Introduction
A total of 3 out of 1,000,000 people are diagnosed
with osteosarcoma, which is the most common primary malignant bone
tumor in children and adolescents (1). Osteosarcoma typically occurs in the
metaphysis of long bones such as the femur, tibia and humerus
(2). Although cytotoxic neoadjuvant
chemotherapy drugs including cisplatin, doxorubicin, methotrexate
and ifosfamide combined with surgery and postoperative chemotherapy
are feasible treatments for osteosarcoma, the survival rate has not
increased significantly in the past decades (3,4).
The focus of osteosarcoma treatment has been shifted
from survival through limb-amputation to improvement of the quality
of life through limb-salvage surgery (5). Radiotherapy can induce cell apoptosis
by breaking DNA double strands to reduce the local recurrence rate
(6). However, it is only used as an
adjuvant treatment in patients who have received limb-salvage
surgery as osteosarcoma is not sensitive to radiotherapy (7). Several studies have shown that
radiotherapy can be used as an alternative surgical treatment for
some staged patients (8,9). After adjuvant chemotherapy,
radiotherapy is effective in 56% of patients with limb tumors
(10).
The proto-oncogene c-myc is a transformed member of
the myc family. Approximately 20% of human cancers may be
associated with c-myc overexpression (11). As a transcription activator, it
regulates cell growth, differentiation, programmed cell death and
apoptosis (12). C-myc-interacting
zinc finger protein-1 (Miz-1) is a poly-Cys2His2 zinc finger (ZF)
activator of cell cycle regulator genes, such as the
cyclin-dependent kinase inhibitor p21 (13). Some studies have found that c-myc
combined with zinc finger transcription factor Miz-1 can upregulate
the expression of p21 by abolishing the interaction between Miz-1
and its co-activators, which activates G2/M phase transition, such
as the cyclinB1/Cdc2 complex (14,15).
Hence, inhibiting c-myc gene expression can restrain tumor cell
growth in the G2/M phase (16,17).
Additionally, some studies have shown that tumors in the G2/M phase
are more sensitive to radiotherapy (18,19).
Therefore, radiotherapy combined with downregulation of the c-myc
gene could be a therapeutic strategy for human osteosarcoma. To the
best of our knowledge, no research has been conducted to
investigate the radiosensitizing effect of the c-myc gene on
osteosarcoma. Therefore, the present study investigated the
radiosensitizing effect of the c-myc gene and the sensitizing
apoptosis pathway, in order to provide a more effective combination
radiotherapy treatment for osteosarcoma.
Materials and methods
Comprehensive analysis
Genetic data (GSE126209) of the corresponding
samples were downloaded from The Cancer Genome Atlas (TCGA)
(https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga)
data portal. The differential expression data of genes were
normalized and analyzed by edgeR, a Bioconductor package based on R
3.53 (https://www.r-project.org/).
Kaplan-Meier survival analyses were utilized to assess the overall
survival of patients with sarcoma who were classified into
high-expression and low-expression groups based on the myc gene
from the University of Alabama web portal (http://ualcan.path.uab.edu/) (20). Available TCGA survival data was used
for Kaplan-Meier survival analysis the overall survival plots were
generated. Functional protein association network of the myc gene
was constructed using the STRING database (https://string-db.org/).
Reagents and antibodies
DMEM, Minimum Essential Medium (MEM), Roswell Park
Memorial Institute (RPMI)-1640 Medium, FBS, penicillin,
streptomycin, PBS and 0.25% trypsin were purchased from Gibco
(Thermo Fisher Scientific, Inc.). Antibodies against caspase-3
(cat. no. 9662), cleaved caspase-3 (cat. no. 9661), poly
(ADP-ribose) polymerase (PARP) (cat. no. 9532), cleaved PARP (cat.
no. 5625), Bax (cat. no. 5023), Bid (cat. no. 2002), Bcl-2 (cat.
no. 4223), Cdc2 (cat. no. 28439), cyclin B1 (cat. no. 12231),
cyclin D1 (cat. no. 55506), p21 (cat. no. 2947) and GAPDH (cat. no.
5174) (all, 1:1,000) were purchased from Cell Signaling Technology,
Inc. Primary antibody dilution buffer (cat. no. FD0040) was
purchased from Fdbio Science.
Cell culture
hHOF1.19, MG63, HOS and U2OS cells from the Shanghai
Cell Bank of the Chinese Academy of Sciences were tested for
mycoplasma contamination. Cells were routinely cultured in a cell
incubator (37°C; 5% CO2; Thermo Fisher Scientific, Inc.)
in high-glucose DMEM supplemented with 100 µl/ml
penicillin-streptomycin and 10% FBS.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was performed to determine the expression of
c-myc in hHOF1.19, MG63, HOS and U2OS cells. TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to
extract total RNA from the cells according to the manufacturer's
instructions. PrimeScript™ RT Reagent kit (Takara Bio, Inc.) was
used to reverse transcribe RNA to cDNA. The reaction conditions
used for RT were as follows: Incubation at 37°C for 15 sec,
followed by 85°C for 30 sec. C-myc levels were quantified using
SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) on an ABI 7500
Fast Sequence Detection System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The following thermocycling conditions were used
for the qPCR: Initial denaturation at 95°C for 1 min, followed by
40 cycles of 95°C for 5 sec and 60°C for 34 sec. GAPDH was used as
the internal reference gene and the relative expression levels of
c-myc in hFOB, MG63, U2OS and HOS osteosarcoma cells were analyzed
using the 2−ΔΔCq method (21) The following primer pairs were used
for the qPCR: c-myc forward, 5′-GTCAAGAGGCGAACACACAAC-3′ and
reverse, 3′-TTGGACGGACAGGATGTATGC-5′ and GAPDH forward,
5′-TGTTCGTCATGGGTGTGAAC-3′ and reverse,
5′-ATGGCATGGACTGTGGTCAT-3′.
Cell transfection
MG63 cells were seeded into six-well plates at a
density of 2×105 cells/well. After reaching 70–90%
confluence, complete medium was replaced with MEM without FBS or
penicillin-streptomycin. Subsequently, small interfering RNA
(siRNA) targeting c-myc was transfected in MG63 cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at a concentration of 50 nM. After 6 h, the
medium was replaced with complete medium to continue cultivation.
To identify the transfection efficiency of siRNA, transfected MG-63
cells were observed by fluorescence imaging and transfection
efficiency was compared with non-transfected MG-63 cells. The siRNA
inhibitor sequence was as follows: Forward,
5′-GGAAGAAAUCGAUGUUGUUTT-3′ and reverse,
3′-AACAACAUCGAUUUCUUCCTT-5′. The si-negative control inhibitor
sequence was as follows: Forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and
reverse, 3′-ACGUGACACGUUCGGAGAATT-5′.
Cell viability assay
A Cell Counting Kit-8 (CCK-8; MedChemExpress) assay
was used to determine cell viability according to the
manufacturer's instructions. Firstly, cells transfected with
si-c-myc and si-negative control (NC) were seeded into 96-well
plates at a density of 5×103 cells/well. A total of 100
ml CCK-8 solution mixed with incomplete MEM (1:10) was added into
each well and cell viability was measured at 24, 48, 72 and 96 h on
a MR7000 microplate reader (Dynatech) at 450 nm absorbance 30 min
later.
Cell cycle analysis
Cells transfected with siRNA after 0, 24, 48, 72 and
96 h were digested and washed twice with PBS and then fixed with
75% ethanol at −20°C overnight. Propidium iodide (PI; BD
Biosciences) was added to cells and incubated for 15 min. Cells
cycle analysis was performed with a FACSCalibur flow cytometer (BD
Biosciences) and data were analyzed with CellQuest software 3.1 (BD
Biosciences).
Apoptosis analysis
The FITC Annexin V apoptosis Detection Kit I (BD
Pharmingen; BD Biosciences) was used for apoptosis analysis. First,
the cells were digested, washed with PBS and resuspended in 1X
Binding Buffer. Subsequently, cells were transferred in 100 µl
resuspended mixture solution (1×105 cells) into a 5 ml
culture tube. A total of 5 µl V-FITC and 5 µl PI were added into
the tube and incubated at room temperature for 15 min. Finally, 400
µl 1X Binding Buffer was added to each tube. Cell apoptosis
analysis was performed with a FACSCalibur flow cytometer (BD
Biosciences), and data were analyzed with CellQuest software 3.1
(BD Biosciences).
Clone formation assay
Cells in the logarithmic growth phase were seeded in
a six-well plate with 200–800 cells/well and incubated at 37°C for
2 weeks until visible colonies appeared in the wells. Following
washing with PBS twice, a mixture of 0.1% crystal violet and 4%
polymethanol was added to fix and stain for 30 min at 37°C. The dye
was washed away slowly with running water. Clones with >50 cells
were counted under an optical microscope (magnification, ×200).
Radiotherapy
MG63 cells transfected with si-NC and si-c-myc were
seeded in a six-well plate with 200–800 cells/well and cultured in
a 37°C and 5% CO2 incubator for 24 h. An X-ray
instrument (Precision X-Ray, Inc.) was used to irradiate the cells
at dose rates of 2, 4, 6 and 8 Gy. Subsequently, MG63 cells were
cultured in the incubator for 10 days.
Fluorescence assays
Cell apoptosis was detected by fluorescence
microscopy using DAPI reagent (Beyotime Institute of Biotechnology)
for nuclear staining. In brief, cells fixed with 4% polymethanol
were stained with DAPI and incubated in the dark for 15 min at
37°C. After washing twice with PBS, cells were observed under a
fluorescence microscope (magnification, ×200) (Olympus Corporation)
to identify nuclear fragmentation and chromatin condensation.
Western blot analysis
MG63 cells in six-well plates treated with si-NC,
si-c-myc or radiotherapy were digested and lysed in RIPA lysis
buffer containing protease inhibitor cocktail (Sigma-Aldrich; Merck
KGaA). Protein concentration was measured using a BCA protein assay
kit (Pierce; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions on a MR7000 microplate reader
(Dynatech) at a wavelength of 570 nm 30 min later. Equal amounts of
protein (40 µg) were separated via SDS-PAGE (12% polyacrylamide
gels). Electrophoresis and membrane transfer were performed at 100
V for 1.5 h and 300 mA for 1 h, respectively. Separated proteins
were transferred to a PVDF membrane, blocked with 5% non-fat milk
in TBS-0.1% Tween 20 (TBS-T) at 37°C for 1 h and then incubated in
primary target antibodies (Cell Signaling Technology, Inc.)
overnight at 4°C. Following three washes with TBS-T, membranes were
incubated with HRP anti-rabbit (cat. no. FD0128) or anti-mouse IgG
(cat. no. FD0142) antibodies (Fdbio Science) diluted in 5% non-fat
milk (1:5,000 working dilution) for 1 h at room temperature.
Protein bands were visualized using the Westar Supernova kit
(Cyanagen) and a molecular imager (Bio-Rad Laboratories, Inc.).
Densitometric analysis was performed using ImageJ 1.8.0 (National
Institutes of Health).
Statistical analysis
The data were expressed as mean ± SD and analyzed by
SPSS 17.0 (SPSS Inc.). The t-test was used to calculate the
difference between two groups. One-way ANOVA to compare differences
among three or more groups followed by Tukey's post hoc test. Tests
were two-tailed and P<0.05 was considered to indicate a
statistically significant difference.
Results
Comprehensive analysis of the myc
gene
Comprehensive analysis using a heat map with
bidirectional hierarchical clustering of genes and RT-qPCR was
conducted. It was found that myc was overexpressed in tumor samples
compared with normal samples (Fig. 1A
and B). Kaplan-Meier curve analysis of myc for overall survival
showed that myc was negatively associated with patient overall
survival (Fig. 1C). STRING database
analysis showed that G2/M cell cycle target proteins such as cdc2,
p21 and cyclin B1 may be associated with the myc gene (Fig. 1D).
Relative c-myc gene expression in
osteosarcoma and siRNA knockdown efficiency
To investigate the relative c-myc expression in
osteosarcoma, RT-qPCR was performed in osteosarcoma cells HOS,
MG63, U-2 and c-myc expression was compared against normal
cartilage cells hHOF1.19 (Fig. 2A).
C-myc was significantly overexpressed in MG-63 cells but not in U-2
and HOS cells. The present data demonstrated that the levels of
c-myc were higher in MG-63 and U-2 tumor cells compared with
hFOB1.19 cells, particularly in MG-63 cells. Thus, MG-63 cells were
used for subsequent experiments. Subsequently, siRNA was
transfected to downregulate c-myc expression in MG-63 osteosarcoma
cells. The transfection efficiency of siRNA was observed using
fluorescence imaging. As shown in Fig.
2B, >80% of MG-63 cells expressed green fluorescence protein
48 h after transfection. The knockdown efficiency of si-c-myc was
verified by RT-qPCR and western blotting. As shown in Fig. 2C and D, the expression levels of the
c-myc gene was significantly decreased in MG-63 cells transfected
with si-c-myc compared with the si-NC group.
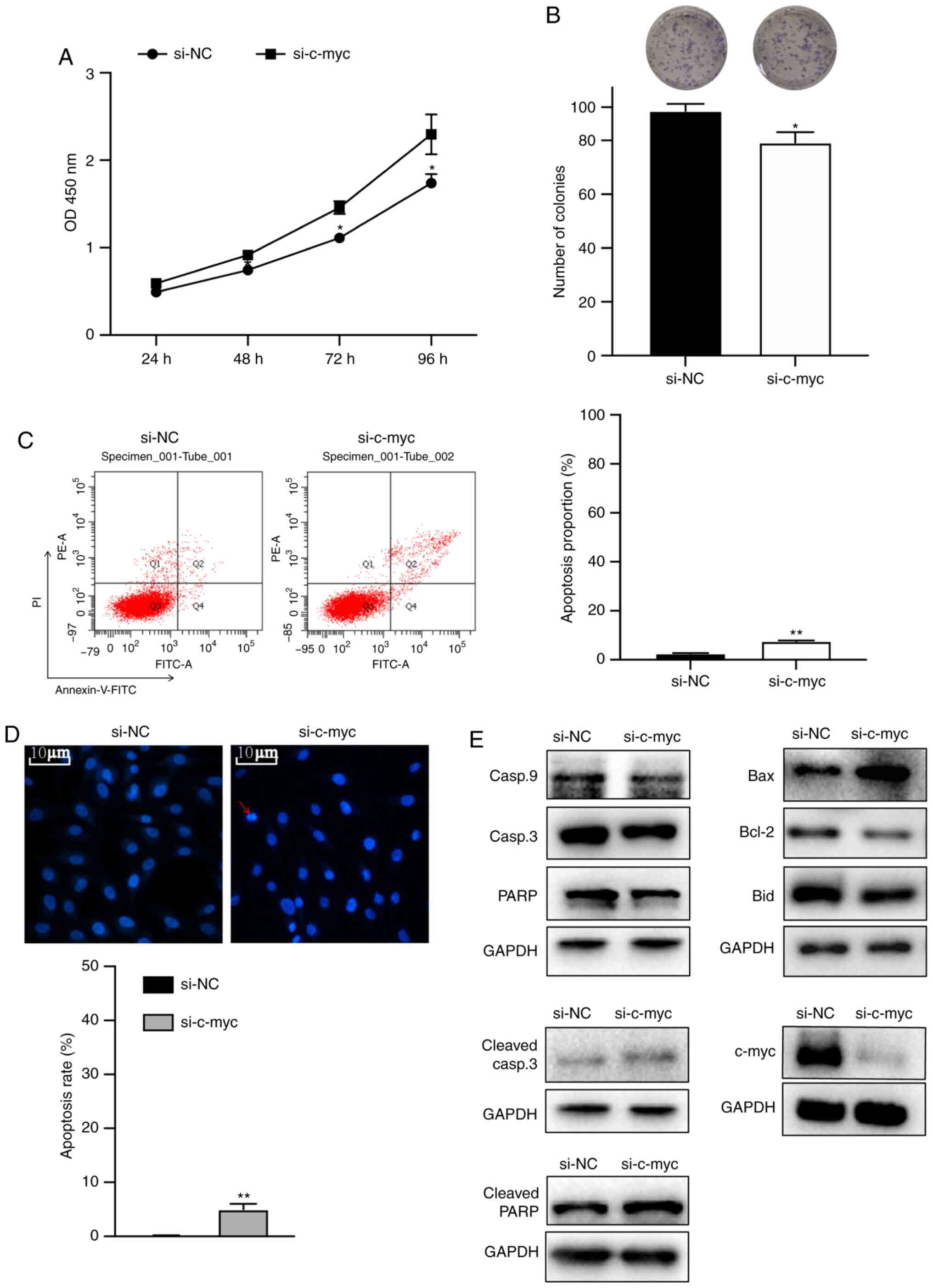
c-myc knockdown inhibits the
proliferation of osteosarcoma cells and induced caspase-dependent
apoptosis via intrinsic pathways
To investigate the effects of c-myc on
proliferation, MG63 cells were transfected with siRNA. Colony
formation and cell viability assays were performed. The cell
viability and colony formation assays showed that knockdown of the
c-myc gene significantly inhibited the proliferation of
osteosarcoma cells (Fig. 3A and B).
To determine whether inhibition of cell growth could be attributed
to apoptosis, DAPI staining and flow cytometry assay were
performed. Fig. 3C and D showed
that c-myc knockdown increased early and late apoptotic cell
proportion, chromatin condensation and DNA fragmentation. Next,
western blotting was performed to identify the pathways involved.
As shown in Fig. 3E, knockdown of
c-myc markedly increased the expression of Bax, cleaved PARP and
cleaved caspase-3 proteins and decreased the expression of c-myc,
caspase-9, Bid, Bcl-2, caspase-3 and PARP proteins.
c-myc knockdown induces G2/M phase
arrest by regulating cell cycle-regulated proteins
To determine whether knockdown of the c-myc gene
inhibited cell proliferation by inducing cell cycle arrest, the
present study examined the cell cycle distribution at 0, 24, 48, 72
and 96 h after si-c-myc transfection in MG-63 cells. As shown in
Fig. 4A, c-myc knockdown increased
G2/M phase cell proportion and decreased G0/G1 and S phases in
MG-63 cells. The arrest rate reached the highest levels at 48 h.
The cell cycle-regulated proteins cyclin B1 and p21 was upregulated
and Cdc2 and cyclin D1 were downregulated (Fig. 4B). The data suggested that knockdown
of the c-myc gene induced G2/M phase arrest by regulating G2/M cell
cycle target markers.
Assessing the radiosensitivity of
MG-63 cells
MG63 cells were irradiated at 0, 2, 4, 6 ND 8 Gy to
determine the radiosensitivity. As shown in Fig. 4C, the colony-forming efficiency was
46.3, 22.2, 2.8, 0.6 and 0.0%, respectively. Cell survival rates
were significantly reduced in a dose-dependent manner.
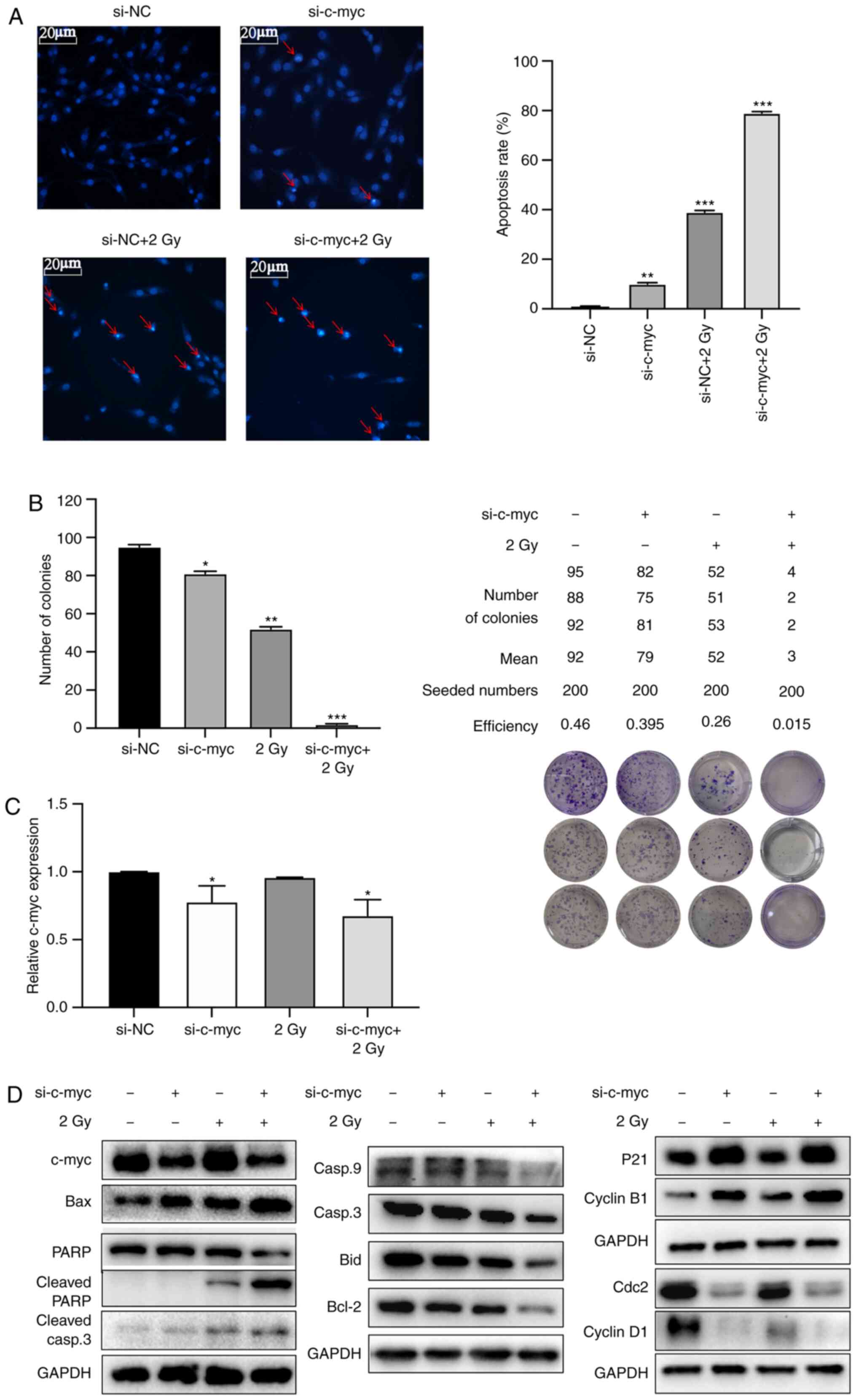
Radiosensitizing effects of c-myc
knockdown is determined by inducing G2/M phase arrest
Radiosensitivity was investigated by reducing c-myc
gene expression via inducing G2/M phase arrest. Fig. 5A shows that apoptotic chromatin
condensation and DNA fragmentation were significantly increased in
the si-c-myc + 2 Gy group compared with other groups. The mean
numbers of colonies formed were 92, 79, 52 and 3, respectively, in
si-NC, si-c-myc, 2 Gy and si-c-myc + 2 Gy groups with a seeding
density of 200 cells/well. The colony-forming efficiency was
significantly reduced in the 2 Gy group with c-myc knockdown group
(Fig. 5B). The knockdown efficiency
of si-c-myc was verified by RT-qPCR (Fig. 5C) and the expression levels the of
c-myc gene was significantly decreased in si-c-myc and si-c-myc + 2
Gy groups. Western blot analysis showed that the cell
cycle-regulated proteins cyclin B1 and p21 were upregulated and
Cdc2 and cyclin D1 was downregulated in both si-c-myc and si-c-myc
+ 2 Gy groups. The apoptosis-related proteins caspase-9, c-myc,
cdc2, cyclin D1, Bid, Bcl-2, caspase-3 and PARP were decreased
while p21, cyclin B1, Bax, cleaved PARP and cleaved caspase-3 were
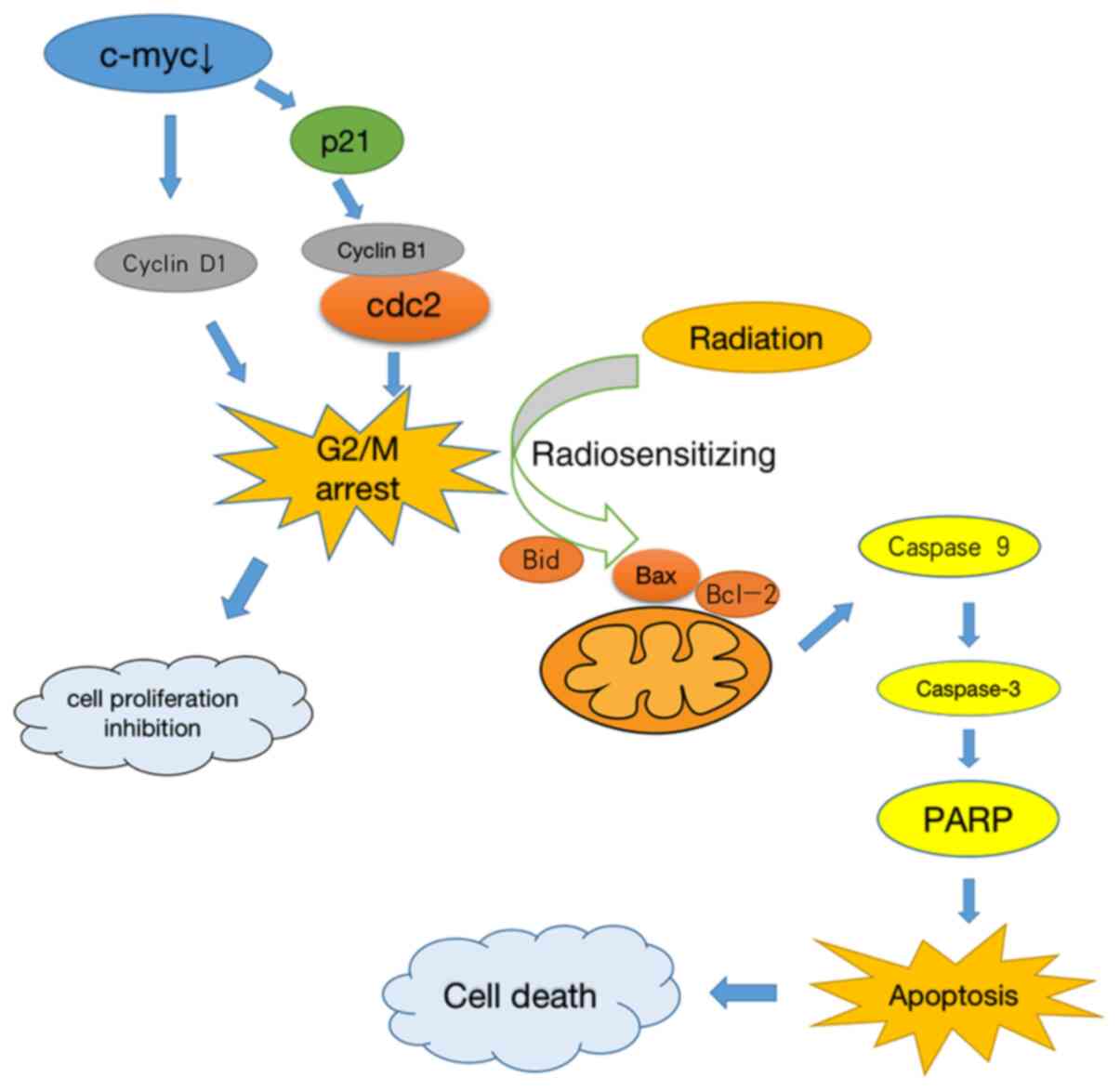
increased in the si-c-myc + 2 Gy group (Fig. 5D). These results suggested that
downregulating c-myc expression induced the radiosensitizing
effects involved in G2/M phase arrest, which increased the
apoptosis of osteosarcoma cells by intrinsic stimuli via the
mitochondrial signaling pathway (Fig.
6).
Discussion
Osteosarcoma is insensitive to radiotherapy
(22,23). Some studies have shown that tumor
cells in the G2/M phase have good sensitivity of radiotherapy
(24–26). Inhibiting c-myc expression can
effectively suppress the proliferation of tumor cells and induce
G2/M phase arrest (27,28) as well as apoptosis of sarcoma cells
(18,29). The present study aimed to assess the
radiosensitizing effects of c-myc knockdown-induced G2/M phase
arrest. The present study showed that downregulating c-myc
significantly inhibited the proliferation of osteosarcoma cells and
induced G2/M phase arrest. The combination of radiotherapy and
c-myc inhibition resulted in a significantly higher apoptosis rate
of osteosarcoma cells compared with using either therapy alone.
Previous analysis showed that the myc gene was
overexpressed in osteosarcoma and negatively associated with
patients' overall survival (20,30).
The present study demonstrated that the c-myc gene was
overexpressed in MG63 osteosarcoma cells compared to osteoblasts.
Tumor proliferation was significantly inhibited after inhibiting
the expression of the c-myc gene in osteosarcoma cells. The G2/M
phase has become a key cell cycle target marker for the inhibition
of tumor proliferation (31,32).
During the initiation of the M phase, Cdc25 binds to proliferating
cell nuclear antigen (PCNA) to catalyze the dephosphorylation of
Cdc2 Y14/Y15, which activates the cyclin B1/Cdc2 complex and
subsequently cell mitosis. Therefore, the cyclin B1/Cdc2 complex
plays a key role in inducing cell G2/M phase transition (33). C-myc and zinc finger transcription
factor Miz-1 can inhibit cell cycle transcription factors, such as
p21 (34). P21, the inhibitor of
cdc2, can form a quaternary complex with Cdc2, cyclinB1 and PCNA,
then competitively inhibits cdc25 binding to PCNA and blocks cdc25
from catalyzing the cdc2 dephosphorylation (35). The present study identified that
c-myc gene downregulation could induce p21 protein upregulation
with the increase of G2/M phase-associated protein cyclin B1 and
the decrease of cdc2 protein, which leads to G2/M phase arrest and
inhibition of osteosarcoma cell proliferation.
To further determine whether cell cycle arrest was
time-dependent, cell cycle distribution tests were performed at
five time points (0, 24, 48, 72 and 96 h). The present study found
that the cycle arrest was time-dependent: It peaked at 48 h and the
arrest capacity of the G2/M phase gradually decreased. Studies have
also shown that c-myc is most effective in blocking tumor growth in
48 h (26). Radiotherapy performed
at 0, 2, 4, 6 and 8 Gy doses showed that the cell growth inhibition
rate was ~50% at 2 Gy, similar to the result of a previous report
(36).
Radiotherapy induces cell apoptosis by damaging DNA
double strands and inhibiting cell cycle checkpoint activation
(37). A previous study found that
radiation sensitivity is highly correlated with cells G2/M phase
(38). The present study
hypothesized that inducing G2/M phase arrest with drugs will
increase the sensitivity of the cells to radiotherapy. Therefore,
radiotherapy was performed on c-myc knockdown cells and verified
the radiosensitizing effects of c-myc gene knockdown-induced G2/M
phase arrest and the activation of mitochondrial-mediated apoptosis
pathway in this process.
In summary, the present study revealed that
inhibiting c-myc gene expression combined with radiotherapy could
significantly increase the apoptosis rate of osteosarcoma cells.
The present study verified the radiosensitizing effects of c-myc
gene knockdown-induced G2/M phase arrest, which was achieved by
intrinsic stimuli via the mitochondrial signaling pathway. The
results of the present study may provide an effective and novel
therapeutic strategy for radiotherapy of osteosarcoma.
Acknowledgements
Not applicable.
Funding
This research was supported by the Agricultural and
Social Development Research Independent Application Project of
Hangzhou City (grant no. 20191203B88).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YF conceived and designed the study and wrote the
manuscript. LZ searched the database and reviewed studies. TX and
XJ performed data analysis and prepared the initial draft of the
manuscript. FH participated in interpretation of data, helped in
drafting the manuscript and critically reviewed the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kansara M, Teng MW, Smyth MJ and Thomas
DM: Translational biology of osteosarcoma. Nat Rev Cancer.
14:722–735. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kager L, Tamamyan G and Bielack S: Novel
insights and therapeutic interventions for pediatric osteosarcoma.
Future Oncol. 13:357–368. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kelleher FC and O'Sullivan H: Monocytes,
macrophages, and osteoclasts in osteosarcoma. J Adolesc Young Adult
Oncol. 6:396–405. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferrari S, Mercuri M and Bacci G: Comment
on ‘Prognostic factors in high-grade osteosarcoma of the
extremities or trunk: An analysis of 1,702 patients treated on
neoadjuvant Cooperative Osteosarcoma Study Group protocols’. J Clin
Oncol. 20:2910–2911. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bishop MW, Janeway KA and Gorlick R:
Future directions in the treatment of osteosarcoma. Curr Opin
Pediatr. 28:26–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bugris V, Harmat V, Ferenc G, Brockhauser
S, Carmichael I and Garman EF: Radiation-damage investigation of a
DNA 16-mer. J Synchrotron Radiat. 26:998–1009. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li QC, Xu H, Wang X, Wang T and Wu J:
miR-34a increases cisplatin sensitivity of osteosarcoma cells in
vitro through up-regulation of c-Myc and Bim signal. Cancer
Biomark. 21:135–144. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Macaeva E, Saeys Y, Tabury K, Janssen A,
Michaux A, Benotmane MA, De Vos WH, Baatout S and Quintens R:
Radiation-induced alternative transcription and splicing events and
their applicability to practical biodosimetry. Sci Rep.
6:192512016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cruse MJ, Kucharik CJ and Norman JM: Using
a simple apparatus to measure direct and diffuse photosynthetically
active radiation at remote locations. PLoS One. 10:e01156332015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li S, Hu T, Yuan T, Cheng D and Yang Q:
Nucleoside diphosphate kinase B promotes osteosarcoma proliferation
through c-Myc. Cancer Biol Ther. 19:565–572. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Machak GN, Tkachev SI, Solovyev YN,
Sinyukov PA, Ivanov SM, Kochergina NV, Ryjkov AD, Tepliakov VV,
Bokhian BY and Glebovskaya VV: Neoadjuvant chemotherapy and local
radiotherapy for high-grade osteosarcoma of the extremities. Mayo
Clin Proc. 78:147–155. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cartee L, Vrana JA, Wang Z, Park JS,
Birrer M, Fisher PB, Grant S and Dent P: Inhibition of the mitogen
activated protein kinase pathway potentiates radiation-induced cell
killing via cell cycle arrest at the G2/M transition and
independently of increased signaling by the JNK/c-Jun pathway. Int
J Oncol. 16:413–422. 2000.PubMed/NCBI
|
|
13
|
Noh HJ, Koh DI, Lee KO, Jeon BN, Kim MK,
Snead ML and Hur MW: Role of MIZ-1 in AMELX gene expression.
Biochem Biophys Rep. 8:340–345. 2016.PubMed/NCBI
|
|
14
|
Bédard M, Maltais L, Montagne M and
Lavigne P: Miz-1 and Max compete to engage c-Myc: Implication for
the mechanism of inhibition of c-Myc transcriptional activity by
Miz-1. Proteins. 85:199–206. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Licchesi JD, Van Neste L, Tiwari VK, Cope
L, Lin X, Baylin SB and Herman JG: Transcriptional regulation of
Wnt inhibitory factor-1 by Miz-1/c-Myc. Oncogene. 29:5923–5934.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Concin N, Stimpfl M, Zeillinger C, Wolff
U, Hefler L, Sedlak J, Leodolter S and Zeillinger R: Role of p53 in
G2/M cell cycle arrest and apoptosis in response to
gamma-irradiation in ovarian carcinoma cell lines. Int J Oncol.
22:51–57. 2003.PubMed/NCBI
|
|
17
|
Suen DF, Norris KL and Youle RJ:
Mitochondrial dynamics and apoptosis. Genes Dev. 22:1577–1590.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen BJ, Wu YL, Tanaka Y and Zhang W:
Small molecules targeting c-Myc oncogene: Promising Anti-cancer
therapeutics. Int J Biol Sci. 10:1084–1096. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee JA, Paik EK, Seo J, Kim DH, Lim JS,
Yoo JY and Kim MS: Radiotherapy and gemcitabine-docetaxel
chemotherapy in children and adolescents with unresectable
recurrent or refractory osteosarcoma. Jpn J Clin Oncol. 46:138–143.
2015.PubMed/NCBI
|
|
20
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Unni KK and Dahlin DC: Osteosarcoma:
Pathology and classification. Semin Roentgenol. 24:143–152. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim E, Kim MS, Lee KH, Sai S, Jeong YK,
Koh JS and Kong CB: Effect of low- and high-linear energy transfer
radiation on in vitro and orthotopic in vivo models
of osteosarcoma by activation of caspase-3 and −9. Int J Oncol.
51:1124–1134. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Miyata H, Doki Y, Yamamoto H, Kishi K,
Takemoto H, Fujiwara Y, Yasuda T, Yano M, Inoue M, Shiozaki H, et
al: Overexpression of CDC25B overrides radiation-induced G2-M
arrest and results in increased apoptosis in esophageal cancer
cells. Cancer Res. 61:3188–3193. 2001.PubMed/NCBI
|
|
25
|
Fei H, Zhou Y, Li R, Yang M, Ma J and Wang
F: HBXIP, a binding protein of HBx, regulates maintenance of the
G2/M phase checkpoint induced by DNA damage and enhances
sensitivity to doxorubicin-induced cytotoxicity. Cell Cycle.
16:468–476. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li YX, Weber-Johnson K, Sun LQ, Paschoud
N, Mirimanoff RO and Coucke PA: Effect of pentoxifylline on
radiation-induced G2-phase delay and radiosensitivity of human
colon and cervical cancer cells. Radiat Res. 149:338–342. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cui F, Hou J, Huang C, Sun X, Zeng Y,
Cheng H, Wang H and Li C: C-Myc regulates radiation-induced G2/M
cell cycle arrest and cell death in human cervical cancer cells. J
Obstet Gynaecol Res. 43:729–735. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang Y, Xue K, Li Z, Zheng W, Dong W, Song
J, Sun S, Ma T and Li W: c-Myc regulates the CDK1/cyclin B1
dependent-G2/M cell cycle progression by histone H4 acetylation in
Raji cells. Int J Mol Med. 41:3366–3378. 2018.PubMed/NCBI
|
|
29
|
Shang Y: LncRNA THOR acts as a
retinoblastoma promoter through enhancing the combination of c-myc
mRNA and IGF2BP1 protein. Biomed Pharmacother. 106:1243–1249. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Group NP: Translocations involving c-myc
and c-myc function. Oncogene. 20:5595–5610. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Androutsopoulos V and Spandidos D:
Anticancer pyridines induce G2/M arrest and apoptosis via p53 and
JNK upregulation in liver and breast cancer cells. Oncol Rep.
39:519–524. 2018.PubMed/NCBI
|
|
33
|
Nurse P: Universal control mechanism
regulating onset of M-phase. Nature. 344:503–508. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Charrier-Savournin FB, Château MT, Gire V,
Sedivy J, Piette J and Dulic V: p21-mediated nuclear retention of
Cyclin B1-Cdk1 in response to Genotoxic Stress. Molr Biol Cell.
15:3965–3976. 2004. View Article : Google Scholar
|
|
35
|
Dang C: MYC on the path to cancer. Cell.
149:22–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang H, Zhang C and Wu D: Activation of
insulin-like growth factor 1 receptor regulates the
radiation-induced lung cancer cell apoptosis. Immunobiology.
220:1136–1140. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Speidel D: Transcription-independent p53
apoptosis: An alternative route to death. Trends in Cell Biology.
20:14–24. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen W, Liu Q, Fu B, Liu K and Jiang W:
Overexpression of GRIM-19 accelerates radiation-induced
osteosarcoma cells apoptosis by p53 stabilization. Life Sci.
208:232–238. 2018. View Article : Google Scholar : PubMed/NCBI
|