Introduction
Exosomes are membrane vesicles with a diameter of
30–100 nm and constitute a subset of extracellular vesicles. As
exosomes carry various bioactive molecules, such as enzymes,
cytokines, eicosanoids and small RNAs, they serve a key role in
intercellular communication (1).
Moreover, exosomes are unique in that they are formed and secreted
by the cellular endosomal pathway. They are subsequently sorted
based on different membrane-trafficking routes that involve
recycling of exosomes back to the plasma membrane and formation of
late endosomes and their degradation in lysosomes or integration
into exosomes in the multi-vesicular body (2). Exosomes also serve critical roles in
cancer progression, intercellular communication, tumor-stromal
interactions, activation of signaling pathways and
immunomodulation, and may have crucial functions that are currently
unknown (1,3).
Reactive oxygen species (ROS) levels are increased
in many types of cancer cell, such as breast and colon cancer cells
(4,5)
compared with normal cells (6). While
a moderate increase in ROS levels can promote cell proliferation
and differentiation (7), high ROS
levels can cause oxidative damage to lipids, proteins and DNA;
therefore, maintaining ROS homeostasis is crucial. Cells maintain
ROS homeostasis by balancing ROS generation and elimination via
antioxidant molecules, such as superoxide dismutase (SOD) (8). Radiotherapy potently induces massive
cell death by triggering apoptosis in cancer cells via the
generation of ROS (9), such as
superoxide anions and hydroxyl radicals and hydrogen peroxide
(H2O2) following the radiolysis of water in
the extracellular environment; these highly reactive entities are
toxic to both cancer cells and surrounding normal tissue (10). In our previous study (11), MIAPaCa-2 cells were found to be more
radio-resistant than other pancreatic cancer cell lines. Moreover,
Doskey et al (12) reported
that the capabilities or rate constants for ROS reduction differed
between 15 tumor and 10 normal cell lines of various tissue types,
and that the MIAPaCa-2 cell line had the smallest rate constants
and catalase activity for H2O2 removal.
MicroRNAs (miRNAs or miRs) are small non-coding RNAs
composed of 18–22 nucleotides that perform a regulatory role by
binding to target mRNAs via multiple imperfect base pairings within
3′-untranslated region (3′-UTR). miRNAs have a wide range of
targets that allow them to modulate many pathways involved in
cancer progression, including cell proliferation, apoptosis,
metastasis and angiogenesis (13).
They are differentially expressed in normal and cancer cells;
certain miRNAs act as tumor suppressors while others serve as
oncogenes, thus promoting tumor initiation and progression
(14). Expression levels of miRNAs
are altered in both radiation-exposed cancer cells (15–17) and
normal cells (18,19) and their expression profiles are
modulated in response to DNA damage (20,21).
However, whether these altered miRNAs are delivered to recipient
cells via exosomes, thus contributing to cell-to-cell
communication, remains unclear.
Irradiated cells generate communication signals and
subsequently cause biological changes in neighboring or distant
non-irradiated cells; this phenomenon is referred to as the
radiation-induced bystander effect (RIBE). A variety of signaling
molecules, such as ROS (22),
cytokines (23,24) and exosomes (25), are initiators of such bystander
responses. However, the role of exosomes in RIBE and the
association between ROS and exosomes remain unclear.
The present study evaluated the role of exosomes in
the radiation response by investigating intracellular ROS and
antioxidant levels, DNA damage, and cell survival using the human
pancreatic cancer cell line MIAPaCa-2.
Materials and methods
Cell culture
The MIAPaCa-2 human pancreatic cancer cell line was
obtained from Japanese Collection of Research Bioresources Cell
Bank (Tokyo, Japan) and maintained in minimal essential medium
(MEM) supplemented with 10% (vol/vol) fetal bovine serum (both
Sigma-Aldrich; Merck KGaA), 1% penicillin-streptomycin mix and 1%
MEM non-essential amino acid solutions (100X; both Nacalai Tesque,
Inc.) in a humidified atmosphere containing 5% CO2 at
37°C. The doubling time of MIAPaCa-2 cells was 20–23 h (26).
Reagents
The following antibodies were purchased:
Anti-cytochrome c and anti-phosphorylated histone H2AX
(γ-H2AX) from Cell Signaling Technology, Inc.; anti-CD63 from BD
Biosciences; donkey anti-goat IgG, F(ab′)2-horseradish
peroxidase (HRP), HRP-conjugated mouse IgGκ light chain binding
protein (m-IgGκ BP), anti-actin and anti-CD9 from Santa Cruz
Biotechnology, Inc.; anti-SOD1 and anti-SOD2 from Merck KGaA; and
rabbit anti-sheep IgG-HRP and tetramethyl rhodamine isothiocyanate
(TRITC)-conjugated anti-rabbit secondary antibody from Dako
(Agilent Technologies, Inc.). The following reagents were
purchased: PKH-67, a lipophilic dye, and N-acetyl-L-cysteine (NAC)
from Sigma-Aldrich; Merck KGaA; Hoechst 33342 and
2′,7′-dichlorodihydrofluorescein diacetate (C-H2DCF)
from Thermo Fisher Scientific, Inc.; methylene blue from FUJIFILM
Wako Pure Chemical Corporation; and DAPI and wheat germ agglutinin
(WGA), Alexa Fluor 594 conjugate from Invitrogen (Thermo Fisher
Scientific, Inc.).
Isolation and morphological evaluation
of exosomes
Exosomes were isolated from media-conditioned cells
by ultracentrifugation, as previously described (27). Briefly, MIAPaCa-2 cells were seeded at
1.5×106 cells per T75 cm2 flask and
irradiated after substituting the media with exosome-depleted 10%
FBS cell culture media (Sigma-Aldrich; Merck KGaA). The cell
culture media was centrifuged at 2,000 × g for 10 min at 4°C and
the supernatant was filtered through a 0.22-µm Minisart®
syringe filter (Sartorius AG). The supernatant was centrifuged at
150,000 × g for 90 min at 4°C. The pellet was washed with PBS and
again centrifuged at 150,000 × g for 90 min at 4°C and resuspended
in 50 µl PBS. Then, the total amount of protein in the exosomes was
measured using a Qubit™ Protein Assay kit (Invitrogen; Thermo
Fisher Scientific, Inc.).
The exosomes isolated from non-irradiated (0 Gy-Exo)
and 5 Gy irradiated cells (5 Gy-Exo) were evaluated by transmission
electron microscopy (TEM). Briefly, 4 µl PBS suspension of isolated
exosomes was loaded onto carbon-coated 200-mesh copper grids for 1
min at 25°C. Excessive fluid was removed using filter paper. The
adsorbed exosomes were negatively stained with 2% uranyl acetate
for 30 sec at 25°C. Finally, the air-dried exosome-containing grids
were observed under a TEM microscope (JEM-1400 plus; JEOL, Ltd.) at
120 kV (magnification, ×50,000). Exosome size, concentration and
distribution were analyzed by nanoparticle tracking analysis (NTA)
software using NanoSight NS300 (Malvern Panalytical). The software
was optimized to identify and track each particle on a
frame-by-frame basis and Brownian movement was tracked and measured
from frame to frame.
Cellular internalization analysis
Exosomes were labeled with the green fluorescent dye
PKH-67 using the PKH67 Green Fluorescent Cell Linker kit for
General Cell Membrane Labeling (Sigma-Aldrich; Merck KGaA) as
previously described (28). Briefly,
6 µg 0 and 5 Gy-Exo were labeled with 2 µM PKH-67 for 5 min at
25°C. Then, free PKH-67 was removed by centrifugation 14,000 × g
for 2 min at 25°C using the VIVACON 500 ultracentrifugation device
(100,000 MWCO; Sartorius Stedim Biotech; Sartorius AG). MIAPaCa-2
cells were cultured for 24 h at 37°C, after which the culture media
was replaced with that containing the labeled exosomes. Following
incubation at 37°C overnight, cells were gently washed twice with
PBS and fixed by 4% paraformaldehyde solution (Nacalai Tesque) for
20 min at 25°C. After washing with Hanks' Balanced Salt Solution
(Gibco; Thermo Fisher Scientific, Inc.), cells were incubated with
10 µg/ml WGA, Alexa Fluor 594 conjugate (Invitrogen, Thermo Fisher
Scientific, Inc.) for 10 min at 25°C. Finally, samples were
incubated with Hoechst 33342 (1:2,000; Invitrogen; Thermo Fisher
Scientific, Inc.) for 5 min at 37°C.
Images were captured using a confocal microscope
(LSM700; Carl Zeiss AG) equipped with an oil immersion objective
lens (magnification, ×40). Images were analyzed with ZEN 2012 (Carl
Zeiss AG) and processed using ImageJ software ver.1.51 (National
Institutes of Health) (29).
Irradiation
Cells were exposed to 5 or 8 Gy 150 kV X-rays
delivered at 0.57 Gy/min using an MBR-1505R2 generator (Hitachi
Ltd.). The beam was filtered through a 1-mm aluminum board and the
accuracy of irradiation was checked, as previously described
(30).
Colony forming assay
Cell survival following irradiation was evaluated by
performing a colony forming assay in the presence or absence of 10
or 20 µg exosomes generated following 5 Gy irradiation (5 Gy-Exo).
Cells were reseeded into 6-well cell culture plates (Corning, Inc.)
at a density of 200–4,000 cells/well and incubated for 7–10 days
(7–10 cell cycles) at 37°C. The number of seeded cells was
different in each group [control, 100; 5 Gy, 500; 5 Gy + 5 Gy-Exo
(5 µg/ml), 1,000 and 5 Gy + 5 Gy-Exo (10 µg/ml), 2,000 cells/ml]
depending on the dose of irradiation. At the end of each experiment
using non-irradiated exosomes (0 Gy-Exo) or 5 Gy-Exo, the cells
were fixed with a solution of 10% methanol and 20% acetic acid for
30 min and stained at 25°C. With methylene blue for 30 min as
previously described (31). Colonies
(≥50 cells) were counted and the surviving fraction was determined
based on the number of colonies per seeded cell.
Determination of intracellular ROS
levels
Intracellular ROS levels were determined using the
oxidation-sensitive fluorescent probe dye C-H2DCF
(Invitrogen; Thermo Fisher Scientific, Inc.) as described
previously (32). Cells were seeded
in 6-well plates (1.5×105 cells/well) overnight at 37°C
and treated with 5 Gy radiation in the presence or absence of 10
µg/ml 5 Gy-Exo and 1 mM NAC (Sigma Aldrich; Merck KGaA) for 24 h at
37°C, as previously described (33).
After washing twice with FBS-free media (MEM; Sigma Aldrich; Merck
KGaA) the cells were stained with 50 µM C-H2DCF for 1 h
at 37°C. The nuclei of cells were then stained with Hoechst 33342
(1:2,000; Invitrogen; Thermo Fisher Scientific, Inc.) for 5 min at
37°C. The fluorescence of C-H2DCF was visualized using a
fluorescence microscope (magnification, ×20) (BZ-9000; Keyence
Corporation).
Detection of DNA damage following
exosome uptake
Induction of DNA damage was investigated by
detecting γ-H2AX foci using immunocytochemistry, as described
previously (34). Cells were seeded
on 35-mm dishes and treated with 10 µg/ml 5 Gy-Exo and 1 mM NAC for
24 h and/or 5 Gy irradiation at 25°C. Then, the cells were fixed in
4% paraformaldehyde in PBS for 20 min, permeabilized with 0.1%
Triton X-100 in PBS for 5 min and blocked in 5% BSA (Sigma-Aldrich;
Merck KGaA) in PBS for 60 min at 25°C. The cells were incubated
with rabbit anti γ-H2AX antibody (1:200; Cell Signaling Technology,
Inc.) overnight at 4°C. The cells were then incubated with
TRITC-conjugated secondary antibody (1:20; Dako; Agilent
Technologies, Inc.) for 90 min at 25°C. The nuclei were stained
with DAPI (1:300; Invitrogen; Thermo Fisher Scientific, Inc.) for
10 min at 25°C. The stained cells were observed using a
fluorescence microscope (magnification, ×40). The number of cells
expressing nuclear γ-H2AX foci were then counted manually in 100
cells of each treatment group, as previously described (35).
Immunoblotting
The expression levels of CD9, CD63 and cytochrome
c were analyzed. Briefly, 3 µg exosomes (0 and 5 Gy-Exo)
were separated by 8 (CD9), 12 (CD63) or 15% (cytochrome c)
SDS-PAGE gels in non-reducing conditions. A total of 30 µg whole
cell lysate was separated using RIPA buffer supplemented with
protease inhibitor cocktail (Nacalai Tesque) previously described
(36) in reducing conditions [boiling
for 5 min at 95°C and addition of reducing agent, 5%
2-Mercaptoethanol (FUJIFILM Wako Pure Chemical Corporation) at
25°C] and transferred to a PVDF membrane. Membranes were blocked
using 3 (cytochrome c) or 5% non-fat milk for 30 min at
25°C, incubated with anti-CD9 (1:500), anti-CD63 (1:500) or
anti-cytochrome c (1:1,000) antibodies overnight at 4°C and
washed three times in Tris-buffered saline with 10% Tween-20
(TBS-T). Subsequently, membranes were incubated for 1 h with m-IgGκ
BP-HRP (1:4,000) at 25°C.
The expression levels of SOD1 and SOD2 in MIAPaCa-2
cells were analyzed. Briefly, cells were seeded at
1.0×105 cells/well in 6-well plates and incubated
overnight at 37°C and subjected to irradiation with 8 Gy or
addition of 30 µg 8 Gy-Exo at 25°C. Proteins was collected using
RIPA buffer supplemented with protease inhibitor cocktail (Nacalai
Tesque) at 24 h after treatment. Quantification of proteins was
proceeded using Qubit™ Protein Assay kit (Invitrogen; Thermo Fisher
Scientific, Inc.). A total of 15 µg protein/lane was separated by
10% SDS-PAGE and transferred to a PVDF membrane. Membranes were
blocked using 5% BSA (Sigma-Aldrich; Merch KGaA) blocking buffer
for 40 min at 25°C, incubated with anti-SOD1 and anti-SOD2 (both
1:1,000) antibodies overnight at 4°C and then washed three times in
10% TBS-T. Subsequently, membranes were incubated for 1 h with
rabbit anti-sheep IgG-HRP (1:2,000) secondary antibody at 25°C. The
secondary antibodies were visualized with ECL™ Prime Western
Blotting Detection Reagents (GE Healthcare) using a gel imaging
system with preconfigured Image Lab Touch software 5.2.1 (ChemiDoc
Touch MP; Bio-Rad Laboratories, Inc.). Subsequently, the PVDF
membranes were incubated with anti-actin (1:5,000) antibody
overnight at 4°C. After washing with TBS-T three times, membranes
were incubated for 1 h with donkey anti-goat IgG,
F(ab′)2-HRP secondary antibody at 25°C, which was
visualized as aforementioned. The intensity of each signal was
analyzed using ImageJ software ver.1.51 (National Institutes of
Health) and the ratios of SOD1, SOD2 and actin levels were
calculated.
Following 48 h transfection with miR-6823-5p-mimic,
the expression levels of SOD1 were analyzed, as aforementioned.
Protein concentrations were determined using a Qubit™ Protein Assay
kit.
Total RNA extraction from exosomes and
miRNA microarray analysis
Total RNA was extracted from exosomes using Toray's
3D-Gene RNA extraction reagent from a liquid sample kit (Toray
Industries, Inc.). Comprehensive miRNA expression analysis was
performed using a 3D-Gene miRNA Labeling kit and a 3D-Gene Human
miRNA Oligo Chip Ver. 21 (Toray Industries, Inc.) according to the
manufacturer's protocol to detect 2,565 human miRNA sequences. The
expression levels of each miRNA were expressed as the
background-subtracted signal intensity of all miRNAs in each
microarray. Any signal intensity in both the duplicate spots at
>1.5 SD of the background signal intensity was considered a
valid measurement. The raw data are available in the Gene
Expression Omnibus database (GSE163133).
Database processing analysis and miRNA
identification
miRNAs from exosomes isolated from cells irradiated
with either 5 or 8 Gy were visualized in the form of a heat map
using R software (version 3.5.3; R-project.org)
(37) and heatmap.2 from the gplots
package (version 3.0.1.1; CRAN.R-project.org/pakage=gplots) (38). The heatmap presents Z-score values for
miRNAs with ratios of expression values between control (exosomes
from non-irradiated cells) and 5 or 8 Gy-Exo <0.5 or >1.5. In
addition, TargetScan (targetscan.org/vert_72/) (39) and miRTarBase (mirtarbase.cuhk.edu.cn/php/index.php) (40) were searched for targets of these
miRNAs that result in increased ROS levels. miRNAs data were then
proceeded hierarchical clustering with Euclidean distance and
complete linkage.
Transfection of miR mimics
In order to investigate the effect of miRNAs on
intracellular ROS levels, mirVana™ miRNA (miR-6823-5p) mimics and
negative control (Sigma-Aldrich; Merch KGaA) were used. MIAPaCa-2
cells (2.2×105 per well) were seeded in 24-well plates
and transfected with 5 nM miR-mimic or negative control using
HiPerFect Transfection Reagent (Qiagen GmbH) for 48 h at 25°C.
Cells were harvested at 37°C and the expression levels of mRNA and
protein were examined 48 h after transfection at 25°C. ROS levels
and DNA damage in transfected cells were assessed as
aforementioned. The mimic sequences were as follows: miR-6823-5p,
5′-UCAGGGUUGGUAGGGGUUGCU-3′; siRNA control 1,
5′-GGUUCGUACGUACACUGUUCA-3′; and siRNA control 2,
5′-CGGUACGAUCGCGGCGGGAUAUC-3′.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA for RT-qPCR was obtained from cell samples
using a mirVana™ miRNA Isolation kit (Invitrogen; Thermo Fisher
Scientific, Inc.) and reverse transcribed at 25°C for 30 min, 37°C
for 2 h, 85°C for 5 min and 4°C for 10 min to cDNA using the High
Capacity cDNA Reverse Transcription kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.). RT-qPCR was performed using EagleTaq
Universal Master Mix (ROX) (Roche Diagnostics) and
TaqMan® Gene Expression Assays (Applied Biosystems;
Thermo Fisher Scientific, Inc.) using following TaqMan probe to
SOD1 (cat. no. Hs00533490_m1). The thermocycling conditions were as
follows: initial denaturation at 50°C for 2 min and 95°C for 10
min, followed by 95°C for 15 sec and 40 cycles at 60°C for 1 min.
Expression data of SOD1 was acquired and analyzed by the
2−ΔΔCq method (41) using
a Thermal Cycler Dice® Real Time System III (Takara Bio,
Inc.). All data were normalized to GAPDH (cat. no.
Hs02786624_g1).
Statistical analysis
Data are presented as the mean ± SEM of three
independent experimental repeats. Differences between the means
were compared using one-way ANOVA, followed by post hoc Tukey's
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Characterization of exosomes
The purity, quality, and morphology of exosomes were
analyzed. According to the result of measurement using Qubit
Protein Assay kit, ~1.67 µg/ml exosomes were collected from the
culture medium. The isolated exosomes were shaped in the form of
closed, round vesicles with a diameter of ~100 nm. The
morphological features of 0 and 5 Gy-Exo were similar, as shown by
TEM (Fig. 1A). CD9 and CD63
expression was observed in both whole cell lysate and exosomes,
while cytochrome c expression was detected only in the whole
cell lysate (Fig. 1B). Distribution
profiles for 0 and 5 Gy-Exo revealed peaks at 107 and 101 nm,
respectively, as assessed using NanoSight (Fig. 1C; Videos
S1 and S2). These data indicated
that the exosomes were successfully isolated from the culture media
supernatant without contamination due to cellular components.
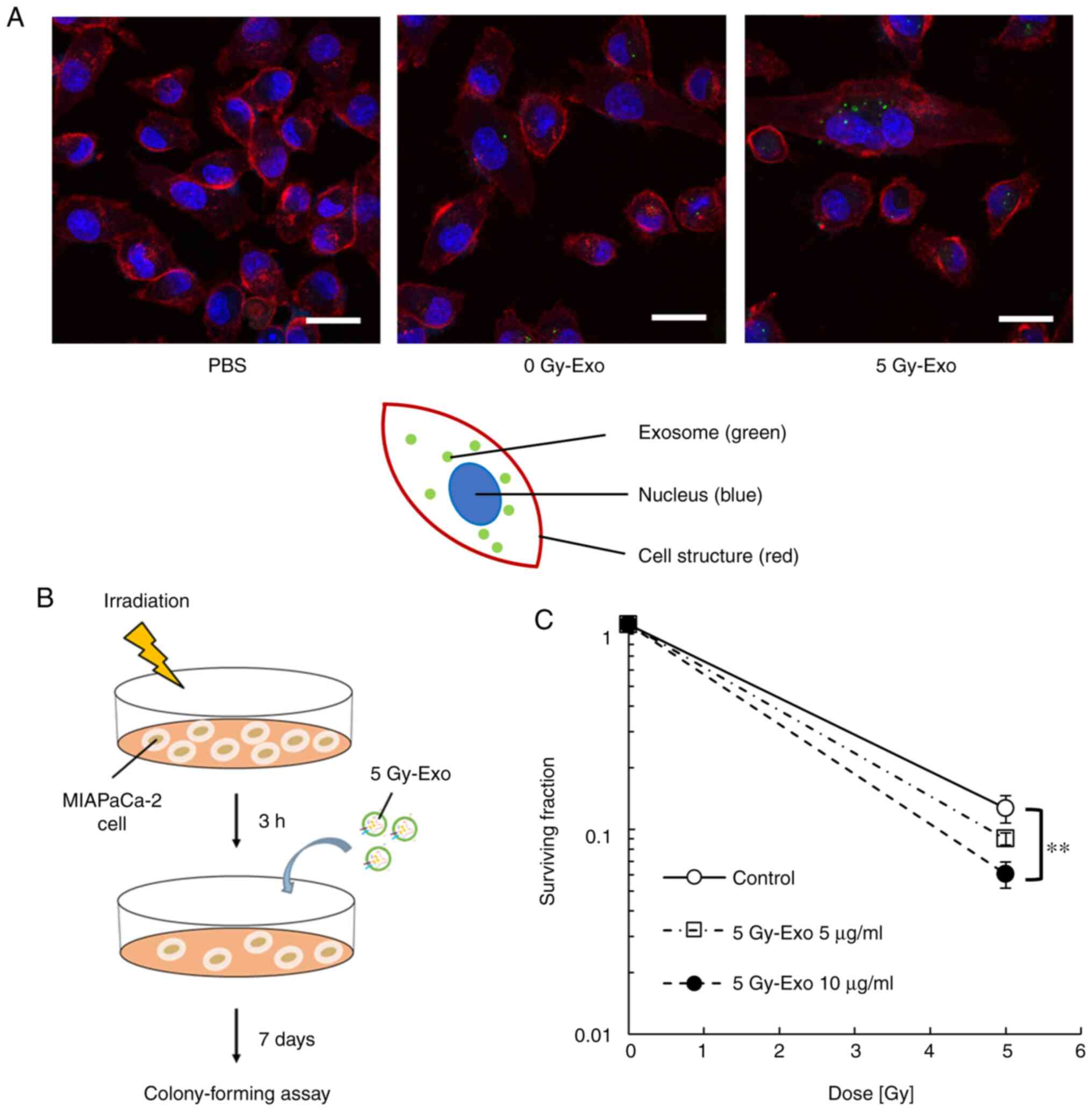
Cellular uptake of exosomes and
survival
The uptake of 5 Gy-Exo was increased compared with
that of 0 Gy-Exo (Fig. 2A). Cells
were irradiated and treated in the presence or absence of 5 Gy-Exo
(Fig. 2B). Cells irradiated with 5
Gy-Exo showed an increased radiosensitizing effect in an exosome
concentration-dependent manner (Fig.
2C; Table SI).
Exosomes increase intracellular ROS
levels
Intracellular ROS levels increased following
addition of 0 or 5 Gy-Exo or irradiation with 5 Gy; the highest
levels were observed following combined irradiation and exosome
treatment. Irradiation (5 Gy) combined with the addition of 0
Gy-Exo resulted in ROS levels similar to those for the
irradiation-alone treatment group (Fig.
3). Intracellular ROS levels were significantly decreased
following addition of the ROS scavenger NAC and these effects were
dose-dependent (Figs. S1 and
S2).
DNA damage is induced following
exosome uptake
Addition of both 0 and 5 Gy-Exo increased the number
of γ-H2AX foci/cell, while combined irradiation and 5 Gy-Exo
treatment significantly increased the number of γ-H2AX foci
further. Irradiation (5 Gy) and treatment with 0 Gy-Exo resulted in
numbers of γ-H2AX foci similar to that of the irradiation-alone
group (Fig. 4). NAC treatment
resulted in fewer γ-H2AX foci/cell, indicating that DNA damage was
induced by an increase in ROS levels and these effects were
dose-dependent (Figs. S2 and
S3).
Identification of miRNA in exosomes
following irradiation
The differential expression levels of miRNAs in 5
and 8 Gy-Exo compared with 0 Gy-Exo were stratified using a heat
map (Fig. 5A). A total of six up- and
five downregulated microRNA in both 5 and 8 Gy-Exo were identified
(Fig. 5B). TargetScan and miRTarBase
were used to identify potential targets of these miRNAs that may be
associated with increased ROS levels. miR-6823-5p was identified as
a potential candidate for SOD1 inhibition and subsequent analysis
confirmed the inhibitory effect of miR-6823-5p (Fig. 6).
Exosomes inhibit SOD1 expression
levels in cancer cells
Cells were treated with 8 Gy irradiation or addition
of 8 Gy-Exo, then the expression levels of antioxidant enzymes
including SOD1 and SOD2 were analyzed. SOD1 expression notably
decreased following addition of 8 Gy-Exo, but SOD2 expression
levels did not change (Fig. 6A).
Using TargetScan, the complementary sequence site of SOD1 was found
to correspond with miR-6823-5p (Fig.
6B). SOD1 expression levels decreased following transfection
with an miR-6823-5p-mimic (Fig. 6C).
Additionally, the relative cDNA level of SOD1 in cells transfected
with miR-6823-5p-mimic significantly decreased compared with that
of non-transfected cells (non-siRNA), although that of cells
transfected with siRNA control 1 and 2 did not change
significantly. (Fig. 6D). Taken
together, these results suggest that miR-6823-5p in exosomes
derived from irradiated cells may contribute to decreased SOD1
expression levels.
miR-6823-5p mimics increase ROS levels
and DNA damage in MIAPaCa-2 cells
ROS levels and DNA damage, which significantly
increased in cells transfected with miR-6823-5p mimic, decreased
significantly following the addition of NAC (Figs. 7 and 8).
These results confirmed that miR-6823-5p increased intracellular
ROS levels, leading to DNA damage in MIAPaCa-2 cells.
Discussion
The aim of the present study was to investigate
whether irradiated exosomes induce intracellular increases in ROS
levels in neighboring cancer cells, leading to amplification of the
radiation effect. The potential underlying mechanisms may be
associated with transportation of certain miRNAs from irradiated
cancer cells via exosomes. To the best of our knowledge, the
present study is the first to report that miR-6823-5p may function
as an inhibitor of SOD1 expression in response to radiation.
SOD1, which is overexpressed in several types of
cancer cell, such as breast (42) and
non-small lung cancer cells (43) may
be essential for the maintenance of cellular ROS levels. Papa et
al (44) reported that SOD1
serves an important role in cancer progression and described a
potential association between SOD1 overexpression and regulation of
the mitochondrial unfolded-protein response. Cells must
continuously contend with extensive intracellular oxidative stress
generated by ROS and reactive nitrogen species (RNS) (45). In order to control the genotoxic
effects of ROS/RNS and their diverse functions (including
signaling), cells regulate their levels via antioxidants, including
SOD (46). Gomes et al
(47) demonstrated that the
expression of SOD1 is decreased in HCT116 colon cancer cells
overexpressing miR-143 or miR-145. Furthermore, miR-143
overexpression increased ROS levels, which was abrogated by the
reintroduction of SOD1. However, an increase in the levels of
miR-143 and miR-145 in exosomes in the MIAPaCa-2 human pancreatic
cancer cell line was not observed in the present study. This may be
because the present study investigated miRNAs in exosomes obtained
following radiation exposure. Moreover, the different sources of
miRNAs may impact the results as the aforementioned study used a
cancer cell line, whereas, here, exosomes were used in addition to
a cancer cell line.
To the best of our knowledge, inhibition of SOD1
expression by exosomes has not been reported previously.
Furthermore, no studies have yet reported that exosomes from
irradiated cells induce DNA damage via increasing ROS levels. These
findings are important for investigating exosome functions in
response to radiation. Glasauer et al (48) showed that inhibition of SOD1
expression, either via small hairpin RNA (shRNA) or a SOD1
inhibitor (ATN-224), notably decreases the ability of the lung
carcinoma cell line A549 to form colonies on soft agar. They
further reported that inhibition of SOD1 expression leads to an
increase, rather than decrease, in H2O2
levels as a result of the inhibition of the glutathione peroxidase
enzymes by superoxide; this suggested that the inhibition of SOD1
induces cell death by apoptosis (48). Taken together, the results of the
present study indicate that SOD1 modulation may be a promising
target for enhancing the radiation effect.
The present findings describe a novel mechanism
associated with RIBE, which is involved in induction of DNA damage
(49) and mutations (50,51), cell
death or apoptosis (52) and altered
gene expression (13,24) and miRNA profiles (53,54). Here,
0 Gy-Exo induced ROS and DNA damage, potentially owing to higher
concentrations of exosomes. The total amount of protein in the
exosomes in the culture media without any treatment was ~1.67
µg/ml. For each ROS/DNA-damage experiment, 10 µg/ml exosomes, a
5-fold higher concentration of exosomes compared with that in the
culture medium, was administered. Therefore, larger amounts of 0
Gy-Exo composites, such as proteins, lipids, cDNA and miRNA, may
have been involved. It was speculated that these composites of 0
Gy-Exo may increase intracellular ROS levels, leading to DNA
damage. Ionizing radiation is frequently accompanied by marked
changes in the miRNA expression profile of cells (55,56).
Although miRNAs have been implicated in regulation of ROS levels
via regulation of enzymes involved in ROS metabolism (57), the association between exogenous ROS
and intercellular communication via exosomes has not been
previously elucidated. In the present study, miR-6823-5p induced
radiosensitive effects via inhibition of SOD1 expression. The
difference in miR-6823-5p expression levels between the 0 and 5
Gy-Exo groups affected the enhancement of ROS level and DNA damage.
ROS and DNA damage induced by X-ray radiation, and they affect cell
survival or death signaling cascades (58,59).
Cancer cell-derived exosomes have demonstrated a role in promoting
cancer cell invasiveness and metastasis, as well as activation of
oncogenic pathways (60,61).
The present study investigated intercellular
communication between cancer cells via exosomes, which may be
involved in other types of communication; between cancer and
non-malignant cells, such as stromal, vascular and immune cells.
Cancer cell-secreted exosomes affect other cancer or host cells and
may lead to the secretion of additional exosomes from non-malignant
cells (62). The present study
confirmed increased uptake of exosomes generated from irradiated
MIAPaCa-2 cells. It was hypothesized that irradiation induces this
uptake by recipient cells although the underlying process remains
unclear. Mutschelknaus et al (63) reported that radiation increases
exosome release and uptake in head and neck squamous carcinoma
cells and confirmed the influence of radiation on the uptake of
exosomes using fluorescence-labeled exosomes. Arscott et al
(64) used glioblastoma cell lines
and reported that cellular irradiation increases exosome release
and that radiation-derived exosomes are more readily taken up by
recipient cells. These results are in accordance with increased
uptake of exosomes by irradiated glioblastoma cells, which is
facilitated by enhancement of cellular attachment to exosomes via
augmented CD29/CD81 complex formation (65). Evidence indicates that exosomes
mediate the delivery of proteins, mRNAs and miRNAs from cancer
cells to recipient or neighboring cells by intercellular
communication, which may assist in the creation of a metastatic
niche and facilitate cancer cell progression and metastasis or
influence the activity and/or behaviors of recipient cells
(66,67). The results of the current study
regarding SOD1 inhibition serve as a basis for further
investigation of novel roles associated with exosomes.
Limitations of the present study include the use of
a single cell line (MIAPaCa-2 cells) and the lack of comparisons
with other cell lines of the same origin. Thus, further
investigations using other cell lines are needed for validation. In
addition, exosome release may be affected by the range of
radiation; effects of radiations in the kV and MV range may be
different. However, radiation apparatus was only available for the
kV range, not the MV range. Therefore, future studies should
evaluate and compare the effects of kV- and MV-range radiation on
exosome release.
In conclusion, the present study identify a novel
function of irradiated exosomes in terms of their ability to
enhance the radiation effect via increasing intracellular ROS
levels in cancer cells. The results contribute to the current
understanding of the bystander effect between neighboring cancer
cells.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Professor Ryo Nitta
and Dr Tsuyoshi Imasaki of Division of Structural Medicine and
Anatomy, Department of Physiology and Cell Biology, Kobe University
Graduate School of Medicine, for their helpful advice regarding TEM
imaging. Parts of the data were presented at the 7th Japan-Taiwan
Radiation Oncology symposium, Tokyo, Japan, May 11 2019.
Funding
The present study was supported by Grants-in-Aid for
Exploratory Research (grant nos. 18K07752, 19K08097 and 08K07676)
from the Ministry of Education, Culture, Sports, Science, and
Technology of Japan.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available in the GEO repository, accession no. GSE163133,
ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE163133.
Authors' contributions
AN, SI, HA and RS conceptualized and designed the
study. AN, MN, MS, YF, HK, MH, NM, RN, TI, DM, TS and RS collected,
analyzed and interpreted the data. AN, HK and RS drafted and
critically revised the manuscript for important intellectual
content. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
miRNA
|
microRNA
|
|
TEM
|
transmission electron microscope
|
|
ROS
|
reactive oxygen species
|
|
C-H2DCF
|
2′,7′-dichlorodihydrofluorescein
diacetate
|
|
γ-H2AX
|
phosphorylated histone 2AX
|
|
SOD1
|
Cu/Zn superoxide dismutase enzyme
|
|
SOD2
|
Mn-superoxide dismutase enzyme
|
|
3′-UTR
|
3′-untranslated region
|
|
RIBE
|
radiation-induced bystander effect
|
|
MEM
|
minimal essential medium
|
|
TRITC
|
tetramethyl rhodamine
isothiocyanate
|
|
NAC
|
N-acetyl-L-cysteine
|
|
NTA
|
nanoparticle tracking analysis
|
|
RNS
|
reactive nitrogen species
|
|
shRNA
|
small hairpin RNA
|
|
RT-q
|
reverse transcription-quantitative
|
|
si
|
small interfering
|
References
|
1
|
Tkach M and Théry C: Communication by
extracellular vesicles: Where we are and where we need to go. Cell.
164:1226–1232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hessvik NP and Llorente A: Current
knowledge on exosome biogenesis and release. Cell Mol Life Sci.
75:193–208. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Greening DW, Gopal SK, Xu R, Simpson RJ
and Chen W: Exosomes and their roles in immune regulation and
cancer. Semin Cell Dev Biol. 40:72–81. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yankovskaya V, Horsefield R, Törnroth S,
Chavez CL, Miyoshi H, Léger C, Byrne B, Cecchini G and Iwata S:
Architecture of succinate dehydrogenase and reactive oxygen species
generation. Science. 299:700–704. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nùkhet AB, Iman MA, Yueming Z, Larry WO
and Douglas RS: Increased levels of superoxide and hydrogen
peroxide mediate the differential susceptibility of cancer cells
vs. normal cells to glucose deprivation. Biochem J. 418:29–37.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Szatrowski TP and Nathan CF: Production of
large amounts of hydrogen peroxide by human tumor cells. Cancer
Res. 51:794–798. 1991.PubMed/NCBI
|
|
7
|
Boonstra J and Post JA: Molecular events
associated with reactive oxygen species and cell cycle progression
in mammalian cells. Gene. 337:1–13. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Miao L and St Clair DK: Regulation of
superoxide dismutase genes: Implications in disease. Free Radic
Biol Med. 47:344–356. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Srinivas US, Tan BWQ, Vellayappan BA and
Jeyasekharan AD: ROS and the DNA damage response in cancer. Redox
Biol. 25:1010842019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zou Z, Chang H, Li H and Wang S: Induction
of reactive oxygen species: An emerging approach for cancer
therapy. Apoptosis. 22:1321–1335. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mukubou H, Tsujimura T, Sasaki R and Ku Y:
The role of autophagy in the treatment of pancreatic cancer with
gemcitabine and ionizing radiation. Int J Oncol. 37:821–828.
2010.PubMed/NCBI
|
|
12
|
Doskey CM, Buranasudja V, Wagner BA,
Wilkes JG, Du J, Cullen JJ and Buettner GR: Tumor cells have
decreased ability to metabolize H2O2:
Implications for pharmacological ascorbate in cancer therapy. Redox
Biol. 10:274–284. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shea A, Harish V, Afzal Z, Chijioke J,
Kedir H, Dusmatova S, Roy A, Ramalinga M, Harris B, Blancato J, et
al: MicroRNAs in glioblastoma multiforme pathogenesis and
therapeutics. Cancer Med. 5:1917–1946. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bracken CP, Scott HS and Goodall GJ: A
network-biology perspective of microRNA function and dysfunction in
cancer. Nat Rev Genet. 17:719–732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Di Francesco A, De Pittà C, Moret F,
Barbieri V, Celotti L and Mognato M: The DNA-damage response to
γ-radiation is affected by miR-27a in A549 cells. Int J Mol Sci.
14:17881–17896. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang B, Chen J, Ren Z, Chen Y, Li J, Miao
X, Song Y, Zhao T, Li Y, Shi Y, et al: A specific miRNA signature
promotes radioresistance of human cervical cancer cells. Cancer
Cell Int. 13:1182013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shin S, Cha HJ, Lee EM, Lee SJ, Seo SK,
Jin HO, Park IC, Jin YW and An S: Alteration of miRNA profiles by
ionizing radiation in A549 human non-small cell lung cancer cells.
Int J Oncol. 35:81–86. 2009.PubMed/NCBI
|
|
18
|
Simone BA, Ly D, Savage JE, Hewitt SM, Dan
TD, Ylaya K, Shankavaram U, Lim M, Jin L, Camphausen K, et al:
MicroRNA alterations driving acute and late stages of
radiation-induced fibrosis in a murine skin model. Int J Radiat
Oncol Biol Phys. 90:44–52. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hou J, Wang F, Kong P, Yu PK, Wang H and
Han W: Gene profiling characteristics of radioadaptive response in
AG01522 normal human fibroblasts. PLoS One. 10:e01233162015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sharma V and Misteli T: Non-coding RNAs in
DNA damage and repair. FEBS Lett. 587:1832–1839. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang C and Peng G: Non-coding RNAs: An
emerging player in DNA damage response. Mutat Res. 763:202–211.
2015. View Article : Google Scholar
|
|
22
|
Lyng FM, Maguire P, McClean B, Seymour C
and Mothersill C: The involvement of calcium and MAP kinase
signaling pathways in the production of radiation-induced bystander
effects. Radiat Res. 165:400–409. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iyer R, Lehnert BE and Svensson R: Factors
underlying the cell growth-related bystander responses to alpha
particles. Cancer Res. 60:1290–1298. 2000.PubMed/NCBI
|
|
24
|
Gow MD, Seymour CB, Ryan LA and Mothersill
CE: Induction of bystander response in human glioma cells using
high-energy electrons: A role for TGF-beta1. Radiat Res.
173:769–778. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Al-Mayah AHJ, Irons SL, Pink RC, Carter
DRF and Kadhim MA: Possible role of exosomes containing RNA in
mediating nontargeted effect of ionizing radiation. Radiat Res.
177:539–545. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
JCRB Cell Bank, . Cell information.
Available from. cellbank.nibiohn.go.jp/~cellbank/cgi-bin/search_res_det.cgi?ID=245
|
|
27
|
Thery C, Amigorena S, Raposo G and Clayton
A: Isolation and characterization of exosomes from cell culture
supernatants and biological fluids. Curr Protoc Cell Biol Chapter.
3:Unit 3.22. 2006.
|
|
28
|
Pegtel DM, Cosmopoulos K, Thorley-Lawson
DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD,
Würdinger T and Middeldorp JA: Functional delivery of viral miRNAs
via exosomes. Proc Natl Acad Sci USA. 107:6328–6333. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rasband WS: ImageJ, U.S. Available from.
https://imagej.nih.gov/ij/1997-2018
|
|
30
|
Shimizu Y, Mukumoto N, Idrus N, Akasaka H,
Inubushi S, Yoshida K, Mikawaki D, Ishihara T, Okamoto Y, Yasuda T,
et al: Amelioration of radiation enteropathy by dietary
supplementation with reduced coenzyme Q10. Adv Radiat Oncol.
4:237–245. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Achanta G, Sasaki R, Feng L, Carew JS, Lu
W, Pelicano H, Keating MJ and Huang P: Novel role of p53 in
maintaining mitochondrial genetic stability through interaction
with DNA Pol gamma. EMBO J. 24:3482–3492. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dutta S, Warshall C, Bandyopadhyay C,
Dutta D and Chandran B: Interactions between exosomes from breast
cancer cells and primary mammary epithelial cells leads to
generation of reactive oxygen species which induce DNA damage
response, stabilization of p53 and autophagy in epithelial cells.
PLoS One. 9:e975802014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sasaki R, Suzuki Y, Yonezawa Y, Ota Y,
Okamoto Y, Demizu Y, Huang P, Yoshida H, Sugimura K and Mizushina
Y: DNA polymerase gamma inhibition by vitamin K3 induces
mitochondria-mediated cytotoxicity in human cancer cells. Cancer
Sci. 99:1040–1048. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nakayama M, Sasaki R, Ogino C, Tanaka T,
Morita K, Umetsu M, Ohara S, Tan Z, Nishimura Y, Akasaka H, et al:
Titanium peroxide nanoparticles enhanced cytotoxic effects of X-ray
irradiation against pancreatic cancer model through reactive oxygen
species generation in vitro and in vivo. Radiat Oncol. 11:912016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bonner WM, Redon CE, Dickey JS, Nakamura
AJ, Sedelnikova OA, Solier S and Pommier Y: GammaH2AX and cancer.
Nat Rev Cancer. 8:957–967. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Inubushi S, Kawaguchi H, Mizumoto S,
Kunihisa T, Baba M, Kitayama Y, Takeuchi T, Hoffman RM, Tanino H
and Sasaki R: Oncogenic miRNAs identified in tear exosomes from
metastatic breast cancer patients. Anticancer Res. 40:3091–3096.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
R Core Team (2018), . R: A language and
environment for statical computing. 2018.
|
|
38
|
Warnes GR, Bolker B, Bonebakker L,
Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A and
Moeller S: gplots: Various R programming tools for plotting data.
2018.
|
|
39
|
Agarwal V, Bell GW, Nam J and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
ELife. 4:e050052015. View Article : Google Scholar
|
|
40
|
Hsu SD, Lin FM, Wu WY, Liang C, Huang WC,
Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al: miRTarBase: A
database curates experimentally validated microRNA-target
interactions. Nucleic Acids Rese. 39:163–169. 2011. View Article : Google Scholar
|
|
41
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gomez ML, Shah N, Kenny CT, Jenkins CE Jr
and Germain D: SOD1 is essential for oncogene-driven mammary tumor
formation but dispensable for normal development and proliferation.
Oncogene. 38:5751–5765. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu S, Li B, Xu J, Hu S, Zhan N, Wang H,
Gao C, Li J and Xu X: SOD1 promotes cell proliferation and
metastasis in non-small cell lung cancer via an
miR-409-3p/SOD1/SETDB1 epigenetic regulatory feedforward loop.
Front Cell Dev Biol. 8:2132020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Papa L, Manfredi G and Germain D: SOD1, an
unexpected novel target for cancer therapy. Genes Cancer. 5:15–21.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Valko M, Rhodes CJ, Moncol J, Izakovic M
and Mazur M: Free radicals, metals and antioxidants in oxidative
stress-induced cancer. Chem Biol Interact. 160:1–40. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Koskenkorva-Frank ST, Weiss G, Koppenol HW
and Burckhardt S: The complex interplay of iron metabolism,
reactive oxygen species, and reactive nitrogen species: Insights
into the potential of various iron therapies to induce oxidative
and nitrosative stress. Free Radic Biol Med. 65:1174–1194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gomes SE, Pereira DM, Roma-Rodrigues C,
Fernandes AR, Borralho PM and Rodrigues CM: Convergence of miR-143
overexpression, oxidative stress and cell death in HCT116 human
colon cancer cells. PLoS One. 13:e01916072018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Glasauer A, Sena LA, Diebold LP, Mazar AP
and Chandel NS: Targeting SOD1 reduces experimental non-small-cell
lung cancer. J Clin Invest. 124:117–128. 2013. View Article : Google Scholar
|
|
49
|
Yang H, Assad N and Held KD:
Medium-mediated intercellular communication is involved in
bystander responses of X-ray-irradiated normal human fibroblasts.
Oncogene. 24:2096–2103. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhou H, Randers-Pehrson G, Waldren CA,
Vannais D, Hall EJ and Hei TH: Induction of a bystander mutagenic
effect of alpha particles in mammalian cells. Proc Natl Acad Sci
USA. 97:2099–2104. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huo L, Nagasawa H and Little JB: HPRT
mutants induced in bystander cells by very low fluences of alpha
particles result primarily from point mutations. Radiat Res.
156:521–525. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lyng FM, Seymour CB and Mothersill C:
Initiation of apoptosis in cells exposed to medium from the progeny
of irradiated cells: A possible mechanism for bystander-induced
genomic instability? Radiat Res. 157:365–370. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kovalchuk O, Zemp FJ, Filkowski JN,
Altamirano AM, Dickey JS, Jenkins-Baker G, Marino SA, Brenner DJ,
Bonner WM and Sedelnikova OA: MicroRNAome changes in bystander
three-dimensional human tissue models suggest priming of apoptotic
pathways. Carcinogenesis. 31:1882–1888. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Koturbash I, Zemp F, Kolb B and Kovalchuk
O: Sex-specific radiation-induced microRNAome responses in the
hippocampus, cerebellum and frontal cortex in a mouse model. Mutat
Res. 722:114–118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Maes OC, An J, Sarojini H, Wu HL and Wang
E: Changes in microRNA expression patterns in human fibroblasts
after low-LET radiation. J Cell Biochem. 105:824–834. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Niemoeller OM, Niyazi M, Corradini S,
Zehentmayr F, Li M, Lauber K and Belka C: MicroRNA expression
profiles in human cancer cells after ionizing radiation. Radiat
Oncol. 6:292011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang X, Ng WL, Wang P, Tian L, Werner E,
Wang H, Doetsch P and Wang Y: MicroRNA-21 modulates the levels of
reactive oxygen species by targeting SOD3 and TNFα. Cancer Res.
72:4707–4713. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Fan PC, Zhang Y, Wang Y, Wei W, Zhou YX,
Xie Y, Wang X, Qi YZ, Chang L, Jia ZP, et al: Quantitative
proteomics reveals mitochondrial respiratory chain as a dominant
target for carbon ion radiation: Delayed reactive oxygen species
generation caused DNA damage. Free Radic Biol Med. 130:436–445.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zulato E, Ciccarese F, Agnusdei V, Pinazza
M, Nardo G, Iorio E, Curtarello M, Silic-Benussi M, Rossi E,
Venturoli C, et al: LKB1 loss is associated with glutathione
deficiency under oxidative stress and sensitivity of cancer cells
to cytotoxic drugs and γ-irradiation. Biochem Pharmacol.
156:479–490. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Urbanelli L, Magini A, Buratta S, Brozzi
A, Sagini K and Polchi A: Signaling pathways in exosomes
biogenesis, secretion and fate. Genes. 4:152–170. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Cardiello C, Cavallini L, Spinelli C, Yang
J, Reis-Sobreiro M, de Candia P, Minciacchi VR and Di VD: Focus on
extracellular vesicles: New frontiers of cell-to-cell communication
in cancer. Int J Mol Sci. 17:1752016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liu LS, Sun P, Li Y, Liu SS and Lu Y:
Exosomes as critical mediators of cell-to-cell communication in
cancer pathogenesis and their potential clinical application.
Transl Cancer Res. 8:298–311. 2019. View Article : Google Scholar
|
|
63
|
Mutschelknaus L, Peters C, Winkler K,
Yentrapalli R, Heider T, Atkinson MJ and Moertl S: Exosomes derived
from squamous head and neck cancer promote cell survival after
ionizing radiation. PLoS One. 11:e01522132016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Arscott WT, Tandle AT, Zhao S, Shabason
JE, Gordon IK, Schlaff CD, Zhang G, Tofilon PJ and Camphausen KA:
Ionizing radiation and glioblastoma exosomes: Implications in tumor
biology and cell migration. Transl Oncol. 6:638–648. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hazawa M, Tomiyama K, Saotome-Nakamura A,
Obara C, Yasuda T, Gotoh T, Tanaka I, Yakumaru H, Ishihara H and
Tajima K: Radiation increases the cellular uptake of exosomes
through CD29/CD81 complex formation. Biochem Biophys Res Commun.
446:1165–1171. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
O'Brien K, Rani S, Corcoran C, Wallace R,
Hughes L, Friel AM, McDonnell S, Crown J, Radomski MW and
O'Driscoll L: Exosomes from triple-negative breast cancer cells can
transfer phenotypic traits representing their cells of origin to
secondary cells. Eur J Cancer. 49:1845–1859. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Costa-Silva B, Aiello NM, Ocean AJ, Singh
S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, et
al: Pancreatic cancer exosomes initiate pre-metastatic niche
formation in the liver. Nat Cell Biol. 17:816–826. 2015. View Article : Google Scholar : PubMed/NCBI
|