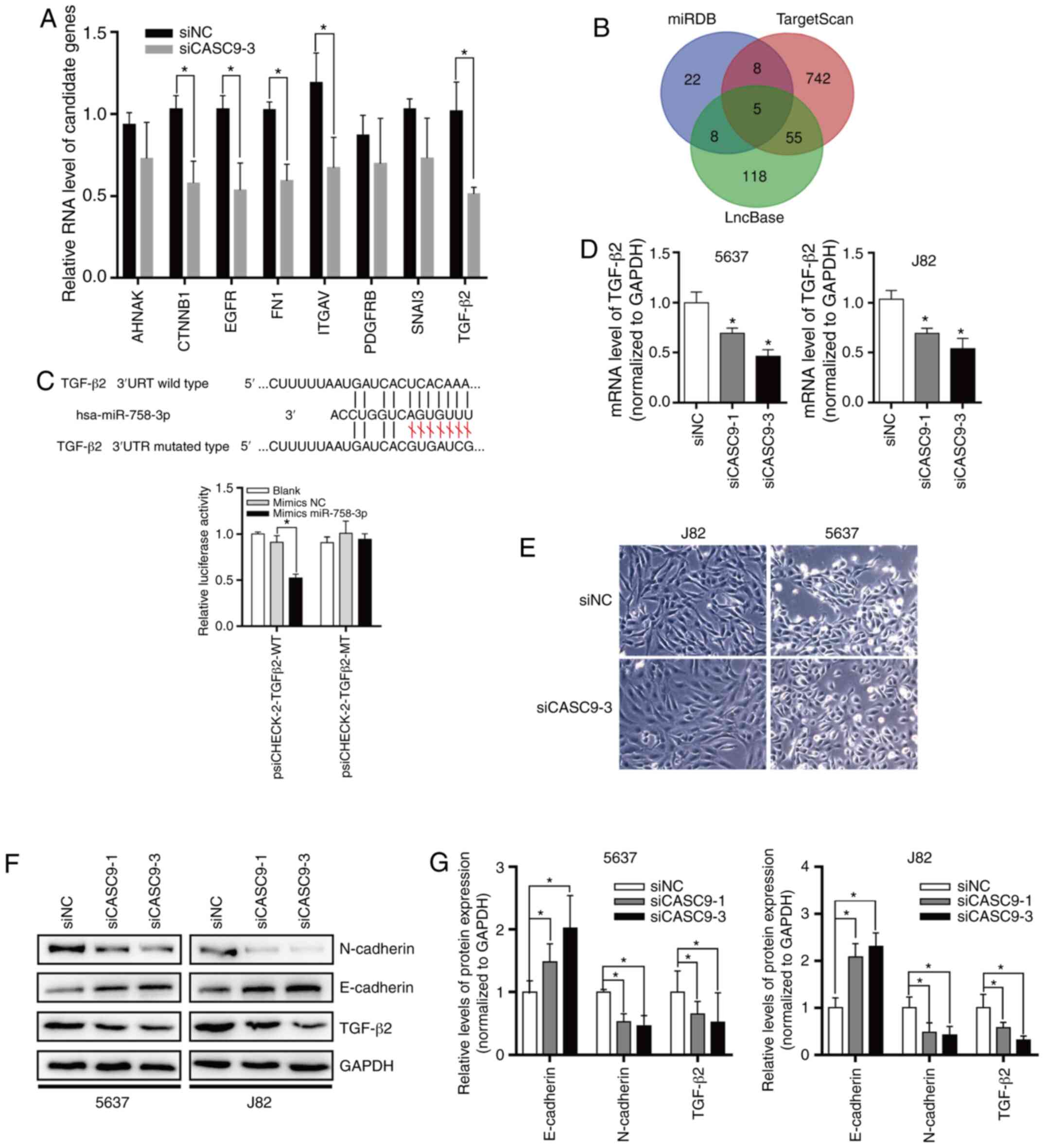
|
1
|
Richters A, Aben KKH and Kiemeney L: The
global burden of urinary bladder cancer: An update. World J Urol.
38:1895–1904. 2020. View Article : Google Scholar
|
|
2
|
Grayson M: Bladder cancer. Nature. 551
(Suppl 1):S332017. View
Article : Google Scholar
|
|
3
|
Smith ZL and Guzzo TJ: Urinary markers for
bladder cancer. F1000Prime Rep. 5:212013. View Article : Google Scholar
|
|
4
|
Kamat AM, Hegarty PK, Gee JR, Clark PE,
Svatek RS, Hegarty N, Shariat SF, Xylinas E, Schmitz-Dräger BJ,
Lotan Y, et al: ICUD-EAU international consultation on bladder
cancer 2012: Screening, diagnosis, and molecular markers. Eur Urol.
63:4–15. 2013. View Article : Google Scholar
|
|
5
|
Santamaria PG, Moreno-Bueno G, Portillo F
and Cano A: EMT: Present and future in clinical oncology. Mol
Oncol. 11:718–738. 2017. View Article : Google Scholar
|
|
6
|
Singh M, Yelle N, Venugopal C and Singh
SK: EMT: Mechanisms and therapeutic implications. Pharmacol Ther.
182:80–94. 2018. View Article : Google Scholar
|
|
7
|
Du B and Shim JS: Targeting
epithelial-mesenchymal transition (EMT) to overcome drug resistance
in cancer. Molecules. 21:9652016. View Article : Google Scholar
|
|
8
|
Pastushenko I, Brisebarre A, Sifrim A,
Fioramonti M, Revenco T, Boumahdi S, Van Keymeulen A, Brown D,
Moers V, Lemaire S, et al: Identification of the tumour transition
states occurring during EMT. Nature. 556:463–468. 2018. View Article : Google Scholar
|
|
9
|
Dongre A and Weinberg RA: New insights
into the mechanisms of epithelial-mesenchymal transition and
implications for cancer. Nat Rev Mol Cell Biol. 20:69–84. 2019.
View Article : Google Scholar
|
|
10
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar
|
|
11
|
Xu J, Lamouille S and Derynck R:
TGF-beta-induced epithelial to mesenchymal transition. Cell Res.
19:156–172. 2009. View Article : Google Scholar
|
|
12
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: TGF-β-induced upregulation of malat1 promotes bladder
cancer metastasis by associating with suz12. Clin Cancer Res.
20:1531–1541. 2014. View Article : Google Scholar
|
|
13
|
Calon A, Espinet E, Palomo-Ponce S,
Tauriello DV, Iglesias M, Céspedes MV, Sevillano M, Nadal C, Jung
P, Zhang XH, et al: Dependency of colorectal cancer on a
TGF-β-driven program in stromal cells for metastasis initiation.
Cancer Cell. 22:571–584. 2012. View Article : Google Scholar
|
|
14
|
Papageorgis P: TGFβ signaling in tumor
initiation, epithelial-to-mesenchymal transition, and metastasis. J
Oncol. 2015:5871932015. View Article : Google Scholar
|
|
15
|
Kapranov P, Cheng J, Dike S, Nix DA,
Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J,
Hofacker IL, et al: RNA maps reveal new RNA classes and a possible
function for pervasive transcription. Science. 316:1484–1488. 2007.
View Article : Google Scholar
|
|
16
|
Schmitz SU, Grote P and Herrmann BG:
Mechanisms of long noncoding RNA function in development and
disease. Cell Mol Life Sci. 73:2491–2509. 2016. View Article : Google Scholar
|
|
17
|
Xiong T, Li J, Chen F and Zhang F: PCAT-1:
A novel oncogenic Long Non-coding RNA in human cancers. Int J Biol
Sci. 15:847–856. 2019. View Article : Google Scholar
|
|
18
|
Flynn RA and Chang HY: Long noncoding RNAs
in cell-fate programming and reprogramming. Cell Stem Cell.
14:752–761. 2014. View Article : Google Scholar
|
|
19
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar
|
|
20
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar
|
|
21
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar
|
|
22
|
Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao
SD, Zhang Y, Yang L, Shan W, He Q, et al: Comprehensive genomic
characterization of long non-coding RNAs across human cancers.
Cancer Cell. 28:529–540. 2015. View Article : Google Scholar
|
|
23
|
Luo M, Li Z, Wang W, Zeng Y, Liu Z and Qiu
J: Long non-coding RNA H19 increases bladder cancer metastasis by
associating with EZH2 and inhibiting E-cadherin expression. Cancer
Lett. 333:213–221. 2013. View Article : Google Scholar
|
|
24
|
Han Y, Liu Y, Nie L, Gui Y and Cai Z:
Inducing cell proliferation inhibition, apoptosis, and motility
reduction by silencing long noncoding ribonucleic acid
metastasis-associated lung adenocarcinoma transcript 1 in
urothelial carcinoma of the bladder. Urology. 81:209.e1–e7. 2013.
View Article : Google Scholar
|
|
25
|
Han Y, Liu Y, Gui Y and Cai Z: Long
intergenic non-coding RNA TUG1 is overexpressed in urothelial
carcinoma of the bladder. J Surg Oncol. 107:555–559. 2013.
View Article : Google Scholar
|
|
26
|
Yang C, Li X, Wang Y, Zhao L and Chen W:
Long non-coding RNA UCA1 regulated cell cycle distribution via CREB
through PI3-K dependent pathway in bladder carcinoma cells. Gene.
496:8–16. 2012. View Article : Google Scholar
|
|
27
|
Yan TH, Lu SW, Huang YQ, Que GB, Chen JH,
Chen YP, Zhang HB, Liang XL and Jiang JH: Upregulation of the long
noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder
cancer. Tumour Biol. 35:10249–10257. 2014. View Article : Google Scholar
|
|
28
|
Pan Z, Mao W, Bao Y, Zhang M, Su X and Xu
X: The long noncoding RNA CASC9 regulates migration and invasion in
esophageal cancer. Cancer Med. 5:2442–2447. 2016. View Article : Google Scholar
|
|
29
|
Wu Y, Hu L, Liang Y, Li J, Wang K, Chen X,
Meng H, Guan X, Yang K and Bai Y: Up-regulation of lncRNA CASC9
promotes esophageal squamous cell carcinoma growth by negatively
regulating PDCD4 expression through EZH2. Mol Cancer. 16:1502017.
View Article : Google Scholar
|
|
30
|
Liang Y, Chen X, Wu Y, Li J, Zhang S, Wang
K, Guan X, Yang K and Bai Y: LncRNA CASC9 promotes esophageal
squamous cell carcinoma metastasis through upregulating LAMC2
expression by interacting with the CREB-binding protein. Cell Death
Differ. 25:1980–1995. 2018. View Article : Google Scholar
|
|
31
|
Yu X, Lin Y, Sui W, Zou Y and Lv Z:
Analysis of distinct long noncoding RNA transcriptional
fingerprints in pancreatic ductal adenocarcinoma. Cancer Med.
6:673–680. 2017. View Article : Google Scholar
|
|
32
|
Shang C, Sun L, Zhang J, Zhao B, Chen X,
Xu H and Huang B: Silence of cancer susceptibility candidate 9
inhibits gastric cancer and reverses chemoresistance. Oncotarget.
8:15393–15398. 2017. View Article : Google Scholar
|
|
33
|
Su X, Li G and Liu W: The long noncoding
RNA cancer susceptibility candidate 9 promotes nasopharyngeal
carcinogenesis via stabilizing HIF1α. DNA Cell Biol. 36:394–400.
2017. View Article : Google Scholar
|
|
34
|
Ma P, Zhang M, Nie F, Huang Z, He J, Li W
and Han L: Transcriptome analysis of EGFR tyrosine kinase
inhibitors resistance associated long noncoding RNA in non-small
cell lung cancer. Biomed Pharmacother. 87:20–26. 2017. View Article : Google Scholar
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative pcr and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
36
|
Li Y, Quan J, Chen F, Pan X, Zhuang C,
Xiong T, Zhuang C, Li J, Huang X, Ye J, et al: MiR-31-5p acts as a
tumor suppressor in renal cell carcinoma by targeting
cyclin-dependent kinase 1 (CDK1). Biomed Pharmacother. 111:517–526.
2019. View Article : Google Scholar
|
|
37
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: Gepia: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar
|
|
38
|
Paraskevopoulou MD, Vlachos IS, Karagkouni
D, Georgakilas G, Kanellos I, Vergoulis T, Zagganas K, Tsanakas P,
Floros E, Dalamagas T and Hatzigeorgiou AG: Diana-lncbase v2:
Indexing microrna targets on non-coding transcripts. Nucleic Acids
Res. 44:D231–D238. 2016. View Article : Google Scholar
|
|
39
|
Chen Y and Wang X: MiRDB: An online
database for prediction of functional microRNA targets. Nucleic
Acids Res. 48:D127–D131. 2020. View Article : Google Scholar
|
|
40
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar
|
|
41
|
McConkey DJ, Choi W, Marquis L, Martin F,
Williams MB, Shah J, Svatek R, Das A, Adam L, Kamat A, et al: Role
of epithelial-to-mesenchymal transition (EMT) in drug sensitivity
and metastasis in bladder cancer. Cancer Metastasis Rev.
28:335–344. 2009. View Article : Google Scholar
|
|
42
|
Chandra Gupta S and Nandan Tripathi Y:
Potential of long non-coding RNAs in cancer patients: From
biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967.
2017. View Article : Google Scholar
|
|
43
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar
|
|
44
|
Kumar MS, Armenteros-Monterroso E, East P,
Chakravorty P, Matthews N, Winslow MM and Downward J: HMGA2
functions as a competing endogenous RNA to promote lung cancer
progression. Nature. 505:212–217. 2014. View Article : Google Scholar
|
|
45
|
Jeyapalan Z, Deng Z, Shatseva T, Fang L,
He C and Yang BB: Expression of CD44 3′-untranslated region
regulates endogenous microRNA functions in tumorigenesis and
angiogenesis. Nucleic Acids Res. 39:3026–3041. 2011. View Article : Google Scholar
|
|
46
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar
|
|
47
|
Chen J, Xu Z, Yu C, Wu Z, Yin Z, Fang F
and Chen B: MiR-758-3p regulates papillary thyroid cancer cell
proliferation and migration by targeting TAB1. Pharmazie.
74:235–238. 2019.
|
|
48
|
Guo J, Zhang Z, Pan L and Zhou Y:
Identification of miR-758-3p as potential modulator of CBX5
expression in gastric cancer. Technol Cancer Res Treat.
17:15330338188160612018. View Article : Google Scholar
|
|
49
|
Wang S and Jiang M: The long non-coding
RNA-DANCR exerts oncogenic functions in non-small cell lung cancer
via miR-758-3p. Biomed Pharmacother. 103:94–100. 2018. View Article : Google Scholar
|
|
50
|
Gupta S, Hau AM, Al-Ahmadie HA, Harwalkar
J, Shoskes AC, Elson P, Beach JR, Hussey GS, Schiemann WP, Egelhoff
TT, et al: Transforming growth factor-beta is an upstream regulator
of mammalian target of rapamycin complex 2-dependent bladder cancer
cell migration and invasion. Am J Pathol. 186:1351–1360. 2016.
View Article : Google Scholar
|
|
51
|
Zhang C, Zhang X, Xu R, Huang B, Chen AJ,
Li C, Wang J and Li XG: TGF-β2 initiates autophagy via Smad and
non-Smad pathway to promote glioma cells' invasion. J Exp Clin
Cancer Res. 36:1622017. View Article : Google Scholar
|