Introduction
Pancreatic cancer is a highly malignant tumor of the
digestive tract and the fourth leading cause of cancer-related
death worldwide (1,2), In recent years, its morbidity and
mortality have exhibited an upward trend worldwide, due to
difficulties in detection and lack of effective treatment, with a
≤5% 5-year survival rate (2). In
order to effectively diagnose, prevent, and treat this disease,
further study of the molecular mechanism of pancreatic cancer is
needed.
DNA methylation is a form of epigenetic modification
and an important mechanism of gene expression regulation (3,4). The
occurrence and development of tumors are closely related to DNA
methylation abnormalities, which are mainly manifested as a
decrease in the overall genomic methylation level in tumor cells
and an increase in the methylation level of the promoter region of
specific genes (5–8). Previous studies have shown that
methylation abnormalities in multiple promoter regions are closely
related to the development of pancreatic cancer (5,6).
As a member of the DNA methyltransferases (DNMTs),
DNMT1 plays an important role in mediating gene expression and
chromatin structure, by preserving existing DNA methylation during
DNA replication (7). DNMT1 was found
to be upregulated in various types of cancer, including pancreatic
cancer (8). A study of pancreatic
cancer demonstrated that lower expression of DNMT1 reversed the
resistance to 5-azadeoxycytidine (9).
In addition, siRNA targeting DNMT1 led to a reduction in cell
viability and induced cell apoptosis in pancreatic cancer cells
(10). Abnormal DNA methylation in
tumors includes overall hypomethylation of the genome and
hypermethylation of certain gene promoter regions. Abnormally
elevated methylation of the promoter region may lead to
transcriptional silencing of important regulatory genes such as
cell cycle regulatory genes, tumor-suppressor genes, and apoptotic
genes, resulting in decreased expression or loss of expression of
related genes, and thereby promoting tumor formation (11). This hypermethylation is another
mechanism leading to the inactivation of tumor-suppressor
genes.
MicroRNAs (miRNAs) are a class of small non-coding
RNAs with about 21–25 nucleotides, which can regulate the
expression of post-transcriptional target genes (12,13). A
large number of researches also confirmed that miRNAs have the dual
role of oncogene or tumor suppressor gene, and its expression
changes are closely related to tumor formation (14–16).
miR-29b-3p is a member of the miR-29 family and is closely related
to the behavior of various tumors (17–21). In
this study, we investigated the effects of the miR-29b-3p promoter
methylation status on angiogenesis, invasion, and migration of
pancreatic cancer cells. This study provides information concerning
the role of the methylation of miR-29b in pancreatic cancer, and
this may be a target for pancreatic cancer therapy.
Materials and methods
Patients and tissue collection
A total of 18 pairs of tissues from pancreatic
cancer patients (mean age, 68.65±14.23 years ranging from 39 to 91;
8 female patients and 10 male patients) were collected at the
Yantai Yuhuangding Hospital of Qingdao University from March to
November 2019. Patients who had received chemotherapy or
radiotherapy were excluded in this study. The tissues were
collected and transferred into liquid nitrogen and then were stored
at −80°C. This study was approved by the Ethics Committee of the
Yantai Yuhuangding Hospital of Qingdao University, and the ethics
approval number is QDU-201902-3. All patients have provided written
informed consent to participate in the study.
Antibodies, reagents, plasmids, miRNA,
and siRNA
Antibodies against DNMT1 (dilution 1:1,000, cat. no.
ab13537), DNMT2 (dilution 1:1,000, cat. no. ab272620), DNMT3a
(dilution 1:1,000, cat. no. ab228691), DNMT3b (dilution 1:1,000,
cat. no. ab122932), zonula occludens-1 (ZO-1) (dilution 1:1,000,
cat. no. ab191143), occludin (dilution 1:1,000, cat. no. ab242202),
claudin-5 (dilution 1:1,000, cat. no. ab15106), GAPDH (dilution
1:1,000, cat. no. ab9485) were from Abcam. HRP-labeled secondary
antibodies (dilution 1:10,000, cat. no. sc-2370 or sc-2371) were
from Santa Cruz Biotechnology, Inc. Fetal bovine serum (FBS, Gibco;
Thermo Fisher Scientific, Inc.), Dulbecco's modified Eagle's medium
(DMEM, Sigma; Merck KGaA, cat. no. 5796), Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc.), and NuPAGE 4–12%
Bis-Tris Gels were purchased from Thermo Fisher Scientific, Inc.
Hydroquinone and sodium bisulfite were obtained from Sigma-Aldrich
(Merck KGaA). Wizard DNA purification resin was obtained from
Promega Corp. The CpGenome DNA Modification Kit was purchased from
Intergen. The Vector and pcDNA3.1-DNMT1 were all designed and
purchased from Invitrogen; Thermo Fisher Scientific, Inc.
miR-29b-3p mimic, negative control mimics and all siRNA
oligonucleotides were synthesized by GenePharma. All mimics and
plasmids were transfected into cells using Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc). At 48-h post
transfection, the transfected cells were collected for the next
analysis.
Cell culture
Normal human pancreatic duct epithelial cells
(HPDE6-C7) and 5 pancreatic cancer cell lines (BxPC3, PANC1, CFPAC,
Capan-2, and AsPC-3) were purchased from Clontech. HUVECs were
obtained from the American Type Culture Collection (ATCC). All
cells were maintained and propagated in DMEM with 10% FBS and 1%
penicillin streptomycin in 5% CO2 at 37°C.
Western blotting
Cytoplasmic and nuclear protein fractions were
extracted with the NE-PER Reagent Kit (Pierce; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Cell or tumor tissue lysates were separated by NuPAGE 4–12%
Bis-Tris Gels, under 60 V electrophoresis for 30 min, followed by
120 V electrophoresis for 120 min. After electrophoresis, proteins
were transferred to PVDF membranes (Millipore), under 300 mA for 30
min. The membrane was then blocked with 5% defatted milk powder for
60 min at room temperature. Mouse anti-human antibodies against
DNMT1, DNMT2, DNMT3a, DNMT3b, ZO-1, occludin, claudin-5 and GAPDH
(all diluted at 1:1,000, Santa Cruz Biotechnology, Inc.) were added
at 4°C room temperature incubation overnight. The membrane was then
washed with phosphate-buffered solution Tween (PBST) for 30 min,
followed by incubation with horseradish peroxidase (HRP)-conjugated
secondary antibody for 60 min (dilution, 1:5,000, Santa Cruz
Biotechnology, Inc.). After the membrane was washed three times
with PBST, chemiluminescence detection reagent was used to develop
the film. Gel image system was used to analyzed the band density
(Bio-Rad Laboratories, Inc.).
Methylation-specific PCR
Genomic DNA was treated with bisulfite, and all
cytosines that were not methylated were converted to uracil, while
methylated cytosines were unchanged. Subsequently, the primers were
designed for PCR at both ends of the CPG island to purify the
target product. Primer pairs for PCR amplification were purchased
from Thermo Fisher Scientific, Inc. After TA cloning, each clone
was selected for positive clone sequencing, and finally the
sequence was compared with the original sequence, the methylation
site and number were counted, and the degree of methylation was
analyzed.
Transient expression of DNMT1 in Bxpc3
and Capan-2 cells
The empty plasmid vector pcDNA3.1 (Invitrogen;
Thermo Fisher Scientific, Inc.) or the plasmid vector containing
DNMT1 cDNA was transfected into Bxpc3 and Capan-2 cells using
Lipofectamine 3000 for subsequent experiments.
Transient transfection for functional
analysis of miR-29b-3p and DNMT1
The cells were seeded in 6-well plates at 1×
105 cells/well followed by culturing for 24 h and then
transfected with 30 nM of the miR-29b-3p mimic and the negative
control mimics (NC) (GenePharma) using Lipofectamine 3000. The
miR-29b-3p mimic and negative control mimic sequences were designed
and synthesized by Gene pharma. The DNMT1 gene was knocked down by
DNMT1 interfering small RNA (siRNA) obtained from Generay and
transfected into the Bxpc3 and Capan-2 cells by Lipofectamine 3000.
The siRNA target sequence was as follows: DNMT1,
5′-TGTTAAGCTGTCTCTTTCCAA-3′ and negative control,
5′-TAGATACTATGAATTCGTCCAA-3′. Medium was replaced with fresh medium
after transfection for 6 h, and the cells were cultured for another
48 h before further analysis.
Quantitative real-time PCR (qPCR)
The total RNA was isolated from cells using TRIzol
reagent (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. RNA (1 µg) was converted into cDNA
using the RevertAid™ First Strand cDNA Synthesis Kit (Fermentas;
Thermo Fisher Scientific, Inc.). After 10-fold dilution, 4 µl of
cDNA was subjected to PCR amplification using SYBR Premix Ex Taq™
II (Takara) according to the manufacturer's protocol in a
StepOnePlus™ Real-Time PCR System (ABI; Thermo Fisher Scientific,
Inc.). The following thermocycling conditions were used for qPCR:
95°C for 10 sec, then 40 cycles with 95°C for 5 sec, 60°C for 34
sec. β-actin served as the internal control. The primer sequences
were as follows: DNMT1 forward, 5′-CCTAGCCCCAGGATTACAAGG-3′ and
reverse, 5′-ACTCATCCGATTTGGCTCTTTC-3′; miR-29b-3p forward,
5′-ACACTCCAGCTGGGTAGCACCATTTGAAATCA-3′, reverse,
5′-CTCAACTGGTGTCGTGGA-3′ and reverse transcription,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAACACTGA-3′; β-actin
forward, 5′-TGTTCGTCATGGGTGTGAAC-3′ and reverse,
5′-ATGGCATGGACTGTGGTCAT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
U6 reverse, 5′-AACGCTTCACGAATTTGCGT-3′. β-actin and U6 were used as
internal references for measuring relative expression of DNMT1 and
miR-29b-3p, respectively. The expressions of genes were quantified
using the 2−∆∆Cq method (22).
Cell migration and invasion assays
(Transwell)
Uncoated or Matrigel-coated chambers Transwells (BD
Biosciences) containing 8-µm pores were used for the assays. Cells
(200 µl) (1×105 cells/ml) were seeded into the upper
chamber in serum-free DMEM medium. A total of 600 µl conditioned
DMEM media from target cells containing 10% FBS was added to the
lower chamber. Cells were fixed in 100% methanol 72 h later and
stained with a 1:5 dilution of Giemsa (Sigma-Aldrich; Merck KGaA)
for 40 min at room temperature. Cells remaining on the upper side
of the filter were removed with a cotton swab. The filters were
then mounted onto slides and images were captured under a
microscope (Wetzlar, Germany, cat. no. DMI 1, Leica) at ×200
magnification. From these images, the number of migratory or
invasive cells was counted.
In vitro angiogenesis experiment of
target cells co-cultured with HUVECs
The target cells (1×105 cells/ml) were
inoculated in a cell culture flask at the same density. After 6 h
of culture, the culture medium was discarded and replaced with
DMEM. After further culturing for 8 h, the culture solution was
collected and centrifuged at 1,000 × g for 10 min to collect the
cell supernatant culture solution. Then 50 µl of Matrigel was added
to each well of a 96-well plate and incubated for 1 h at 37°C.
HUVECs were then added to the upper layer of Matrigel at
5×103 cells per well, and then incubated with the
collected tumor cell culture supernatant. After 12–18 h, the
formation of blood vessel-like structures of the HUVECs was
observed and photographed under a fluorescence microscope (Keyence,
cat. no. BZ-9000).
Statistics
All the quantitative data are represented as mean ±
SEM of at least three independent experiments. The difference
between two groups was evaluated with the 2-tailed Student's
t-test. One-way ANOVA and Tukey post hoc test were used to evaluate
differences of multiple comparisons. All statistical analyses were
conducted using GraphPad Prism software (version 7; GraphPad
Software, Inc.). Differences were considered significant at
P<0.05.
Results
The miR-29b-3p gene promoter region
methylation levels are increased, the DNMT1 expression levels are
increased, and miR-29b-3p expression levels are decreased in
pancreatic cancer
We identified one CpG-rich region for each genomic
locus of the miR-29b-3p promoter using MethPrimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi)
and designed primer sets to analyze the CpG-rich regions. Based on
a database comparison, we predicted that the methylation level of
the miR-29b-3p gene promoter region (−3,000 bp) is increased in
pancreatic cancer tissues (Fig.
S1).
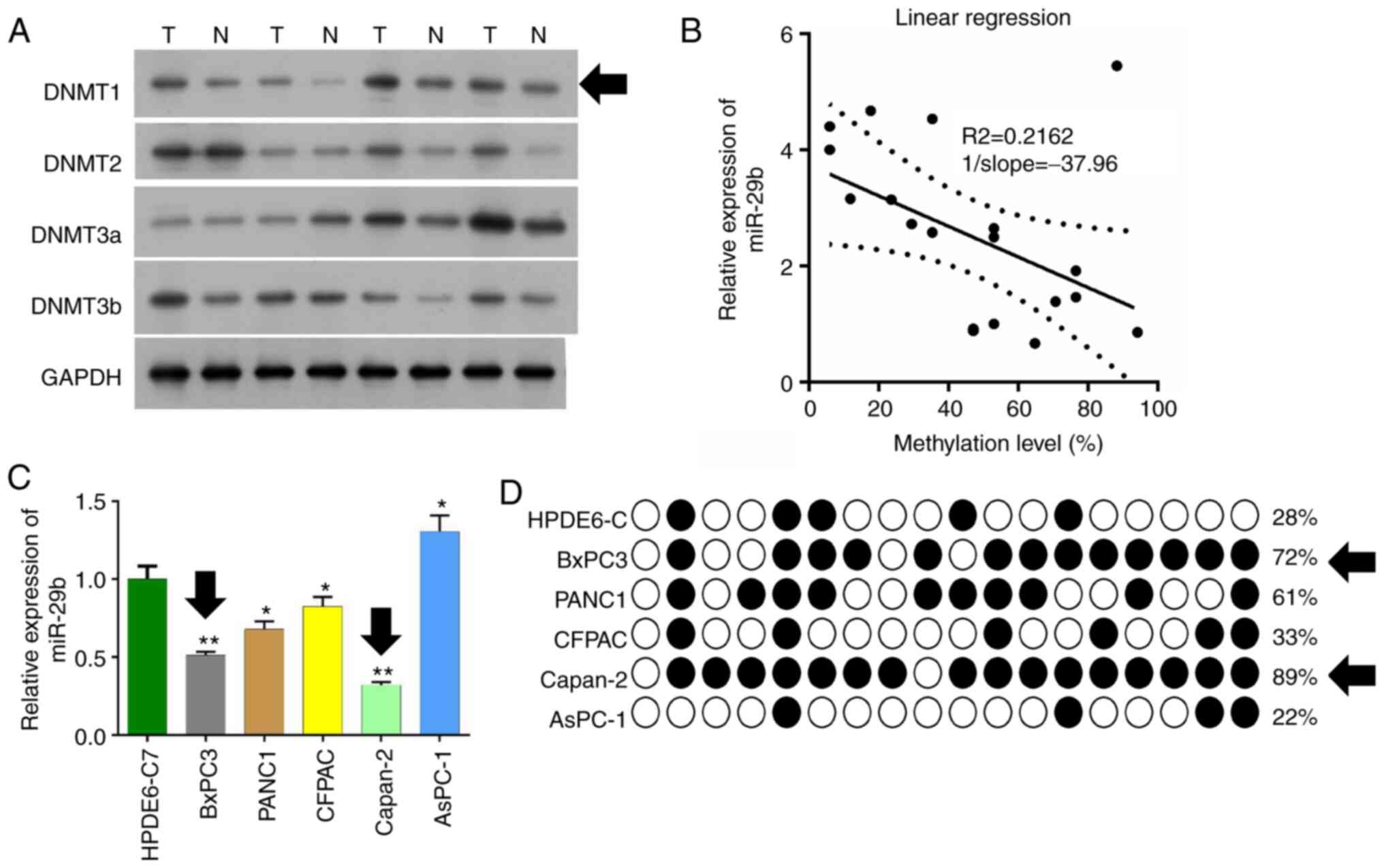
In the present study, we detected expression of
DNMTs in pancreatic cancer tissues and adjacent tissues by western
blot analysis. It was found that the expression level of DNMT1 in
pancreatic cancer tissues was markedly higher than that in the
adjacent tissues (Fig. 1A). DNMT1
expression was significant downregulated in adjacent tissues
compared with tumor tissues, and this was used for later
experiments. qPCR was used to detect expression of miR-29b-3p in
the pancreatic cancer tissues, and the methylation levels of
promoter regions were detected by pyrosequencing in pancreatic
cancer tissues and adjacent tissues. It was found that the
expression level of miR-29b-3p was decreased and this was
negatively correlated with the methylation level of the miR-29b-3p
promoter. (R2=0.2162, 1/slope=37.96) (Fig. 1B).
Six pancreatic cancer cell lines: HPDE6-C, BxPC3,
PANC1, CFPAC, Capan-2, and AsPC-1 were cultured, and qPCR was used
to detect the miR-29b-3p expression levels. The expression level of
miR-29b-3p in BxPC3 and Capan-2 was found to be significantly lower
than that of the other cell lines (P<0.01) (Fig. 1C). The methylation level of the
miR-29b-3p gene promoter in these six pancreatic cancer cells was
detected by BSP sequencing. It was found that BxPC3 and Capan-2 had
more methylation sites and higher methylation levels (Fig. 1D).
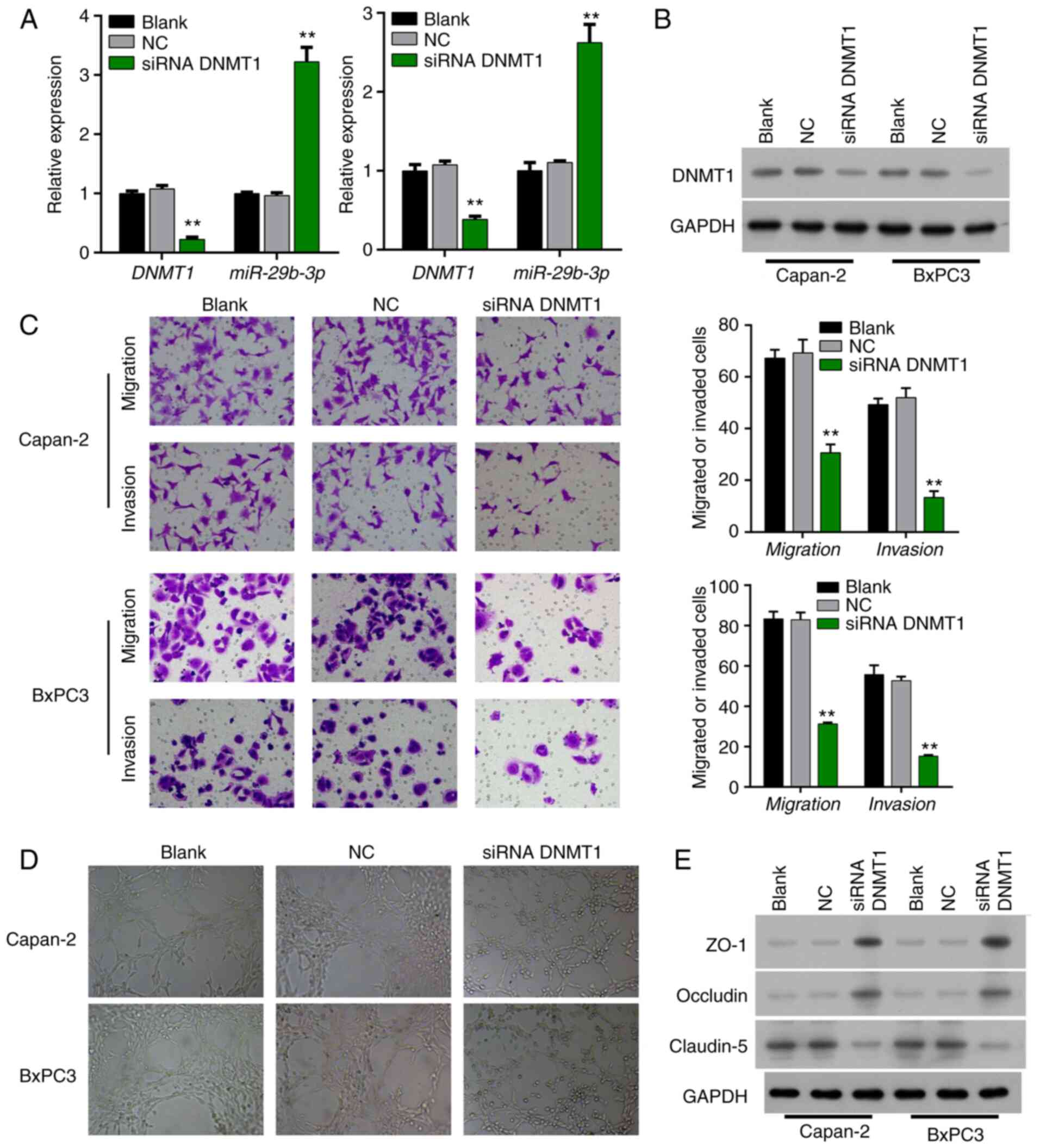
Interference with expression of DNMT1
in Bxpc3 and Capan-2 cells in order to detect angiogenesis,
invasion, and migration of pancreatic cancer cells
siRNA was utilized to interfere with DNMT1 in Bxpc3
and Capan-2 cells. qPCR revealed that the expression level of the
miR-29b-3p gene in the siRNA DNMT1 group was significantly
increased (P<0.001 and P<0.01) (Fig. 2A). Western blot analysis revealed that
expression of DNMT1 was decreased in the DNMT1 siRNA-transfected
Bxpc3 and Capan-2 cells relative to that in the NC transfected
Bxpc3 and Capan-2 cells, indicating that the interference effect
was obvious (Fig. 2B).
Transwell assay showed that the migration and
invasion abilities of pancreatic cancer cells in the siRNA DNMT1
group in Bxpc3 and Capan-2 cells were weakened, and the difference
was statistically significant (P<0.01) (Fig. 2C). Co-culture with HUVECs revealed
that the angiogenic ability of the HUVECs was markedly attenuated
after siRNA interference of DNMT1 expression (Fig. 2D). Western blotting found that the
expression levels of ZO-1 and occludin were increased, and
claudin-5 expression was decreased in the DNMT1 siRNA-transfected
Bxpc3 and Capan-2 cells compared to that in the NC-transfected
Bxpc3 and Capan-2 cells (Fig.
2E).
DNMT1-overexpressing Bxpc3 and Capan-2
cells were cultured, and miR-29b-3p mimic transfection was utilized
in order to detect angiogenesis, invasion, and migration of
pancreatic cancer cells
Results of the qPCR found that DNMT1 expression was
significantly increased (P<0.001), miR-29b-3p expression was
significantly decreased (P<0.05, P<0.001) in the
DNMT1-overexpressed group compared with vector group; while the
expression of DNMT1 was not significantly different in the
DNMT1+miR-29b-3p group from that in the DNMT1 group, the expression
of miR-29b-3p was significantly increased in DNMT1 and miR-29b-3p
co-transfection group relative to that in DNMT1-overexpressed group
(P<0.001) (Fig. 3A). Western
blotting showed that DNMT1 expression in the DNMT1 group was higher
than that in the NC group and the blank group, and the
DNMT1+miR-29b-3p group had lower DNMT1 expression than the DNMT1
group (Fig. 3B). The results
indicated that overexpression and interference were effective.
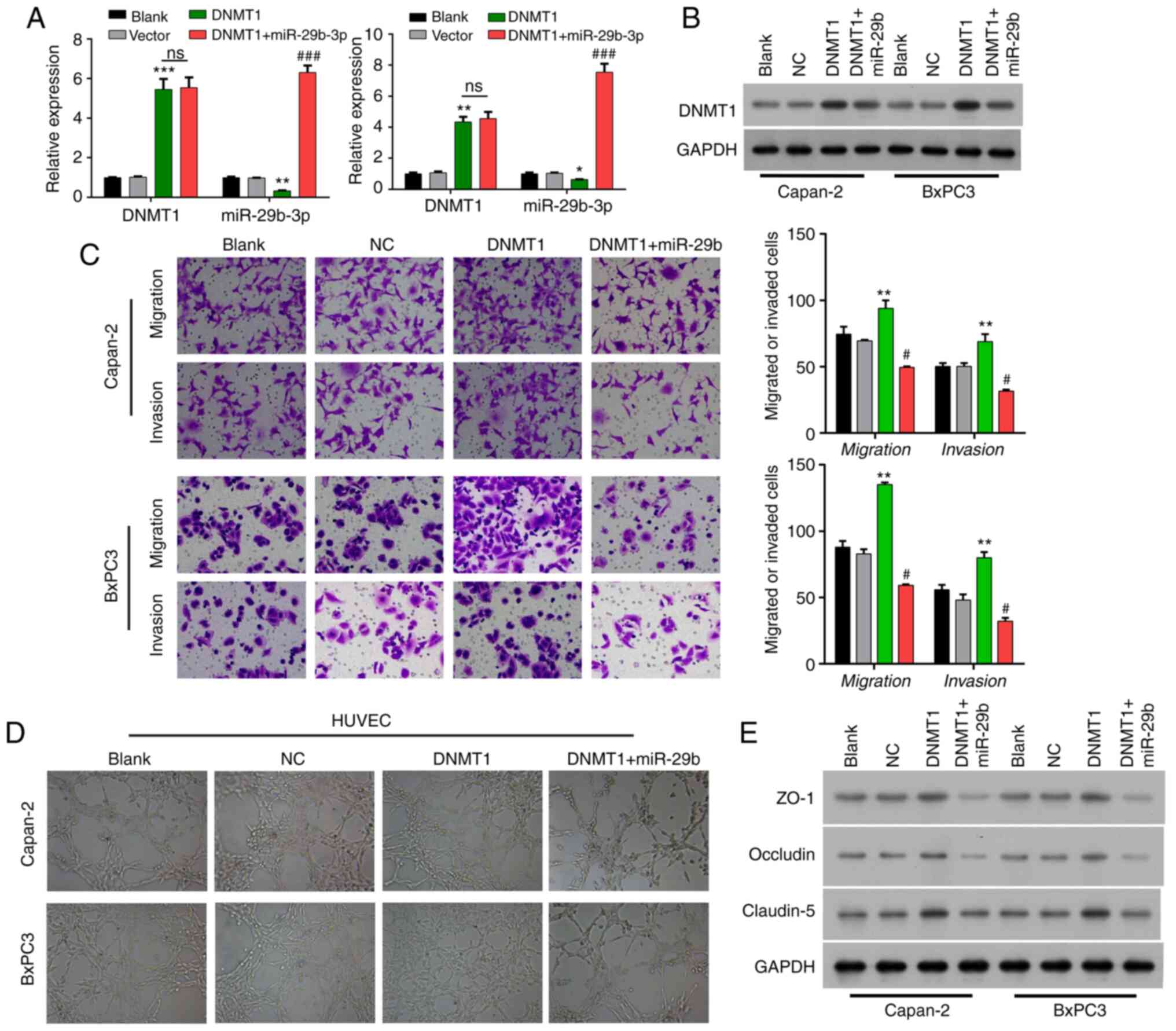 | Figure 3.Methylation of the miR-29b-3p
promoter contributes to angiogenesis, invasion, and migration in
pancreatic cancer. (A) Expression of DNMT1 and miR-29b-3p in cells
transfected with the DNMT1 expression plasmid and co-transfected
with the DNMT1 expression plasmid and miR-29b-3p mimics in BxPC3
and Capan-2 cells. (B) Expression of DNMT1 in cells transfected
with the DNMT1 expression plasmid and co-transfected with the DNMT1
expression plasmid and miR-29b-3p mimics. (C) The migration and
invasive ability of pancreatic cancer cells in cells transfected
with the DNMT1 expression plasmid and co-transfected with the DNMT1
expression plasmid and miR-29b-3p mimics. (D) The angiogenic
ability of HUVECs co-cultured with pancreatic cancer cells
transfected with the DNMT1 expression plasmid and co-transfected
with the DNMT1 expression plasmid and miR-29b-3p mimics. (E)
Expression of ZO-1, claudin-5, and occludin in cells transfected
with the DNMT1 expression plasmid and co-transfected with the DNMT1
expression plasmid and miR-29b-3p mimics. HUVECs, human umbilical
vein endothelial cells; DNMT1, DNA methyltransferase 1; ZO-1,
zonula occludens-1. *P<0.05, **P<0.01, ***P<0.001 vs. the
Vector group; #P<0.05, ###P<0.001 vs.
the DNMT1 group; ns, not significant. |
Transwell assay showed that the migration and
invasive abilities of Bxpc3 and Capan-2 cells were significantly
enhanced in the DNMT1 group vs. that in the vector group, while the
enhancement of migration and invasion capacities mediated by DNMT1
overexpression were significantly weakened by miR-29b-3p in Capan-2
and BxPC3 cells (P<0.01) (Fig.
3C). Co-culture with HUVECs showed that the angiogenic ability
of the HUVECs was enhanced in the DNMT1 group compared with that in
the vector group, which also could be attenuated by miR-29b-3p
addition in Capan-2 and BxPC3 cells (Fig.
3D). Western blotting analysis also discovered that ZO-1 and
occludin expressions were markedly reduced, and claudin-5
expression was dramatically elevated in the DNMT1 overexpression
group relative to that in the vector group, while the addition of
miR-29b-3p then could prominently reverse the expression changes of
ZO-1, occludin and claudin-5 in Bxpc3 and Capan-2 cells (Fig. 3E).
Discussion
DNA methylation in mammals means that methyl (−CH3)
is covalently bound to the carbon atom of the cytosine (C) base of
the DNA molecule under the catalysis of DNA methyltransferases
(DNMTs) (23,24). This usually occurs at the 5-position
carbon atom of cytosine, forming 5-methylcytosine (5Mc), which is
an epigenetic covalent modification process and the main way to
inhibit gene expression and loss of function (24,25).
Promoter methylation is involved in the early stage
of cancer, and the degree of methylation increases with the
increase in structural anomalies (26). Abnormal methylation of specific genes
can be used as an indicator to judge the progression of pancreatic
tumors (27). Numerous studies have
shown that multiple gene methylation abnormalities are often
detected in pancreatic cancer (28–31). It
was also found that in precancerous lesions of pancreatic cancer,
the methylation of the NPTX2 promoter increases with the degree of
abnormal proliferation, suggesting that NPTX2 promoter regional
hypermethylation is associated with early tumorigenesis in
pancreatic cancer (32,33). One study also found that pENK is
highly methylated in pancreatic cancer tissue samples and
pancreatic juice in pancreatic cancer patients, and its methylation
to some extent promotes the formation of pancreatic cancer
(34). In the present study, we found
that the methylation of the miR-29 promoter was involved with
malignant activities of pancreatic cancer cell lines.
Overexpression of DNMT1 resulted in lower expression of miR-29,
which led to cell migration, invasion, and angiogenesis.
MicroRNAs (miRNAs) are a family of non-coding RNAs
that are very conservative and are approximately 15 to 25 nt in
length. In tumor research, according to the target gene of its
downstream action, there are two major types of mircoRNAs, which
are similar to the properties of oncogenes or tumor-suppressor
genes (35). DNA aberrant methylation
causes epigenetic silencing of some microRNAs and plays an
important role in tumorigenesis and development (36). The human microRNA-29 (miRNA-29,
miR-29) family is a group of small RNAs with the same seed sequence
‘AGCACCA’, including miR-29a, miR-29b, and miR-29c. There are
miR-29 expression disorders in various tumor tissues, which are
involved in expression of genes involved in tumor cell metabolism,
proliferation, differentiation, and apoptosis through
post-transcriptional regulation, and have the dual role of oncogene
or tumor-suppressor gene (37).
miR-29b-3p is a member of the miR-29 family and is involved in the
development of pancreatic cancer (38,39),
colorectal cancer (40), lung cancer
(41), bladder cancer (19), and multiple myeloma (42). The relationship between miR-29b-3p
promoter methylation and pancreatic cancer has not yet been
reported. In the present study, we investigated the methylation of
the miR-29b-3p promoter in pancreatic cancer and its expression
level, and explored the effect of miR-29b-3p promoter methylation
on angiogenesis, invasion, and migration of pancreatic cancer, thus
providing a new theoretical basis for the treatment of pancreatic
cancer.
It was found that the methylation level of the
miR-29b-3p promoter region in pancreatic cancer tissues was
significantly higher than that in adjacent tissues. In addition,
the expression level of miR-29b-3p was significantly decreased,
which was negatively correlated with the methylation level of its
promoter. CpG methyltransferases (DNMTs) play a key role in DNA
methylation, including DNMT1, DNMT2 DNMT3a, and DNMT3b. DNMT1 is
the most important catalytic enzyme in the DNMT family. DNMT1 is
associated with abnormal methylation of DNA and both are closely
related to the occurrence and development of tumors (43,44). The
expression level of the DNMT1 protein in pancreatic cancer tissues
was higher than that in adjacent tissues, suggesting that DNMT1
promotes promoter region methylation of the miR-29b-3p gene. siRNA
was used to interfere with DNMT1 in Bxpc3 and Capan-2 cell lines,
and expression of miR-29b-3p was significantly increased. We
cultured DNMT1-overexpressing Bxpc3 and Capan-2, and expression of
miR-29b-3p was significantly decreased. The above experiments
proved that the methylation degree of the miR-29b-3p gene in
pancreatic cancer leads to a change in its gene expression level,
and the hypermethylation of the miR-29b-3p gene leads to its low
expression.
Angiogenesis is the budding and subsequent
stabilization of existing vascular wall cells (45). In 1973, FoIkman first discovered that
tumor cells induce angiogenesis and rapid growth, and since then,
more and more attention has been paid to solid tumor angiogenesis
(46). The vascular endothelial
growth factor (VEGF), insulin-like growth factor 1 (IGF1), and
other factors can play a role in promoting tumor angiogenesis,
which is the basis of malignant tumor growth and metastasis
(47–49). Zhang et al (50) found that exogenous low expression of
miR-29a/c can increase expression and release of VEGF in gastric
cancer cells, and promote the growth of vascular endothelial cells.
Melo and Kalluri (51) found that
miR-29b can inhibit the signaling molecules involved in
angiogenesis and the extracellular matrix, such as VEGF, MMP9,
ANGPTL4, and lysyloxidase (LOX), thereby inhibiting tumor
angiogenesis and metastasis. This study investigated the role of
miR-29b-3p in angiogenesis in pancreatic cancer cells, and found
that miR-29b-3p inhibits angiogenesis and pancreatic cancer cell
migration and invasion, and after inhibition of miR-29b-3p, the
migration and invasive ability of pancreatic cancer cells
increased. In this study, we aimed to investigate the role of DNMT1
and miR-29b-3p in pancreatic cancer, on cell migration, invasion
and angiogenesis. However, the effect of DNMT1/miR-29b-3p on cell
apoptosis and cycle was not investigated in the present study.
Based on previous research, DNMT1 siRNA induces a significant cell
viability decrease, leads to a G2-phase block and cell apoptosis in
pancreatic cancer (10,52), indicating that this axis may promote
cell survival. Further study will focus on this aspect.
In conclusion, methylation of the miR-29b-3p
promoter contributes to angiogenesis, invasion, and migration in
pancreatic cancer. Its molecular mechanisms of regulating
tumorigenesis and development need to be further studied.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LW designed the experiments. LW and NM performed the
experiments, collected the data and analyzed the data. LW drafted
the manuscript, and NQ validated the data analysis and revised the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Yantai Yuhuangding Hospital of Qingdao University, and the ethics
approval number is QDU-201902-3. Written informed consent was
obtained from each participant.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhu H, Li T, Du Y and Li M: Pancreatic
cancer: Challenges and opportunities. BMC Med. 16:2142018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bogdanović O and Lister R: DNA methylation
and the preservation of cell identity. Curr Opin Genet Dev.
46:9–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang MW, Fujiwara K, Che X, Zheng S and
Zheng L: DNA methylation in the tumor microenvironment. J Zhejiang
Univ Sci B. 18:365–372. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Matsubayashi H, Canto M, Sato N, Klein A,
Abe T, Yamashita K, Yeo CJ, Kalloo A, Hruban R and Goggins M: DNA
methylation alterations in the pancreatic juice of patients with
suspected pancreatic disease. Cancer Res. 66:1208–1217. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pizzi S, Azzoni C, Bottarelli L, Campanini
N, D'Adda T, Pasquali C, Rossi G, Rindi G and Bordi C: RASSF1A
promoter methylation and 3p21.3 loss of heterozygosity are features
of foregut, but not midgut and hindgut, malignant endocrine
tumours. J Pathol. 206:409–416. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bird A: DNA methylation patterns and
epigenetic memory. Genes Dev. 16:6–21. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hong L, Sun G, Peng L, Tu Y, Wan Z, Xiong
H, Li Y and Xiao W: The interaction between miR-148a and DNMT1
suppresses cell migration and invasion by reactivating tumor
suppressor genes in pancreatic cancer. Oncol Rep. 40:2916–2925.
2018.PubMed/NCBI
|
|
9
|
Li A, Omura N, Hong SM and Goggins M:
Pancreatic cancer DNMT1 expression and sensitivity to DNMT1
inhibitors. Cancer Biol Ther. 9:321–329. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu M, Gao J, Du YQ, Gao DJ, Zhang YQ, Li
ZS, Zhang YL, Gong YF and Xu P: Reduction of pancreatic cancer cell
viability and induction of apoptosis mediated by siRNA targeting
DNMT1 through suppression of total DNA methyltransferase activity.
Mol Med Rep. 3:699–704. 2010.PubMed/NCBI
|
|
11
|
Ikegami K, Ohgane J, Tanaka S, Yagi S and
Shiota K: Interplay between DNA methylation, histone modification
and chromatin remodeling in stem cells and during development. Int
J Dev Biol. 53:203–214. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cui J, Zhou B, Ross SA and Zempleni J:
Nutrition, microRNAs, and human health. Adv Nutr. 8:105–112. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao J, Dong X, Liu QC and Lu Q:
Expression of plasma miR-106a in epithelial ovarian cancer and its
diagnostic and prognostic significance. Eur J Gynaecol Oncol.
39:769–772. 2018.
|
|
15
|
Ganju A, Khan S, Hafeez BB, Behrman SW,
Yallapu MM, Chauhan SC and Jaggi M: miRNA nanotherapeutics for
cancer. Drug Discov Today. 22:424–432. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qadir MI and Faheem A: miRNA: A diagnostic
and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene
Expr. 27:197–204. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li X, Xiao J, Fan Y, Yang K, Li K, Wang X,
Lu Y and Zhou Y: miR-29 family regulates the puberty onset mediated
by a novel Gnrh1 transcription factor TBX21. J Endocrinol.
242:185–197. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ding D, Li C, Zhao T, Li D, Yang L and
Zhang B: lncRNA H19/miR-29b-3p/PGRN axis promoted
epithelial-mesenchymal transition of colorectal cancer cells by
acting on wnt signaling. Mol Cells. 41:423–435. 2018.PubMed/NCBI
|
|
19
|
Lv M, Zhong Z, Huang M, Tian Q, Jiang R
and Chen J: lncRNA H19 regulates epithelial-mesenchymal transition
and metastasis of bladder cancer by miR-29b-3p as competing
endogenous RNA. Biochim Biophys Acta Mol Cell Res. 1864:1887–1899.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Worst TS, Previti C, Nitschke K, Diessl N,
Gross JC, Hoffmann L, Frey L, Thomas V, Kahlert C, Bieback K, et
al: miR-10a-5p and miR-29b-3p as extracellular vesicle-associated
prostate cancer detection markers. Cancers (Basel). 12:432019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang B, Shetti D, Fan C and Wei K:
miR-29b-3p promotes progression of MDA-MB-231 triple-negative
breast cancer cells through downregulating TRAF3. Biol Res.
52:382019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Feng L and Lou J: DNA methylation
analysis. Methods Mol Biol. 1894:181–227. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kasai H and Kawai K: DNA methylation at
the C-5 position of cytosine by methyl radicals: A possible role
for epigenetic change during carcinogenesis by environmental
agents. Chem Res Toxicol. 22:984–989. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Przybilla J, Hopp L, Lübbert M, Loeffler M
and Galle J: Targeting DNA hypermethylation: Computational modeling
of DNA demethylation treatment of acute myeloid leukemia.
Epigenetics. 12:886–896. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kotandeniya D, Seiler CL, Fernandez J,
Pujari SS, Curwick L, Murphy K, Wickramaratne S, Yan S, Murphy D,
Sham YY and Tretyakova NY: Can 5-methylcytosine analogues with
extended alkyl side chains guide DNA methylation? Chem Commun
(Camb). 54:1061–1064. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yamashita K, Hosoda K, Nishizawa N, Katoh
H and Watanabe M: Epigenetic biomarkers of promoter DNA methylation
in the new era of cancer treatment. Cancer Sci. 109:3695–3706.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Van Tongelen A, Loriot A and De Smet C:
Oncogenic roles of DNA hypomethylation through the activation of
cancer-germline genes. Cancer Lett. 396:130–137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brancaccio M, Natale F, Falco G and
Angrisano T: Cell-free DNA methylation: The new frontiers of
pancreatic cancer biomarkers' discovery. Genes (Basel). 11:142019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mishra NK and Guda C: Genome-wide DNA
methylation analysis reveals molecular subtypes of pancreatic
cancer. Oncotarget. 8:28990–29012. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Natale F, Vivo M, Falco G and Angrisano T:
Deciphering DNA methylation signatures of pancreatic cancer and
pancreatitis. Clin Epigenetics. 11:1322019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou H, Zhu Y, Wei F, Shao Y, Pan J, Wang
G, Xu K and Cheng Y: Significance of MUC2 gene methylation
detection in pancreatic cancer diagnosis. Pancreatology.
19:1049–1053. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Park JK, Ji KR, Kim YT and Yong BY: Early
diagnosis and aberrant methylation of NPTX2 gene in pancreatic
cancer. Cancer Res. 68:55142008.
|
|
33
|
Ling Z, Gao J, Li Z and Gong Y: Neuronal
pentraxin II (NPTX2) is frequently down-regulated by promoter
hypermethylation in pancreatic cancers. Dig Dis Sci. 57:2608–2614.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Singh N, Rashid S, Rashid S, Dash NR,
Gupta S and Saraya A: Clinical significance of promoter methylation
status of tumor suppressor genes in circulating DNA of pancreatic
cancer patients. J Cancer Res Clin Oncol. 146:897–907. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Moutinho C and Esteller M: MicroRNAs and
Epigenetics. Adv Cancer Res. 135:189–220. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jiang H, Zhang G, Wu JH and Jiang CP:
Diverse roles of miR-29 in cancer (Review). Oncol Rep.
31:1509–1516. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao X, Liu Y, Li Z, Zheng S, Wang Z, Li
W, Bi Z, Li L, Jiang Y, Luo Y, et al: Linc00511 acts as a competing
endogenous RNA to regulate VEGFA expression through sponging
hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. J Cell Mol Med.
22:655–667. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun Y, Wang P, Yang W, Shan Y, Zhang Q and
Wu H: The role of lncRNA MSC-AS1/miR-29b-3p axis-mediated CDK14
modulation in pancreatic cancer proliferation and
Gemcitabine-induced apoptosis. Cancer Biol Ther. 20:729–739. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Osawa T, Takeuchi A, Kojima T, Shinohara
N, Eto M and Nishiyama H: Overview of current and future systemic
therapy for metastatic renal cell carcinoma. Jpn J Clin Oncol.
49:395–403. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li L, Liu Z, Jiang YY, Shen WX, Peng YP
and Qiu YH: Acetylcholine suppresses microglial inflammatory
response via α7nAChR to protect hippocampal neurons. J Integr
Neurosci. 18:51–56. 2019.PubMed/NCBI
|
|
42
|
Liu D, Wang J and Liu M: Long noncoding
RNA TUG1 promotes proliferation and inhibits apoptosis in multiple
myeloma by inhibiting miR-29b-3p. Biosci Rep. 39:BSR201824892019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim DH, Kim HM, Huong PTT, Han HJ, Hwang
J, Cha-Molstad H, Lee KH, Ryoo IJ, Kim KE, Huh YH, et al: Enhanced
anticancer effects of a methylation inhibitor by inhibiting a novel
DNMT1 target, CEP 131, in cervical cancer. BMB Rep. 52:342–347.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Somasundaram S, Forrest ME, Moinova H,
Cohen A, Varadan V, LaFramboise T, Markowitz S and Khalil AM: The
DNMT1-associated lincRNA DACOR1 reprograms genome-wide DNA
methylation in colon cancer. Clin Epigenetics. 10:1272018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Risau W: Mechanisms of angiogenesis.
Nature. 386:671–674. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Folkman J: Tumor angiogenesis. Adv Cancer
Res. 19:331–358. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tang X, Zhang Q, Shi S, Yen Y, Li X, Zhang
Y, Zhou K and Le AD: Bisphosphonates suppress insulin-like growth
factor 1-induced angiogenesis via the HIF-1alpha/VEGF signaling
pathways in human breast cancer cells. Int J Cancer. 126:90–103.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhou Y, Li S, Li J, Wang D and Li Q:
Effect of microRNA-135a on cell proliferation, migration, invasion,
apoptosis and tumor angiogenesis through the IGF-1/PI3K/Akt
signaling pathway in non-small cell lung cancer. Cell Physiol
Biochem. 42:1431–1446. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Azuma K, Suzuki S, Ishii Y, Ueda Y,
Fujikawa T, Morinaga K, Shimoyama K and Oda J: Tortuosity of the
brachiocephalic artery complicated with arterial injury after
tracheotomy: A case report. Signa Vitae. 15:77–78. 2019. View Article : Google Scholar
|
|
50
|
Zhang H, Bai M, Deng T, Liu R, Wang X, Qu
Y, Duan J, Zhang L, Ning T, Ge S, et al: Cell-derived microvesicles
mediate the delivery of miR-29a/c to suppress angiogenesis in
gastric carcinoma. Cancer Lett. 375:331–339. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Melo SA and Kalluri R: miR-29b moulds the
tumour microenvironment to repress metastasis. Nat Cell Biol.
15:139–140. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Thakar M, Hu Y, Morreale M, Lerner L, Ying
Lin W, Sen R, Cai Y, Karunasena E, Thakar M, Saggi S, et al: A
novel epigenetic modulating agent sensitizes pancreatic cells to a
chemotherapy agent. PLoS One. 13:e01991302018. View Article : Google Scholar : PubMed/NCBI
|