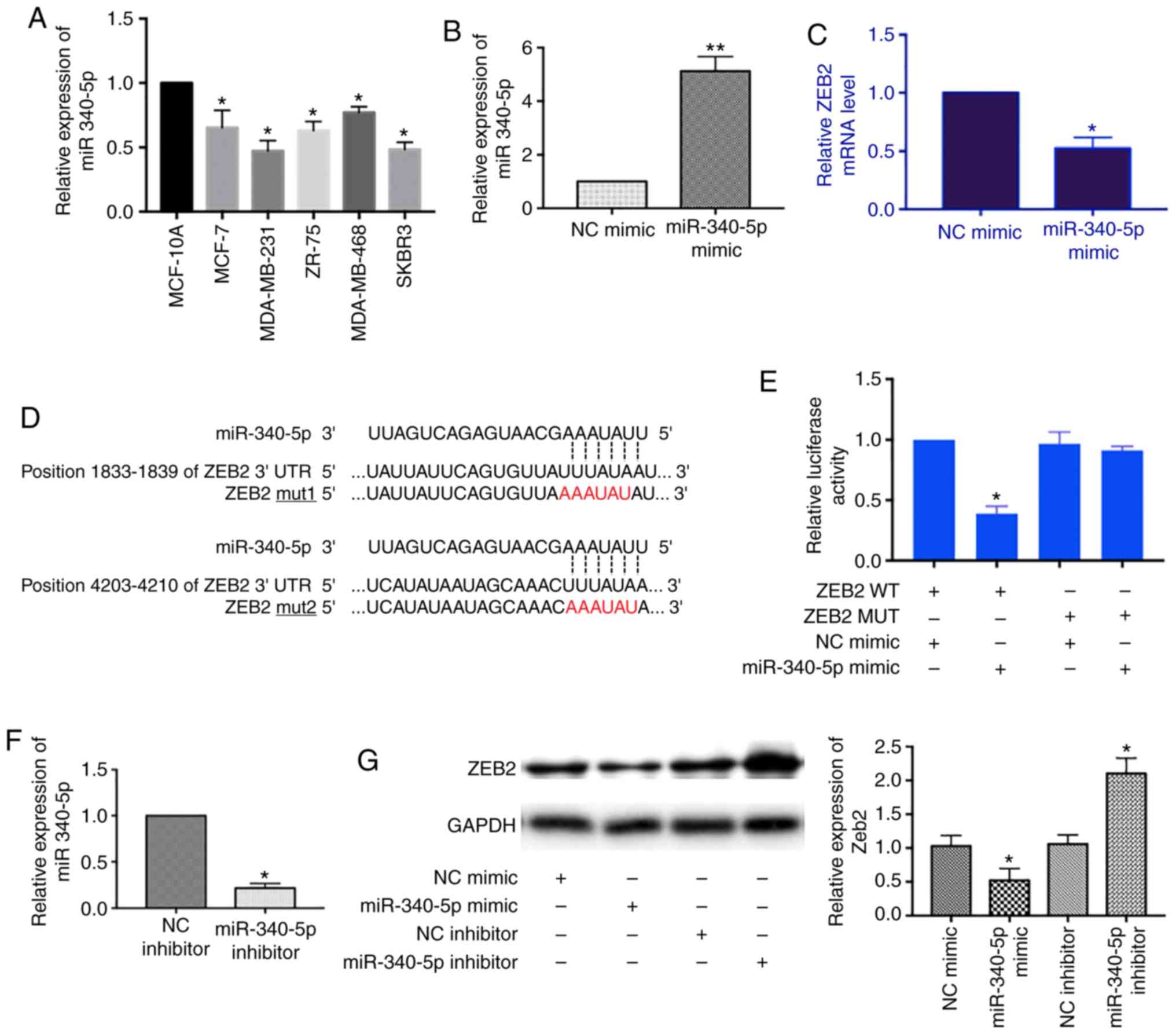
miR-340-5p is downregulated in breast cancer cells
and regulates ZEB2 expression. A recent study reported that
miR-340-5p was negatively associated with distant metastasis in
invasive breast cancers (28).
Thus, we measured the relative expression of miR-340-5p in breast
cancer cell lines. The level of miR-340-5p was decreased in MCF-7,
MDA-MB-231, ZR-75, MDA-MB-468 and SKBR3 cells as compared to human
breast epithelial cell line MCF10A (Fig. 3A).
Next, we screened mRNA targets of miR-340-5p using
TargetScan and Starbase tools and found the 3′UTR of ZEB2 mRNA
contains two binding sites for miR-340-5p. We transfected
miR-340-5p mimics into MCF-7 cells and detected the expression of
ZEB2 mRNA by RT-qPCR. With the miR-340-5p overexpression (Fig. 3B), the level of ZEB2 mRNA was
decreased (Fig. 3C). We further
confirmed the direct binding between ZEB2 mRNA and miR-340-5p by
dual luciferase reporter assay. The ZEB2 3′UTR was constructed and
the mutant form was designed according to the miR-340-5p binding
sequences (Fig. 3D). As shown in
Fig. 3E, the luciferase activity
was only reduced in the ZEB2 3′UTR wild-type and miR-340-5p mimics
co-transfection group which suggested the direct binding between
ZEB2 mRNA and miR-340-5p. In addition, we examined the effects of
miR-340-5p on the protein expression of ZEB2 by overexpression of
either miR-340-5p mimics or inhibitors in MCF-7 cells. Similarly,
the level of ZEB2 was decreased with miR-340-5p mimics transfection
whereas it was increased with miR-340-5p inhibitors transfection
(Fig. 3F and G). Collectively,
these results supported that miR-340-5p regulates ZEB2 expression
by binding to complementary sequences in the 3′UTR of ZEB2
mRNA.
OIP5-AS1 regulates ZEB2 indirectly
through sponging miR-340-5p
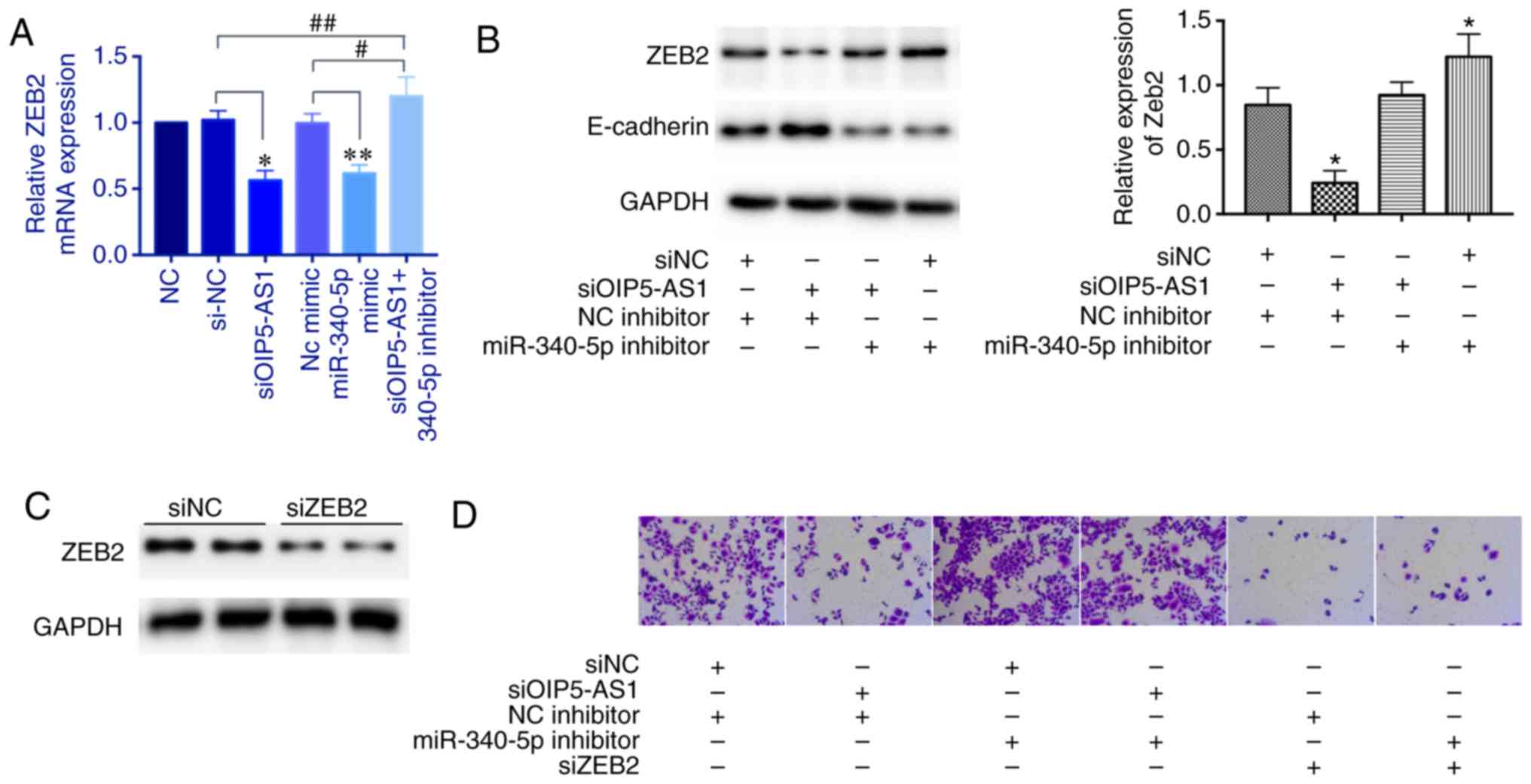
We next explored whether OIP5-AS1 regulates ZEB2
expression through sponging miR-340-5p. The ZEB2 mRNA expression
was decreased with either knockdown of OIP5-AS1 or overexpression
of miR-340-5p mimics; however, this effect was reversed by
miR-340-5p inhibitors (Fig. 4A).
Then, we tested the protein level of ZEB2. The miR-340-5p
inhibitors also reversed the repressed effect of OIP5-AS1 knockdown
and miR-340-5p inhibitors alone upregulated ZEB2 expression. ZEB2
is a known transcriptional repressor of E-cadherin. In this
experiment, we found that the protein level of E-cadherin was
inversely correlated with the ZEB2 level (Fig. 4B). Moreover, we examined the cell
invasion ability. The siOIP5-AS1 group showed a decreased number of
invasive cells which was reversed by miR-340-5p inhibitors.
miR-340-5p inhibitors alone enhanced invasive ability. However,
knockdown of ZEB2 markedly repressed cell invasion even with
miR-340-5p inhibitors, suggesting that ZEB2 is a downstream factor
(Fig. 4C and D). Overall, these
results demonstrated that OIP5-AS1 regulates ZEB2 indirectly
through sponging miR-340-5p.
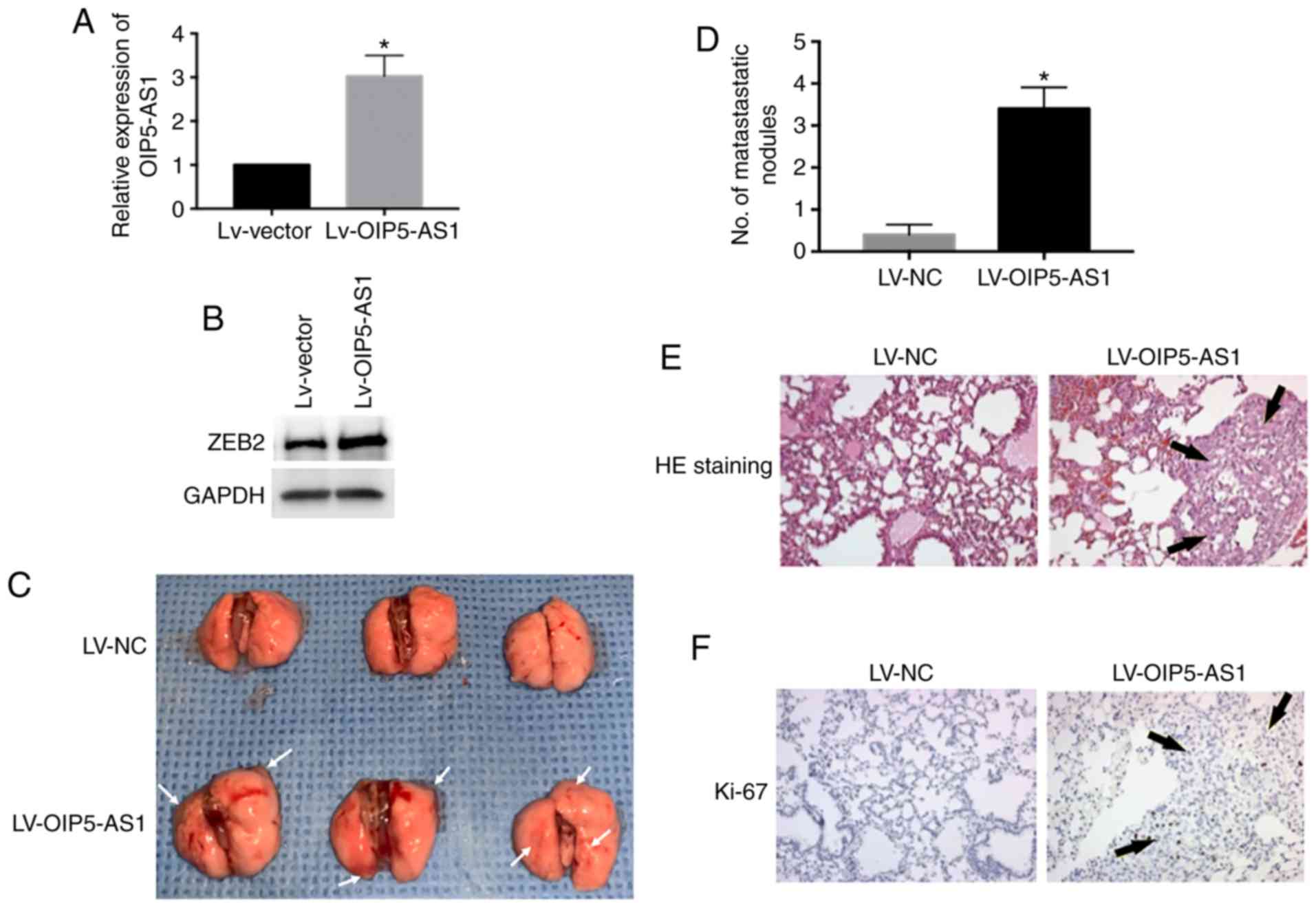
OIP5-AS1 promotes breast cancer cells
into lung metastasis in vivo
To determine whether OIP5-AS1 causes breast cancer
cell metastasis in vivo, the metastasis assay was conducted
and the primary pulmonary metastasis was observed. We overexpressed
OIP5-AS1 by lentivirus infection in MCF-7 cells and then injected
cells into nude mice via tail vein (Fig. 5A). The protein level of ZEB2 was
elevated by OIP5-AS1 overexpression (Fig. 5B). The LV-OIP5-AS1 group showed
marked lung colonization and increased metastatic lung nodules
compared with the LV-NC group (Fig. 5C
and D). We performed H&E staining of the metastatic lung
tissue in the LV-OIP5-AS1 group. The results were consistent with
our observation showing increased metastatic lung nodules (Fig. 5E). Furthermore, the metastatic
tumors were positively stained with Ki-67 the marker of cell
proliferation (Fig. 5F). In
conclusion, these results demonstrate that overexpression of
OIP5-AS1 promotes breast cancer cells into lung metastasis in
vivo.
Discussion
In the present study, we investigated the role of
long non-coding RNA OIP5-AS1 in breast cancer metastasis. We found
that OIP5-AS1 was upregulated in five breast cancer cell lines
which was consistent with earlier studies and in agreement with
supporting evidence from genome-wide analysis of human cancers
indicating the prevalent upregulation of OIP5-AS1 (21,29).
In vivo experiments also confirmed the effects of OIP5-AS1
in breast cancer cells on lung metastasis. Furthermore, knockdown
of OIP5-AS1 markedly weakened cell migration and invasion abilities
and inhibited epithelial-to-mesenchymal transition (EMT). These
results suggest the pivotal role of OIP5-AS1 in breast cancer
metastasis and indicate its potential to be a marker for metastatic
breast cancer or for therapeutic evaluation. Moreover, we provided
evidence that ZEB2 is an important effector of OIP5-AS1
dysregulation and this association was evident through the
regulation of miR-340-5p.
Emerging evidence reveals the role of long
non-coding RNAs (LncRNAs) in tumorigenesis and tumor metastasis as
the regulator for key gene expression at either transcriptional or
translational levels (30). Studies
interfered metastasis-associated lncRNAs, such as MALAT1, NEAT1 and
BCAR4, showed significant metastasis inhibition (14,31,32).
OIP5-AS1 is a newly identified lncRNA, the dysregulation of which
has been found in multiple cancer types including breast cancer
(33). It is involved in cancer
cell proliferation showing a G2/M to G0/G1-phase arrest. Silencing
of OIP5-AS1 has been shown to inhibit cell proliferation in
multiple cancers (20,21,23,24).
In addition, downregulation of OIP5-AS1 has been shown to regulate
EMT markers E-cadherin and to reduce metastasis in lung
adenocarcinoma (23). Similar
results were also obtained in hepatoblastoma demonstrating the
involvement of OIP5-AS1 in EMT progress (34). Together with our findings, the
functions of OIP5-AS1 in cancer metastasis have been verified in
multiple cancer types. Thus, further investigations are needed to
validate the network of OIP5-AS1 with clinical stages in related
cancer types. In our study, we only examined the function of
OIP5-AS1 in the regulation of EMT-related proteins in MCF-7 and
MDA-MB-231 cells. These two breast cancer cell lines represent
different molecular subtypes of breast cancer which show different
metastasis capabilities. Although the regulation of EMT-related
proteins was confirmed in these two cell lines, more experiments
should be performed in multiple subtypes of breast cancer cell
lines due to the different metastatic ability and diversity in the
molecular interactions involved even in the same cancer type.
Moreover, the general upregulation of OIP5-AS1 has been revealed in
different cell lines, but the varying expression values that
correlate to metastatic ability is not clear.
OIP5-AS1 was probed using FISH assay was
predominantly in the cytoplasm which indicates the potential role
of being ceRNAs. Findings have shown that lncRNAs act as ceRNAs
which compete for miRNAs to regulate the expression of target genes
(12). In the present study, we
tested miR-340-5p according to the predicted binding sequences from
TargetScan and Starbase tools. The results of RIP assay and dual
luciferase reporter assay demonstrated the direct binding of
OIP5-AS1 and miR-340-5p. Interestingly, it has been reported that
miR-340-5p is negatively associated with distant metastasis in
invasive breast cancers, which suggests the pivotal role of
miR-340-5p in metastasis (28). Our
results elucidate the ability of miR-340-5p to target ZEB2 which is
a new finding confirmed by regulation at both the mRNA and protein
levels. Long non-coding RNAs have the potential binding ability
with multiple miRNAs through complementary sequences. Several
miRNAs were reported to target OIP5-AS1 in the literature, such as
miR-129-5p, miR-448, miR-378a-3p and miR-498 (21,23,35,36).
Notably, in our results, the siOIP5-AS1-suppressed EMT process in
breast cancer cells was markedly blocked by miR-340-5p inhibitors,
suggesting a specific inhibitory role of miR-340-5p for OIP5-AS1 in
metastasis process. However, the experiments validating the
OIP5-AS1/miR-129-5p/ZEB2 axis was only performed in MCF-7 cells
which is a potential limitation of this study. Therefore, this
molecular mechanism needs to be confirmed in other breast cancer
cell lines. Additionally, the functions of long non-coding RNA as
miRNA sponge allow us to consider its regulatory networks in tumor
biology. More genome-wide analysis and follow-up functional studies
on OIP5-AS1 should carried out to understand its diverse role in
different types of cancer.
In conclusion, we identified the OIP5-AS1/miR-340-
5p/ZEB2 axis in breast cancer cell metastasis. OIP5-AS1 facilitated
breast cancer metastasis by sponging miR-340-5p to upregulate ZEB2
mRNA transcripts. The current results provide a new direction for
the further investigation of molecular mechanism of breast cancer
metastasis. Defining the underlying mechanisms of differentially
expressed lncRNA in cancers may be useful in developing novel
strategies for cancer diagnosis and treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
LM and HL conceived and designed the study. LM, XY
and DZ performed the experiments. LM and HL wrote the manuscript.
All authors read and approved the final manuscript and agreed to be
accountable for all aspects of the work in ensuring that questions
related to the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
Animal experiments conducted in the present study
were approved by Animal Care Committee of China-Japan Union
Hospital Affiliated to Jilin University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li N, Deng Y, Zhou L, Tian T, Yang S, Wu
Y, Zheng Y, Zhai Z, Hao Q, Song D, et al: Global burden of breast
cancer and attributable risk factors in 195 countries and
territories, from 1990 to 2017: Results from the global burden of
disease study 2017. J Hematol Oncol. 12:1402019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis CE, Ma J, Sauer AG, Newman LA and
Jemal A: Breast cancer statistics, 2017, racial disparity in
mortality by state. CA Cancer J Clin. 67:439–448. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cardoso F, Costa A, Senkus E, Aapro M,
André F, Barrios CH, Bergh J, Bhattacharyya G, Biganzoli L, Cardoso
MJ, et al: 3rd ESO-ESMO international consensus guidelines for
advanced breast cancer (ABC 3). Ann Oncol. 28:16–33. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Malhotra GK, Zhao X, Band H and Band V:
Histological, molecular and functional subtypes of breast cancers.
Cancer Biol Ther. 10:955–960. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xiao W, Zheng S, Yang A, Zhang X, Zou Y,
Tang H and Xie X: Breast cancer subtypes and the risk of distant
metastasis at initial diagnosis: A population-based study. Cancer
Manag Res. 10:5329–5338. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kozłowski J, Kozłowska A and Kocki J:
Breast cancer metastasis-insight into selected molecular mechanisms
of the phenomenon. Postepy Hig Med Dosw (Online). 69:447–451. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chaffer CL, Juan BPS, Lim E and Weinberg
RA: EMT, cell plasticity and metastasis. Cancer Metastasis Rev.
35:645–654. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: An alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee JY, Park MK, Park JH, Lee HJ, Shin DH,
Kang Y, Lee CH and Kong G: Loss of the polycomb protein Mel-18
enhances the epithelial-mesenchymal transition by ZEB1 and ZEB2
expression through the downregulation of miR-205 in breast cancer.
Oncogene. 33:1325–1335. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Evans JR, Feng FY and Chinnaiyan AM: The
bright side of dark matter: lncRNAs in cancer. J Clin Invest.
126:2775–2782. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calle AS, Kawamura Y, Yamamoto Y,
Takeshita F and Ochiya T: Emerging roles of long non-coding RNA in
cancer. Cancer Sci. 109:2093–2100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Marchese FP, Raimondi I and Huarte M: The
multidimensional mechanisms of long noncoding RNA function. Genome
Biol. 18:2062017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mendell JT: Targeting a long noncoding RNA
in breast cancer. N Engl J Med. 374:2287–2289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li W, Zhai L, Wang H, Liu C, Zhang J, Chen
W and Wei Q: Downregulation of LncRNA GAS5 causes trastuzumab
resistance in breast cancer. Oncotarget. 7:27778–27786. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang YS, Chang CC, Lee SS, Jou YS and
Shih HM: Xist reduction in breast cancer upregulates AKT
phosphorylation via HDAC3-mediated repression of PHLPP1 expression.
Oncotarget. 7:43256–43266. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tian T, Wang M, Lin S, Guo Y, Dai Z, Liu
K, Yang P, Dai C, Zhu Y, Zheng Y, et al: The impact of lncRNA
dysregulation on clinicopathology and survival of breast cancer: A
systematic review and meta-analysis. Mol Ther Nucleic Acids.
12:359–369. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ulitsky I, Shkumatava A, Jan CH, Sive H
and Bartel DP: Conserved function of lincRNAs in vertebrate
embryonic development despite rapid sequence evolution. Cell.
147:1537–1550. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Meseure D, Alsibai KD, Nicolas A, Bieche I
and Morillon A: Long noncoding RNAs as new architects in cancer
epigenetics, prognostic biomarkers, and potential therapeutic
targets. BioMed Res Int. 2015:3202142015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Naemura M, Kuroki M, Tsunoda T, Arikawa N,
Sawata Y, Shirasawa S and Kotake Y: The long noncoding RNA OIP5-AS1
is involved in the regulation of cell proliferation. Anticancer
Res. 38:77–81. 2018.PubMed/NCBI
|
|
21
|
Zeng H, Wang J, Chen T, Zhang K, Chen J,
Wang L, Li H, Tuluhong D, Li J and Wang S: Downregulation of long
non-coding RNA Opa interacting protein 5-antisense RNA 1 inhibits
breast cancer progression by targeting sex-determining region Y-box
2 by microRNA-129-5p upregulation. Cancer Sci. 110:289–302.
2019.PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Deng J, Deng H, Liu C, Liang Y and Wang S:
Long non-coding RNA OIP5-AS1 functions as an oncogene in lung
adenocarcinoma through targeting miR-448/Bcl-2. Biomed
Pharmacother. 98:102–110. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Y, Shi F, Xia Y and Zhao H: LncRNA
OIP5-AS1 predicts poor prognosis and regulates cell proliferation
and apoptosis in bladder cancer. J Cell Biochem. Nov 18–2018.(Epub
ahead of print).
|
|
25
|
Ren X, He J, Qi L, Li S, Zhang C, Duan Z,
Wang W, Tu C and Li Z: Prognostic and clinicopathologic
significance of long non-coding RNA opa-interacting protein
5-antisense RNA 1 in multiple human cancers. Artif Cells Nanomed
Biotechnol. 48:353–361. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen LL: Linking long noncoding RNA
localization and function. Trends Biochem Sci. 41:761–772. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rohan TE, Wang T, Weinmann S, Wang Y, Lin
J, Ginsberg M and Loudig O: A miRNA expression signature in breast
tumor tissue is associated with risk of distant metastasis. Cancer
Res. 79:1705–1713. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Arunkumar G, Anand S, Raksha P,
Dhamodharan S, Rao HPS, Subbiah S, Murugan AK and Munirajan AK:
LncRNA OIP5-AS1 is overexpressed in undifferentiated oral tumors
and integrated analysis identifies as a downstream effector of
stemness-associated transcription factors. Sci Rep. 8:70182018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang MC, Ni JJ, Cui WY, Wang BY and Zhuo
W: Emerging roles of lncRNA in cancer and therapeutic
opportunities. Am J Cancer Res. 9:1354–1366. 2019.PubMed/NCBI
|
|
31
|
Arun G, Diermeier S, Akerman M, Chang KC,
Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, et
al: Differentiation of mammary tumors and reduction in metastasis
upon Malat1 lncRNA loss. Genes Dev. 30:34–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xing Z, Lin A, Li C, Liang K, Wang S, Liu
Y, Park PK, Qin L, Wei Y, Hawke DH, et al: lncRNA directs
cooperative epigenetic regulation downstream of chemokine signals.
Cell. 159:1110–1125. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Comijn J, Berx G, Vermassen P, Verschueren
K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D and van Roy
F: The two-handed E box binding zinc finger protein SIP1
downregulates E-cadherin and induces invasion. Mol Cell.
7:1267–1278. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang Z, Liu F, Yang F and Liu Y: Kockdown
of OIP5-AS1 expression inhibits proliferation, metastasis and EMT
progress in hepatoblastoma cells through up-regulating miR-186a-5p
and down-regulating ZEB1. Biomed Pharmacother. 101:14–23. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang M, Sun X, Yang Y and Jiao W: Long
non-coding RNA OIP5-AS1 promotes proliferation of lung cancer cells
and leads to poor prognosis by targeting miR-378a-3p. Thorac
Cancer. 9:939–949. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang X, Xu X, Ge G, Zang X, Shao M, Zou
S, Zhang Y, Mao Z, Zhang J, Mao F, et al: miR-498 inhibits the
growth and metastasis of liver cancer by targeting ZEB2. Oncol Rep.
41:1638–1648. 2019.PubMed/NCBI
|