Introduction
High-risk human papillomaviruses (HPVs),
particularly HPV16 and HPV18, are the main cause of cervical
cancer, which ranks as the fourth most frequently diagnosed cancer
and the fourth leading cause of cancer-associated deaths in women,
worldwide (1). Additionally, the
incidence and mortality rates of cervical cancer are increasing in
several countries (2). HPV E6 and E7
are important viral oncogenes that act in concert to maintain the
malignant phenotype of cervical cancer. These genes constitute
attractive therapeutic targets, as it has been demonstrated that
E6/E7 inhibition rapidly induces senescence in HPV-positive cancer
cells (3). However, infection with
oncogenic HPV alone is not sufficient for cancer development;
genetic variations and epigenetic alterations are required for the
development of precancerous lesions and cervical cancer (4,5). Thus,
improving the clinical management of patients and developing an
effective novel treatment strategy for patients with cervical
cancer is a major challenge.
To determine the additional alterations that occur
in cervical cancer, previous studies have demonstrated that E6
inactivates p53 by binding to the cellular ubiquitin ligase
E6-associated protein, thereby preventing the replicative
senescence of cervical cancer cells (6–8). As E6
contains a PDZ binding motif (PBM; including postsynaptic density
protein 95/DLG/zona occludins-1) at the extreme C terminus, it can
further bind to a number of tumor suppressor protein-containing PDZ
domains, including DLG, scribble and membrane associated guanylate
kinase, WW and PDZ domain-containing 1 (9). In addition, the transforming activity of
E7 is strongly increased when E6 is co-expressed; however,
high-risk HPV E7 modulates the expression and degradation of
several host proteins, including retinoblastoma (pRB), p107, p130,
tyrosine-protein phosphatase non- receptor type14 (PTPN14) and p21,
which leads to unscheduled cell cycle progression (10).
Recently, accumulating evidence has further
confirmed that the modulation of long non-coding RNA (lncRNA/lnc)
expression is an important aspect of oncogenic activity in
high-risk HPV E6 and E7 proteins. For example, HPV 16 E6 increased
the expression of lnc-cervical carcinoma expressed PCNA regulatory
(CCEPR) and lnc-family with sequence similarity 83 member H
antisense RNA 1 (FAM83H-AS1) through a mechanism that is not
directly dependent on p53 inactivation, which thereby promoted
proliferation and migration, and inhibited the apoptosis of
cervical cancer cells (11,12). It has been previously demonstrated
that HPV16/18 E6 and E7 proteins were associated with increased
enhancer of zeste homolog 2 (EZH2)-binding lncRNA in cervical
cancer (lnc-EBIC) expression in cervical cancer cells. The study
additionally determined that E6 promoted lnc-EBIC expression to
sequester certain tumor repressor microRNAs (miRs), including
miR-375 and miR-139 that target HPV16/18 E6/E7 mRNA, thus forming a
positive feedback loop that mutually derepressed gene expression in
cervical cancer cells (13). Lnc-EBIC
is a pseudogene that is highly expressed in human cervical cancer
tissues and cell lines, which interacts with EZH2 to repress the
expression of E-cadherin and thus promote cellular proliferation
and invasion (14,15). In addition to E6, a previous study
determined that HPV18 E7 also stimulated lnc-EBIC expression in
HeLa cells (13). However, the
tumorigenic roles and potential molecular mechanism of the
E7/lnc-EBIC axis on cervical cancer has not been fully
elucidated.
Transcriptome sequencing was performed in a previous
study to analyze the effects of lnc-EBIC depletion on the mRNA
levels of certain protein-coding genes (13). Oncogenic Kelch domain-containing 7B
(KLHDC7B) was identified to be correlated with lnc-EBIC expression,
and was determined to interact with Kelch-containing proteins via
the C-terminal Kelch domain (16,17).
Moreover, KLHDC7B was identified to be upregulated in breast cancer
and could promote breast tumorigenesis by modulating genes involved
in the interferon signaling pathway (16,18). In
addition, recent transcriptome analysis further suggested that
KLHDC7B could be used as a biomarker for prognostic prediction and
may be involved in the development and progression of cervical
cancer (19).
In the present study, the relationship between
HPV16/18 E7 and lnc-EBIC in cervical cancer cells was investigated,
and the effects of lnc-EBIC on the proliferation (using CCK-8 and
EdU/DAPI staining assay), apoptosis [utilizing flow cytometer
Annexin V/propidium iodide (PI) assay], cell invasion and migration
(using Transwell assays) of HPV+ and HPV−
cervical cancer cells were assessed, to provide a novel mechanism
and potential therapeutic target for cervical cancer.
Materials and methods
Cell culture and transfection
Human cervical cancer cell lines, including HeLa
(HPV18+), CaSki (HPV16+) and C33A
(HPV16−/18−), were purchased from The Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences.
Cells were cultured in DMEM-low glucose (Gibco; Thermo Fisher
Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin
(Beyotime Institute of Biotechnology), in a humidified atmosphere
at 37°C with 5% CO2. Cells in the exponential phase of
growth were used in the experiments. The protein-coding plasmids
pCMV-Tag2B-HPV18 E7 and pCMV-Tag2B-HPV16 E7 were previously
described (13). The nucleotide
sequences of lnc-EBIC were synthesized and inserted into pcDNA3.1
vectors (Thermo Fisher Scientific, Inc.) to construct the
pcDNA3.1-lnc-EBIC overexpression plasmids. Empty vectors were used
as negative controls (NCs). A total of 4 µg of each plasmid vector
was added to 100 µl of serum-free medium, and the volume of 100 µl
of each mixture was added to 1×105/ml cells in 6-cell
culture plates. TAL1 small interfering (si)RNA duplexes were
designed and synthesized at concentration of 100 nM (Guangzhou
RiboBio Co., Ltd.). The sequences are as follows: siHPV18-E7,
5′-CCTTCTATGTCACGAGCAA-3′; siHPV16-E7, 5′-CACCTACATTGCATGAATA-3′;
silnc-EBIC, 5′-GGGAGTAAAGACTCCAGTA-3′; siTAL1,
5′-AACCATGGAATCAACAAGGAT-3′; siKLHDC7B,
5′-CAGTGACAATGACTGGGATAGTGCT-3′; siRNA NC,
5′-GTTCTCCGAACGTGTCACGT-3′. Plasmids were transiently transfected
into cells using Lipofectamine® 2000 as instructed by
the manufacturer's protocol (Invitrogen; Thermo Fisher Scientific,
Inc.) at 37°C for 48 h. After 48 h transfection, the selective
overexpression and silencing of HPV16 E7, HPV18 E7, lnc-EBIC and
TAL1 were detected by reverse transcription-quantitative (RT-q) PCR
and western blotting.
RT-qPCR analysis
Total RNA (1 µg) was extracted from cells using the
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and reverse transcribed into cDNA using an RT-PCR kit (Thermo
Fisher Scientific, Inc.) in accordance with the manufacturer's
protocol. qPCR was performed using SYBR Green (Takara Biotechnology
Co., Ltd.) according to the manufacturer's instructions. The
following primers were utilized: HPV16E7 forward,
5′-AGCAGAACCGGACAGAGCCCA-3′ and reverse,
5′-TGTACGCACAACCGAAGCGT-3′; HPV18E7 forward,
5′-TGAAATTCCGGTTGACCTTC-3′ and reverse, 5′-TCGGGCTGGTAAATGTTGAT-3′;
lnc-EBIC forward, 5′-AAGGGCGTCGTGGTTCCAACTC-3′ and reverse,
5′-AGCATTGCCGTCCTGGGTGTAG-3′; TAL1 forward,
5′-CAACTGGAAAATCCAAAGGCTATGG-3′ and reverse,
5′-GACGCAATTCCTCCACAGTACACAG-3′; KLHDC7B forward,
5′-TGGGAACGAACACTCTTAC-3′ and reverse, 5′-CAGCAACTGAACACTTGAC-3′. A
total of 20 µl reaction mixture contained 1.5 µl of cDNA, 10 µl of
2× SYBR Primer Ex TagII (TaKaRa), 7.5 µl of ddH2O and 1
µl of primers (10 µM). The ABI 7500 system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) was used to perform the
amplification reaction, using the following thermal cycling
profile: 94°C for 10 min, followed by 40 cycles of amplification
(94°C for 30 sec, 56°C for 30 sec and 72°C for 30 sec), and 72°C
for 10 min. Each experiment was performed in triplicate and was
analyzed using the 2−ΔΔCq method (20).
Western blot analysis
RIPA buffer (cat. no. P0013C; Beyotime Institute of
Biotechnology) was used for the extraction and concentration
determination of total protein from cells. Protein concentrations
were determined with a BCA Protein Assay kit. Protein samples (30
µg) were separated via SDS-PAGE (8–10%) and transferred onto
polyvinylidene fluoride membranes (EMD Millipore). The membranes
were blocked using 5% non-fat milk for 2 h at room temperature, and
then incubated with the following primary antibodies at 4°C
overnight: HPV16 E7 (cat. no. sc-6981; 1:1,000; 21 kDa) and HPV18
E7 (cat. no. sc-365035; 1:1,000; 15 kDa) both from Santa Cruz
Biotechnology, lnc., p21 (product code ab109520; 1:1,000; 21 kDa),
caspase-3 (product code ab32351; 1:5,000; 32 kDa), Bcl-2 (product
code ab32124; 1:1,000; 26 kDa), c-JUN (product code ab32137;
1:5,000; 36 kDa), cysteine-rich 61 (Cyr61; product code ab24448;
1:1,000; 42 kDa), myosin regulatory light polypeptide 9 (MYL9;
product code ab191393; 1:1,000; 20 kDa) and GAPDH (product code
ab9485; 1:2,500; 37 kDa) all from Abcam. The membranes were
subsequently incubated with HRP-conjugated IgG secondary antibodies
(product code ab7090; 1:4,000; Abcam) at room temperature for 2 h.
The chemiluminescence intensity was detected using an ECL kit (EMD
Millipore) according to the manufacturer's protocol and ImageJ
v1.8.0 software (National Institutes of Health) was used to analyze
the gray value of the target band.
Cell Counting Kit-8 (CCK-8) assay
Cells were seeded into 96-well plates (Corning,
Inc.) at a density of 1×104/100 µl and incubated at 37°C
with 5% CO2 for 24 h. CCK-8 (10 µl/ml; Dojindo Molecular
Technologies, Inc.) was subsequently added to each well and
incubated at 37°C with 5% CO2 for a further 4 h, after
which the absorbance was measured using a microplate reader
(Bio-Rad Laboratories, Inc.) at 450 nm.
Flow cytometric assay
The apoptosis of HeLa, CaSki and C33A cells was
detected via flow cytometry. After transfection for 48 h, cells
were collected and stained using an Annexin V-FITC/propidium iodide
(PI) Apoptosis Detection kit (Beyotime Institute of Biotechnology)
according to the manufacturer's protocol. Fluorescence signals were
detected using a FACSCanto II flow cytometer (BD Biosciences) and
analyzed using FlowJo 7.6.5 software (FlowJo LLC).
EDU and DAPI staining assay
Cellular proliferation and apoptosis was assessed
using the BeyoClick™ EdU Cell Proliferation kit with Alexa Fluor
488 (Beyotime Institute of Biotechnology) and DAPI dihydrochloride
(Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. A total of 1×104 cells were
seeded in 24-well plates and incubated at 37°C and 5%
CO2 with 10% FBS-medium for 12 h. Cells were then fixed
in 4% paraformaldehyde for 15 min and permeabilized with 0.3%
Triton X-100 for 20 min at room temperature. Cells were washed
thrice with PBS and cultured at room temperature with 100 µl Click
Reaction Mixture (50 µM) for 20 min in the dark. Cell nuclei were
counterstained with 100 µl DAPI (1 mg/ml) at room temperature for 5
min. A fluorescence microscope of 10×20 (Carl Zeiss AG) was used to
count the number of proliferative/apoptotic cells in three random
fields of view per slide.
Transwell assay
For the cell invasion assay, the upper chamber (8-µm
pore size; Costar; Coring, Inc.) was supplemented with Matrigel,
while an upper chamber without Matrigel was used for the migration
assay. Cells (5×104) were resuspended in serum-free
medium and plated into the upper chamber. Complete medium in the
lower chamber was used as a chemical attractant. After incubation
at 37°C with 5% CO2 for 48 h, the migrated or invasive
cells attached to the lower chamber surface were fixed with 4%
formaldehyde at room temperature for 15 min and stained with 0.5%
crystal violet at room temperature for 30 min. The invasive or
migrated cells in three random fields of view were subsequently
imaged under an inverted light microscope. Experiments were
performed independently and in triplicate.
Bioinformatics analysis
To determine the potential transcription factors
that regulate lnc-EBIC expression, the promoter sequence of
lnc-EBIC was extracted from the UCSC Genome Browser bioinformatics
program (http://www.genome.ucsc.edu) (21) and analyzed via the Gene Transcription
Regulation Database (GTRD; http://gtrd.biouml.org) (22), JASPAR (http://jaspar.genereg.net/) (23) and ChIP-Atlas-Enrichment Analysis
program (http://chip-atlas.org) (24).
Statistical analysis
Data are presented as the mean ± SD and analyzed
using GraphPad Prism V 6.00 software (GraphPad, Inc.). Statistical
significance was determined using a paired Student's t-test or
ANOVA followed by Tukey's post hoc test. P<0.05 was considered
to indicate a statistically significant difference.
Results
HPV16/18 E7 promotes the expression of
lnc-EBIC
To determine whether lnc-EBIC is involved in
cervical cancer progression, Tag2B-HPV18-E7 and Tag2B-HPV16-E7 were
transfected into HeLa and CasKi cells, respectively. As presented
in Fig. 1, the protein expression of
E7 was increased in HeLa (Fig. 1A and
C) and CasKi cells (Fig. 1E and
G). Additionally, the expression of lnc-EBIC was significantly
increased (Fig. 1A and E). To further
confirm whether HPV16/18 E7 regulated the expression of lnc-EBIC,
siRNA specific to HPV18 E7 and HPV16 E7 was transfected into HeLa
and CasKi cells to knockdown endogenous E7 expression. The
interfering efficiency of HPV16/18 E7 is presented in Fig. 1D and H. The results of RT-qPCR
demonstrated that HPV16/18 E7 silencing significantly blocked the
expression of lnc-EBIC (Fig. 1B and
F). The results indicated that the HPV16/18 E7 protein promoted
the excessive expression of lnc-EBIC in cervical cancer cells.
lnc-EBIC overexpression regulates the
proliferation, apoptosis, the cell cycle, migration and invasion of
HPV− C33A cervical cancer cells
To investigate the role of lnc-EBIC in cervical
cancer, the pcDNA3.1-lnc-EBIC overexpression plasmid and
corresponding NC were transfected into HPV− C33A cells
(Fig. 2A). The results of the CCK-8
assay revealed that cell viability was significantly increased in
C33A cells transfected with pcDNA3.1-lnc-EBIC compared with
pcDNA3.1-transfected cells after 72 h (Fig. 2B). Additionally, EdU staining further
confirmed that the upregulation of lnc-EBIC enhanced the
proliferation of C33A cells (Fig.
2C). The effect of this upregulation on the cell cycle was then
assessed via flow cytometry. The results demonstrated a markedly
increased number of cells in the S and G2 phases (Fig. 2D), indicating that the upregulation of
lnc-EBIC enhanced the cell cycle transition of cervical cancer
cells. The results of the Transwell assay further demonstrated that
cell migration and invasion were markedly increased in C33A cells
transfected with pcDNA3.1-lnc-EBIC (Fig.
2E). In addition, an Annexin V-FITC/PI assay was performed to
evaluate the apoptosis of C33A cells, the results of which revealed
that the apoptotic rate was significantly reduced in
lnc-EBIC-overexpression cells compared with the control (Fig. 2F). Moreover, the protein expression of
pro-apoptotic p21 and Cleaved caspase-3 were decreased, and
anti-apoptotic Bcl-2, c-JUN, Cyr61 and MYL9 were increased in
lnc-EBIC-overexpressing C33A cells compared with
pcDNA3.1-transfected C33A cells (Fig.
2G). The results indicated that lnc-EBIC promoted cell
proliferation, cell cycle progression, migration and invasion, and
inhibited the apoptosis of cervical cancer cells in HPV−
cervical cancer cells.
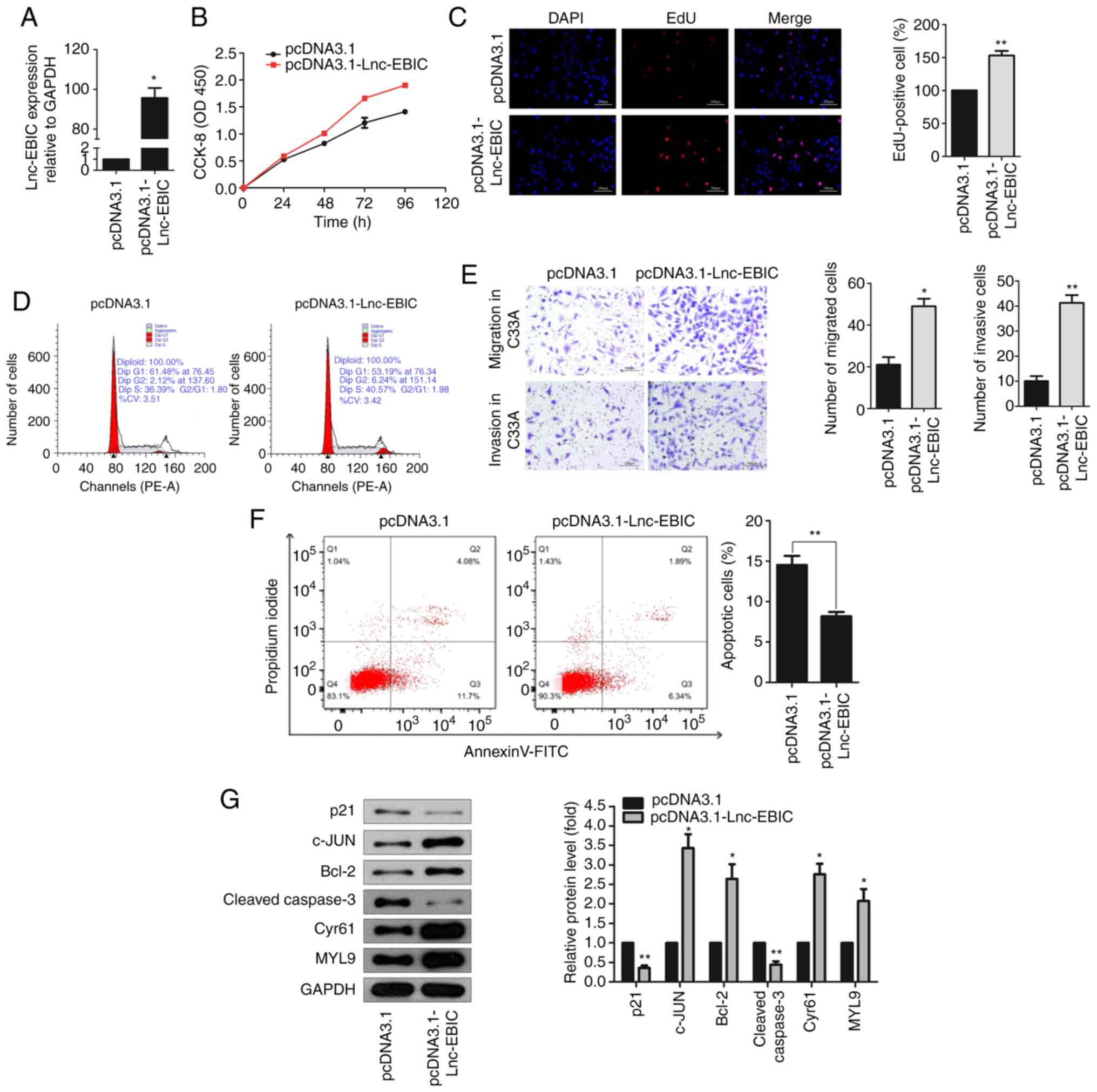 | Figure 2.lnc-EBIC affects the function of
cervical cancer cells. (A) The expression of lnc-EBIC in C33A cells
was detected by RT-qPCR. C33A cells were transfected with pcDNA3.1
and pcDNA3.1-lnc-EBIC, respectively. (B) The proliferation of C33A
cells was increased following lnc-EBIC overexpression. CCK-8 assays
were performed at 24 h intervals as indicated. (C) The fractions of
S-phase C33A cells were increased upon lnc-EBIC-overexpression. EdU
assays were applied to visualize cells in the S-phase of the cell
cycle. (D) Cell cycle analysis of the lnc-EBIC-overexpressed C33A
cells. PI-stained C33A cells were subjected to FACS. (E) Transwell
assays indicate that lnc-EBIC-overexpression increased C33A cell
migration and invasion. (F) Overexpression of lnc-EBIC decreased
the percentage of apoptotic cells. The percentage of cells in each
quadrant is indicated. (G) The western blot analysis of cellular
functional proteins on cell cycle, apoptosis, migration lysates
from lnc-EBIC-overexpressed in C33A cells. The blots revealed
lnc-EBIC increased the expression of c-JUN, Bcl-2, Cyr61, MYL9 and
reduced the expression of p21 and Cleaved caspase-3. Data are
presented as the means ± SD. n=3. *P<0.05 and **P<0.01. lnc,
long non-coding RNA; lnc-EBIC, enhancer of zeste homolog 2-binding
lncRNA in cervical cancer; RT-qPCR, reverse
transcription-quantitative PCR; CCK-8, Cell Counting Kit-8. |
lnc-EBIC is required for the HPV16/18
protein, E7, to serve tumorigenic activities in cervical cancer
cells
To determine the function of lnc-EBIC in E7-mediated
tumorigenesis, siRNA targeting lnc-EBIC was co-transfected with an
E7-overexpression plasmid in HeLa and CasKi cells (Fig. S1A and B). Western blot analysis
revealed that lnc-EBIC knockdown significantly inhibited the
promotive effects of E7 on anti-apoptotic proteins c-JUN, Bcl-2,
Cyr61 and MYL9 in HPV+ cervical cancer cells, and
increased the expression of pro-apoptotic proteins p21 and Cleaved
caspase-3, which were decreased by E7 overexpression (Figs. 3A and S1C). The EdU staining and Annexin V-FITC/PI
assay further confirmed that lnc-EBIC knockdown suppressed the
tumorigenic effects of E7 on the proliferation and apoptosis of
HeLa (Fig. 3B and C) and CasKi
(Fig. S1D and E) cells. A series of
Transwell assays were performed to evaluate the influence of
lnc-EBIC on the migration and invasion of HeLa and CasKi cells.
Compared with HPV16/18 E7 overexpression alone, co-transfection
with lnc-EBIC siRNA significantly inhibited the migration and
invasion of HeLa (Fig. 3D) and CasKi
(Fig. S1F) cells. The results
indicated that lnc-EBIC may be an important mediator of E7 that
enhances tumorigenic activities in cervical cancer cells.
HPV16/18 E7 protein promotes lnc-EBIC
expression by inhibiting TAL1 expression
HPV E7 is not a DNA-binding transcription factor
(3,25). Thus, the effect of HPV E7 on lnc-EBIC
expression may be mediated by cellular transcription factors. TAL1
was identified in the three databases. The present study
demonstrated that E7 overexpression in HeLa (Fig. 4A-C) and CasKi cells (Fig. 4D-F) significantly decreased the
expression of TAL1, with the opposite effect when E7 knockdown. To
further confirm whether E7 depended on TAL1 suppression to enhance
lnc-EBIC expression, lnc-EBIC levels were assessed in cervical
cancer cells transfected with siRNA against TAL1 alone or in the
presence of E7 (Fig. 4G-I). In C33A,
HeLa and CasKi cervical cancer cells, TAL1 knockdown significantly
increased the expression of lnc-EBIC, an effect that was
significantly suppressed following E7 inhibition (Fig. 4J-L). The results suggested that TAL1
inactivation may serve an important role in HPV E7-induced lnc-EBIC
upregulation.
HPV16/18 E7 protein depends on
lnc-EBIC to suppress KLHDC7B expression
Previous studies have demonstrated that KLHDC7B
promotes HPV viral replication and secretion in HPV-infected
cervical intraepithelial neoplasia (26). Furthermore, transcriptome sequencing
analysis conducted in a previous study revealed that KLHDC7B was
decreased in siRNA lnc-EBIC transfected HeLa cells (13). The expression of KLHDC7B in pcDNA3.1
and pcDNA3.1-lnc-EBIC transfected C33A cells was therefore assessed
in the present study. The results revealed that the mRNA expression
of KLHDC7B was significantly increased in pcDNA3.1-lnc-EBIC C33A
cells compared with pcDNA3.1-transfected cells (Fig. 5A). Conversely, lnc-EBIC knockdown
significantly decreased the expression of KLHDC7B in C33A cells
(Fig. 5B). These results were further
confirmed via western blot analysis (Fig.
5C). To determine whether the regulatory role of lnc-EBIC on
KLHDC7B expression was associated with HPV16/18 E7, HeLa cells were
co-transfected with Tag2B-HPV18-E7 and siRNA lnc-EBIC. Western blot
analysis revealed that lnc-EBIC knockdown markedly decreased the
expression of KLHDC7B in HPV18 E7-overexpression cells (Fig. 5D and E). Similar results were obtained
in HPV16 E7-transfected CasKi cells (Fig.
5F and G). The results indicated that KLHDC7B could be
upregulated by lnc-EBIC and that this effect might be further
enhanced by HPV16/18 E7 in cervical cancer cells.
KLHDC7B cooperates with lnc-EBIC to
promote the tumorigenic activities of cervical cancer cells
To elucidate whether KLHDC7B was involved in
lnc-EBIC-mediated tumorigenic activity, C33A cells were transfected
with the lnc-EBIC overexpression plasmid alone or in the presence
of siRNA KLHDC7B. As presented in Fig.
6A, lnc-EBIC overexpression significantly increased the
expression of anti-apoptotic c-JUN, Bcl-2, Cyr61 and MYL9, and
reduced the expression of pro-apoptotic p21 and Cleaved caspase-3
in C33A cells. Moreover, as predicted, KLHDC7B knockdown
significantly suppressed the effects of lnc-EBIC on the expression
of apoptotic proteins (Fig. 6A) and
markedly inhibited the tumor-promotive effects of lnc-EBIC,
including increased cellular proliferation (Fig. 6B), migration and invasion (Fig. 6D), and decreased apoptosis (Fig. 6C) in C33A cells. The results
demonstrated that KLHDC7B served an important role in the
lnc-EBIC-mediated tumorigenic activities of cervical cancer
cells.
Discussion
lncRNAs serve key regulatory roles in the occurrence
and progression of cervical cancer. For example, breast cancer
anti-estrogen resistance 4, a lapatinib-responsive lncRNA (an
EGFR/HER2 inhibitor) enhanced cell proliferation in
estrogen-resistant breast cancer, and serves as a
metastasis-promoting lncRNA in cervical cancer (27). A nine-lncRNA signature composed of
ATXN8 opposite strand lncRNA, chromosome 5 Open Reading Frame 60,
DIO3 opposite strand upstream RNA, EMX2 opposite strand/antisense
RNA, inactivation escape 1, KCNQ1 downstream neighbor protein,
KCNQ1 overlapping transcript 1, loss of heterozygosity on
chromosome 12 region 2 and RFPL1 antisense RNA 1 exhibits great
potency for the prediction of cervical cancer recurrence (28). Moreover, recent studies have indicated
that lncRNAs including growth arrest-specific 5, H19 imprinted
maternally expressed transcript (non-protein coding), FAM83H
antisense RNA 1, metastasis-associated lung adenocarcinoma
transcript 1 and CCEPR (11,12) can be exploited by HPVs to perform
tumorigenic activities. However, these lncRNAs are specifically
regulated by E6; less is known about the lncRNAs that are regulated
by E7. The present study revealed that oncogenic lnc-EBIC could be
exploited by HPV16/18 E7 to accelerate the proliferation, migration
and invasion, and inhibit apoptosis in cervical cancer cells.
lnc-EBIC also exhibited oncogenic activities even in
HPV− cervical cancer cells. Therefore, lnc-EBIC may be a
novel therapeutic target for patients with HPV+ and
HPV− cervical cancers.
lncRNAs regulate a variety of critical cellular
processes by promoting or repressing transcription, serving as
epigenetic regulators or as scaffolds to interact with various
proteins in cervical cancer (29,30).
Lnc-cervical cancer DExH-box helicase 9 (DHX9) suppressive
transcript (lnc-CCDST), a recently identified tumor-suppressive
lncRNA that can be abolished by E7, has been revealed to promote
pro-oncogenic DHX9 degradation by serving as a scaffold to
facilitate the formation of mouse double minute 2 (MDM2) and DHX9
complexes, while not influencing the mRNA expression of DHX9
(31). Lnc-EBIC has been confirmed to
act as a competing endogenous RNA that sequesters tumor repressors
miR-375 and miR-139, which target HPV16/18 E6/E7 mRNA (13), and to serve as a scaffold that
interacts with EZH2, thus repressing E-cadherin expression
(14,15). The present study further demonstrated
that lnc-EBIC promoted cellular proliferation, migration and
invasion, and inhibited apoptosis in cervical cancer cells by
enhancing KLHDC7B expression.
The transcription factor TAL1 is an essential
regulator of hematopoiesis that promotes prostate cancer cell
growth via the MAPK/ERK, PI3K/AKT and AMPK signaling pathways
(32). However, its role in cervical
cancer remains unknown. The present study revealed that TAL1 was
significantly downregulated in HPV E7 cervical cells. Furthermore,
it was demonstrated that the inactivation of TAL1 may serve an
important role in HPV E7-induced lnc-EBIC upregulation.
KLHDC7B is associated with an aggressive subtype of
cancer and predicts a poor prognosis in patients with breast
(16) and laryngeal (33) cancers. Furthermore, KLHDC7B is
upregulated in HPV-induced vulvar intraepithelial neoplasia
(26) and can be used as a biomarker
for the diagnosis and prognostic prediction of patients with
cervical cancer (19). In the present
study, interfering KLHDC7B expression was observed to significantly
inhibit the oncogenic activities of lnc-EBIC. Thus, KLHDC7B may be
a pivotal target of lnc-EBIC in cervical cancer cells. As both mRNA
and protein levels of KLHDC7B were enhanced by lnc-EBIC, the exact
interaction pathway between lnc-EBIC and KLHDC7B requires further
elucidation.
E7 performs oncogenic activities by modulating the
expression of several host proteins, including pRB, p107, p130,
p21, octamer-binding transcription factor 4 and PTPN14 (34,35). The
present study demonstrated that HPV E7 was dependent on the
inhibition of TAL1 to promote lnc-EBIC expression. TAL1 is a
transcription factor that is aberrantly expressed in 60% of cases
of human T-cell acute lymphoblastic leukemia (T-ALL) cases,
activating several important oncogenes, including the MYC, MYB,
Notch1, cyclin E, and tribbles pseudokinase 2 (36,37).
Bioinformatic analysis was performed in the present study to
identify the lncRNAs that are regulated by TAL1 in T-ALL cells
(38). The results revealed that
lnc-EBIC was one such lncRNA. Additionally, a transcriptome
profiling study determined that TAL1 was overexpressed in
gastric-type cervical cancer that was not associated with HPV
infection (39). TAL1 knockdown in
HPV+ (HeLa and CasKi) and HPV− (C33A) cells
in the present study induced a significant increase in lnc-EBIC
expression. TAL1 may therefore represent a novel target for E7 in
HPV infection. However, its role in cervical cancer progression
requires further clarification.
In conclusion, the present study revealed that
oncogenic lnc-EBIC can be exploited by HPV16/18 E7 to increase
cellular proliferation, migration and invasion, and decrease
apoptosis in cervical cancer cells. Molecular analysis revealed
that E7 is dependent on the TAL1/lnc-EBIC/KLHDC7B axis to perform
its tumor-promotive activities. Furthermore, lnc-EBIC exhibited
oncogenic activity by enhancing KLHDC7B expression in
HPV− cervical cancer cells. Thus, the lnc-EBIC/KLHDC7B
axis represents a novel molecular mechanism and potential
therapeutic target for both HPV+ and HPV−
cervical cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (grant no. 81201604), The Open Research
Fund Program of the State Key Laboratory of Virology of China
(grant no. 2015KF010), the Natural Science Foundation of Wuhan
Municipal Health Commission (grant no. WX18Q27), The Top Medical
Young Talents of Hubei Province (2019) and The Yellow Crane Talents
Fund (2016).
Availability of data and materials
All the datasets generated and/or analyzed during
the present study are included in this published article.
Authors' contributions
JW, FX and XL performed the experiments, contributed
to data analysis and wrote the manuscript. XM, XC, XS and YY
analyzed the data. CY, YX and HX conceptualized the study design,
contributed to data analysis and experimental materials. All
authors have read and approved the final version of this
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. Feb 4–2021.(Online
ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Boda D, Neagu M, Constantin C, Voinescu
RN, Caruntu C, Zurac S, Spandidos DA, Drakoulis N, Tsoukalas D and
Tsatsakis AM: HPV strain distribution in patients with genital
warts in a female population sample. Oncol Lett. 12:1779–1782.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hoppe-Seyler K, Bossler F, Braun JA,
Herrmann AL and Hoppe-Seyler F: The HPV E6/E7 oncogenes: Key
factors for viral carcinogenesis and therapeutic targets. Trends
Microbiol. 26:158–168. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dardiotis E, Siokas V, Garas A,
Paraskevaidis E, Kyrgiou M, Xiromerisiou G, Deligeoroglou E,
Galazios G, Kontomanolis EN, Spandidos DA, et al: Genetic
variations in the SULF1 gene alter the risk of cervical cancer and
precancerous lesions. Oncol Lett. 16:3833–3841. 2018.PubMed/NCBI
|
|
5
|
Boda D, Docea AO, Calina D, Ilie MA,
Caruntu C, Zurac S, Neagu M, Constantin C, Branisteanu DE,
Voiculescu V, et al: Human papilloma virus: Apprehending the link
with carcinogenesis and unveiling new research avenues (Review).
Int J Oncol. 52:637–655. 2018.PubMed/NCBI
|
|
6
|
Li S, Hong X, Wei Z, Xie M, Li W, Liu G,
Guo H, Yang J, Wei W and Zhang S: Ubiquitination of the HPV
oncoprotein E6 is critical for E6/E6AP-mediated p53 degradation.
Front Microbiol. 10:24832019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Martinez-Zapien D, Ruiz FX, Poirson J,
Mitschler A, Ramirez J, Forster A, Cousido-Siah A, Masson M, Vande
Pol S, Podjarny A, et al: Structure of the E6/E6AP/p53 complex
required for HPV-mediated degradation of p53. Nature. 529:541–545.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Celegato M, Messa L, Goracci L, Mercorelli
B, Bertagnin C, Spyrakis F, Suarez I, Cousido-Siah A, Trave G,
Banks L, et al: A novel small-molecule inhibitor of the human
papillomavirus E6-p53 interaction that reactivates p53 function and
blocks cancer cells growth. Cancer Lett. 470:115–125. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ganti K, Massimi P, Manzo-Merino J, Tomaic
V, Pim D, Playford MP, Lizano M, Roberts S, Kranjec C, Doorbar J
and Banks L: Interaction of the human papillomavirus E6 oncoprotein
with sorting nexin 27 modulates endocytic cargo transport pathways.
PLoS Pathog. 12:e10058542016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hatterschide J, Bohidar AE, Grace M,
Nulton TJ, Kim HW, Windle B, Morgan IM, Munger K and White EA:
PTPN14 degradation by high-risk human papillomavirus E7 limits
keratinocyte differentiation and contributes to HPV-mediated
oncogenesis. Proc Natl Acad Sci USA. 116:7033–7042. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Barr JA, Hayes KE, Brownmiller T, Harold
AD, Jagannathan R, Lockman PR, Khan S and Martinez I: Long
non-coding RNA FAM83H-AS1 is regulated by human papillomavirus 16
E6 independently of p53 in cervical cancer cells. Sci Rep.
9:36622019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sharma S and Munger K: Expression of the
cervical carcinoma expressed PCNA regulatory (CCEPR) long noncoding
RNA is driven by the human papillomavirus E6 protein and modulates
cell proliferation independent of PCNA. Virology. 518:8–13. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He H, Liu X, Liu Y, Zhang M, Lai Y, Hao Y,
Wang Q, Shi D, Wang N, Luo XG, et al: Human papillomavirus E6/E7
and long noncoding RNA TMPOP2 mutually upregulated gene expression
in cervical cancer cells. J Virol. 93:e01808–18. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dong J, Su M, Chang W, Zhang K, Wu S and
Xu T: Long non-coding RNAs on the stage of cervical cancer
(Review). Oncol Rep. 38:1923–1931. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun NX, Ye C, Zhao Q, Zhang Q, Xu C, Wang
SB, Jin ZJ, Sun SH, Wang F and Li W: Long noncoding RNA-EBIC
promotes tumor cell invasion by binding to EZH2 and repressing
E-cadherin in cervical cancer. PLoS One. 9:e1003402014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jeong G, Bae H, Jeong D, Ham J, Park S,
Kim HW, Kang HS and Kim SJ: A Kelch domain-containing KLHDC7B and a
long non-coding RNA ST8SIA6-AS1 act oppositely on breast cancer
cell proliferation via the interferon signaling pathway. Sci Rep.
8:129222018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Martin-Pardillos A and Cajal SRY:
Characterization of Kelch domain-containing protein 7B in breast
tumours and breast cancer cell lines. Oncol Lett. 18:2853–2860.
2019.PubMed/NCBI
|
|
18
|
Beltrán-Anaya FO, Romero-Córdoba S,
Rebollar-Vega R, Arrieta O, Bautista-Piña V, Dominguez-Reyes C,
Villegas-Carlos F, Tenorio-Torres A, Alfaro-Riuz L, Jiménez-Morales
S, et al: Expression of long non-coding RNA ENSG00000226738
(LncKLHDC7B) is enriched in the immunomodulatory triple-negative
breast cancer subtype and its alteration promotes cell migration,
invasion, and resistance to cell death. Mol Oncol. 13:909–927.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo P, Wang D, Wu J, Yang J, Ren T, Zhu B
and Xiang Y: The landscape of alternative splicing in cervical
squamous cell carcinoma. Onco Targets Ther. 8:73–79.
2015.PubMed/NCBI
|
|
20
|
Wu X, Zheng X, Cheng J, Zhang K and Ma C:
LncRNA TUG1 regulates proliferation and apoptosis by regulating
miR-148b/IGF2 axis in ox-LDL-stimulated VSMC and HUVEC. Life Sci.
243:1172872020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kuhn RM, Haussler D and Kent WJ: The UCSC
genome browser and associated tools. Brief Bioinform. 14:144–161.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mishra GP, Ghosh A, Jha A and Raghav SK:
BedSect: An integrated web server application to perform
intersection, visualization, and functional annotation of genomic
regions from multiple datasets. Front Genet. 11:32020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu Y, Chen X, Cheng R, Yang F, Yu M, Wang
C, Cui S, Hong Y, Liang H, Liu M, et al: The Jun/miR-22/HuR
regulatory axis contributes to tumourigenesis in colorectal cancer.
Mol Cancer. 17:112018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Z, Zhao Z, Cai Z, Sun Y, Li L, Yao F,
Yang L, Zhou Y, Zhu H, Fu Y, et al: Runx2 (Runt-related
transcription factor 2)-mediated microcalcification is a novel
pathological characteristic and potential mediator of abdominal
aortic aneurysm. Arterioscler Thromb Vasc Biol. 40:1352–1369. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tian S, Zhang L, Li Y, Cao D, Quan S, Guo
Y, Yang X and Yang T: Human papillomavirus E7 oncoprotein promotes
proliferation and migration through the transcription factor E2F1
in cervical cancer cells. Anticancer Agents Med Chem. Nov
5–2020.(Epub ahead of print). View Article : Google Scholar
|
|
26
|
Santegoets LA, Seters M, Helmerhorst TJ,
Heijmans-Antonissen C, Hanifi-Moghaddam P, Ewing PC, van Ijcken WF,
van der Spek PJ, van der Meijden WI and Blok LJ: HPV related VIN:
Highly proliferative and diminished responsiveness to extracellular
signals. Int J Cancer. 121:759–766. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Integrated genomic and molecular
characterization of cervical cancer. Nature. 543:378–384. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mao Y, Dong L, Zheng Y, Dong J and Li X:
Prediction of Recurrence in Cervical Cancer Using a Nine-lncRNA
Signature. Front Genet. 10:2842019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hosseini ES, Meryet-Figuiere M,
Sabzalipoor H, Kashani HH, Nikzad H and Asemi Z: Dysregulated
expression of long noncoding RNAs in gynecologic cancers. Mol
Cancer. 16:1072017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Feng S, Liu W, Bai X, Pan W, Jia Z, Zhang
S, Zhu Y and Tan W: LncRNA-CTS promotes metastasis and
epithelial-to-mesenchymal transition through regulating
miR-505/ZEB2 axis in cervical cancer. Cancer Lett. 465:105–117.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ding X, Jia X, Wang C, Xu J, Gao SJ and Lu
C: A DHX9-lncRNA-MDM2 interaction regulates cell invasion and
angiogenesis of cervical cancer. Cell Death Differ. 26:1750–1765.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu X, Xiao Y, Yan W, Ji Z and Zheng G: The
human oncogene SCL/TAL1 interrupting locus (STIL) promotes tumor
growth through MAPK/ERK, PI3K/Akt and AMPK pathways in prostate
cancer. Gene. 686:220–227. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang G, Fan E, Yue G, Zhong Q, Shuai Y,
Wu M, Feng G, Chen Q and Gou X: Five genes as a novel signature for
predicting the prognosis of patients with laryngeal cancer. J Cell
Biochem. Oct 31–2019.(Epub ahead of print).
|
|
34
|
Westrich JA, Warren CJ, Klausner MJ, Guo
K, Liu CW, Santiago ML and Pyeon D: Human papillomavirus 16 E7
stabilizes APOBEC3A protein by inhibiting cullin 2-dependent
protein degradation. J Virol. 92:e01318–17. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Panayiotou T, Michael S, Zaravinos A,
Demirag E, Achilleos C and Strati K: Human papillomavirus E7 binds
Oct4 and regulates its activity in HPV-associated cervical cancers.
PLoS Pathog. 16:e10084682020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sanda T, Lawton LN, Barrasa MI, Fan ZP,
Kohlhammer H, Gutierrez A, Ma W, Tatarek J, Ahn Y, Kelliher MA, et
al: Core transcriptional regulatory circuit controlled by the TAL1
complex in human T cell acute lymphoblastic leukemia. Cancer Cell.
22:209–221. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mansour MR, Sanda T, Lawton LN, Li X,
Kreslavsky T, Novina CD, Brand M, Gutierrez A, Kelliher MA,
Jamieson CH, et al: The TAL1 complex targets the FBXW7 tumor
suppressor by activating miR-223 in human T cell acute
lymphoblastic leukemia. J Exp Med. 210:1545–1557. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ngoc PCT, Tan SH, Tan TK, Chan MM, Li Z,
Yeoh AEJ, Tenen DG and Sanda T: Identification of novel lncRNAs
regulated by the TAL1 complex in T-cell acute lymphoblastic
leukemia. Leukemia. 32:2138–2151. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vasseur D, Lopez J, Croce S, Tondeur G,
Bonin L, Descotes F, Golfier F and Devouassoux-Shisheboran M:
Transcriptome profiling of gastric-type endocervical
adenocarcinomas identifies key signaling pathways for tumor
progression. Gynecol Oncol. 157:775–782. 2020. View Article : Google Scholar : PubMed/NCBI
|




















