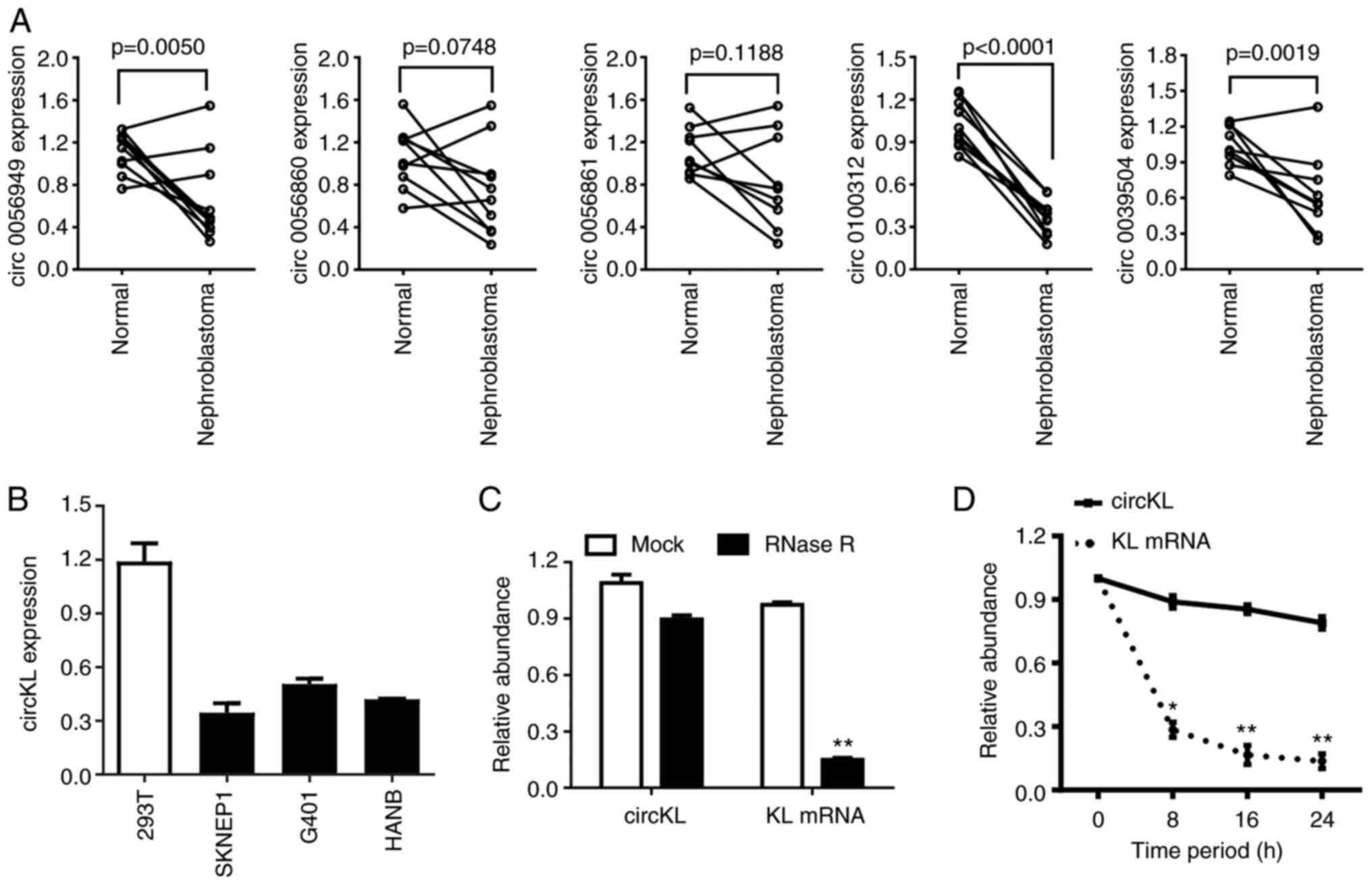
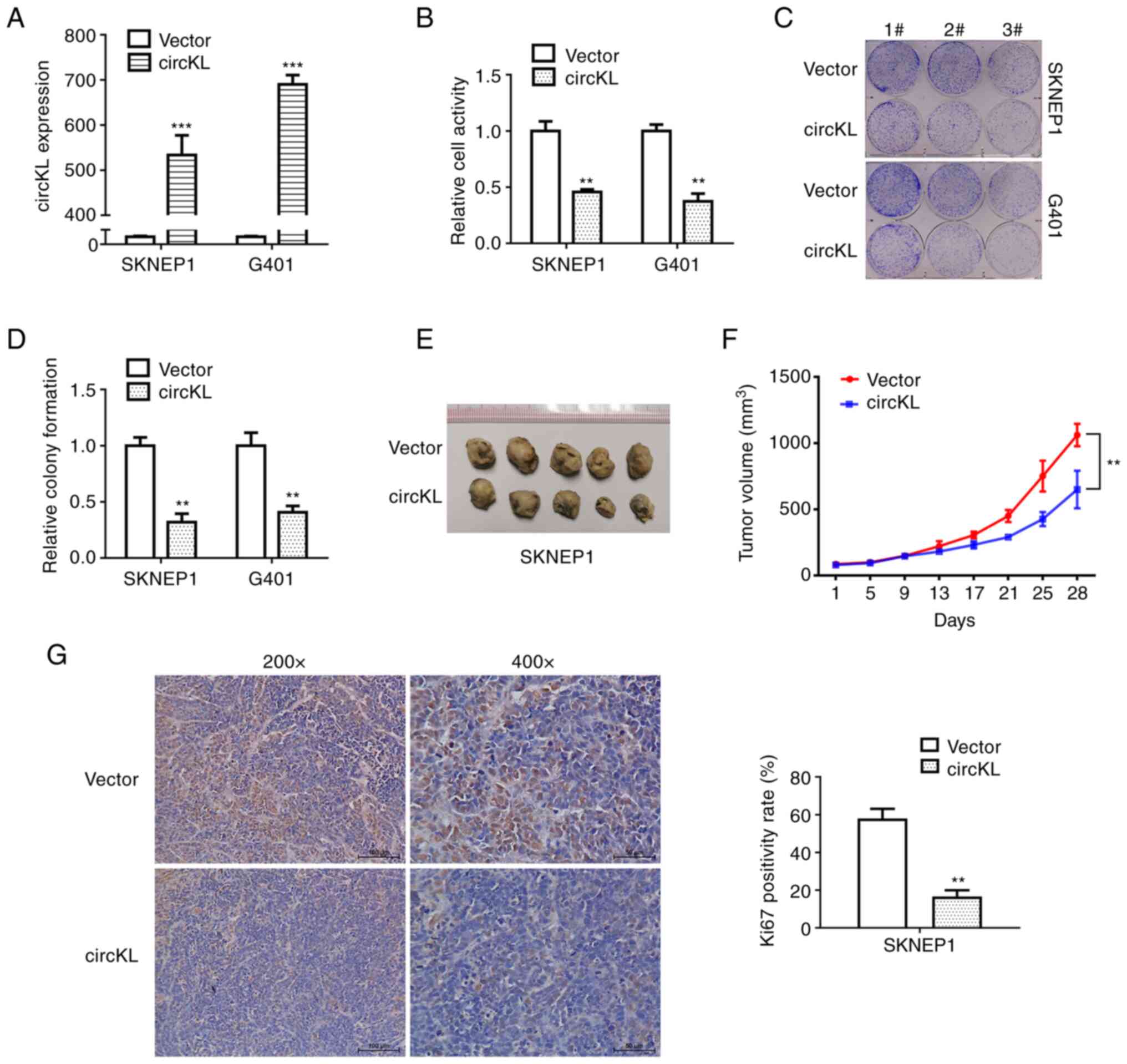
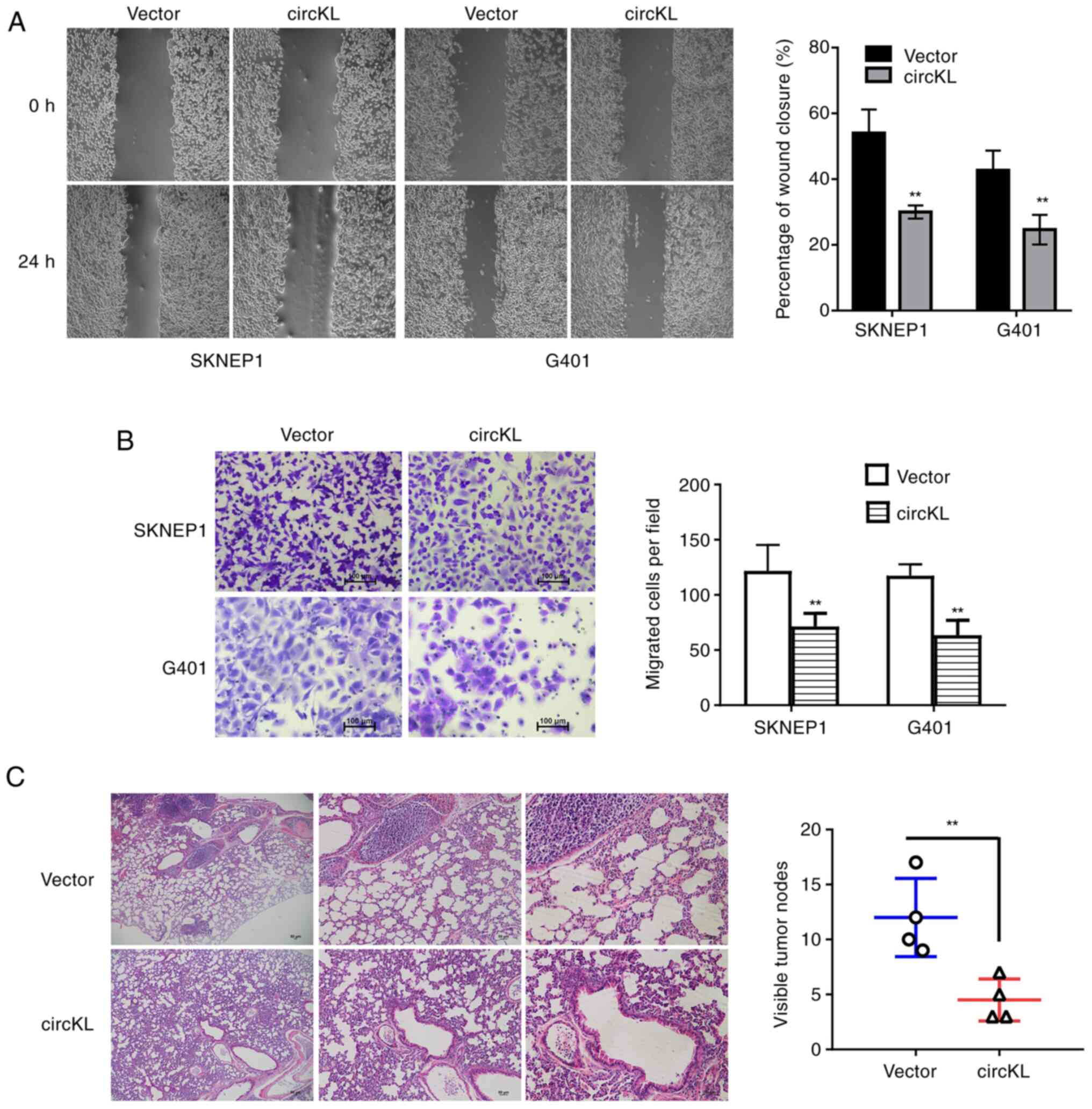
|
1
|
Treger TD, Chowdhury T, Pritchard-Jones K
and Behjati S: The genetic changes of Wilms tumour. Nat Rev
Nephrol. 15:240–251. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chowdhury N and Drake CG: Kidney cancer:
An overview of current therapeutic approaches. Urol Clin North Am.
47:419–431. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Anvar Z, Acurzio B, Roma J, Cerrato F and
Verde G: Origins of DNA methylation defects in Wilms tumors. Cancer
Lett. 457:119–128. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lange J, Peterson SM, Takashima JR,
Grigoriev Y, Ritchey ML, Shamberger RC, Beckwith JB, Perlman E,
Green DM and Breslow NE: Risk factors for end stage renal disease
in non-WT1-syndromic Wilms tumor. J Urol. 186:378–386. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dome JS, Graf N, Geller JI, Fernandez CV,
Mullen EA, Spreafico F, Van den Heuvel-Eibrink M and
Pritchard-Jones K: Advances in Wilms tumor treatment and biology:
Progress through international collaboration. J Clin Oncol.
33:2999–3007. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clericuzio CL and Johnson C: Screening for
Wilms tumor in high-risk individuals. Hematol Oncol Clin North Am.
9:1253–1265. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Goodall GJ and Wickramasinghe VO: RNA in
cancer. Nat Rev Cancer. 21:22–36. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ebbesen KK, Hansen TB and Kjems J:
Insights into circular RNA biology. RNA Biol. 14:1035–1045. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen LL: The expanding regulatory
mechanisms and cellular functions of circular RNAs. Nat Rev Mol
Cell Biol. 21:475–490. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao
L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al: The
landscape of circular RNA in cancer. Cell. 176:869–881.e13. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han B, Chao J and Yao H: Circular RNA and
its mechanisms in disease: From the bench to the clinic. Pharmacol
Ther. 187:31–44. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A
and Qi F: circRNA Cdr1as functions as a competitive endogenous RNA
to promote hepatocellular carcinoma progression. Aging (Albany NY).
11:8182–8203. 2019.
|
|
14
|
Yang W, Yang X, Wang X, Gu J, Zhou D, Wang
Y, Yin B, Guo J and Zhou M: Silencing CDR1as enhances the
sensitivity of breast cancer cells to drug resistance by acting as
a miR-7 sponge to down-regulate REGγ. J Cell Mol Med. 23:4921–4932.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zou Y, Zheng S, Deng X, Yang A and Xie X,
Tang H and Xie X: The role of circular RNA CDR1as/ciRS-7 in
regulating tumor microenvironment: A pan-cancer analysis.
Biomolecules. 9:4292019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zou Y, Zheng S, Deng X, Yang A, Kong Y,
Kohansal M, Hu X and Xie X: Diagnostic and prognostic value of
circular RNA CDR1as/ciRS-7 for solid tumours: A systematic review
and meta-analysis. J Cell Mol Med. 24:9507–9517. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang C, Zeng X, Shan R, Wen W, Li J, Tan
J, Li L and Wan R: The emerging picture of the roles of
circRNA-CDR1as in cancer. Front Cell Dev Biol. 8:5904782020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan
Y, Kong X, Bu J, Liu M and Xu S: circRNA-002178 act as a ceRNA to
promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis.
11:322020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dai X, Zhang N, Cheng Y, Yang T, Chen Y,
Liu Z, Wang Z, Yang C and Jiang Y: RNA-binding protein
trinucleotide repeat-containing 6A regulates the formation of
circular RNA circ0006916, with important functions in lung cancer
cells. Carcinogenesis. 39:981–992. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lin G, Wang S, Zhang X and Wang D:
Circular RNA circPLK1 promotes breast cancer cell proliferation,
migration and invasion by regulating miR-4500/IGF1 axis. Cancer
Cell Int. 20:5932020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kong Y, Yang L, Wei W, Lyu N, Zou Y, Gao
G, Ou X, Xie X and Tang H: CircPLK1 sponges miR-296-5p to
facilitate triple-negative breast cancer progression. Epigenomics.
11:1163–1176. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zou Y, Zheng S, Xiao W and Xie X, Yang A,
Gao G, Xiong Z, Xue Z, Tang H and Xie X: circRAD18 sponges
miR-208a/3164 to promote triple-negative breast cancer progression
through regulating IGF1 and FGF2 expression. Carcinogenesis.
40:1469–1479. 2019.PubMed/NCBI
|
|
24
|
Huang Y, Zhang W, Song H and Sun N: A
nomogram for prediction of distant metastasis in children with
Wilms tumor: A study based on SEER database. J Pediatr Urol.
16:473.e1–473.e9. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chi J, Liu S, Wu Z, Shi Y, Shi C, Zhang T,
Xiong B, Zeng Y and Dong X: circNSUN2 promotes the malignant
biological behavior of colorectal cancer cells via the
miR-181a-5p/ROCK2 axis. Oncol Rep. 46:1422021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cao J, Huang Z, Ou S, Wen F, Yang G, Miao
Q, Zhang H, Wang Y, He X, Shan Y, et al: circ0093740 promotes tumor
growth and metastasis by sponging miR-136/145 and upregulating
DNMT3A in Wilms tumor. Front Oncol. 11:6473522021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dong Y, He D, Peng Z, Peng W, Shi W, Wang
J, Li B, Zhang C and Duan C: Circular RNAs in cancer: An emerging
key player. J Hematol Oncol. 10:22017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bolha L, Ravnik-Glavač M and Glavač D:
Circular RNAs: Biogenesis, function, and a role as possible cancer
biomarkers. Int J Genomic. 2017:62183532017.PubMed/NCBI
|
|
31
|
Li S and Han L: Circular RNAs as promising
biomarkers in cancer: Detection, function, and beyond. Genome Med.
11:152019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen Y, Yang F, Fang E, Xiao W, Mei H, Li
H, Li D, Song H, Wang J, Hong M, et al: Circular RNA circAGO2
drives cancer progression through facilitating HuR-repressed
functions of AGO2-miRNA complexes. Cell Death Differ. 26:1346–1364.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xiao W, Zheng S, Zou Y, Yang A and Xie X,
Tang H and Xie X: CircAHNAK1 inhibits proliferation and metastasis
of triple-negative breast cancer by modulating miR-421 and RASA1.
Aging (Albany NY). 11:12043–12056. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu P, Zou Y, Li X, Yang A, Ye F, Zhang J,
Wei W and Kong Y: circGNB1 facilitates triple-negative breast
cancer progression by regulating miR-141-5p-IGF1R axis. Front
Genet. 11:1932020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu T, Lu Q, Liu J, Xie S, Feng B, Zhu W,
Liu M, Liu Y, Zhou X, Sun W, et al: Circular RNA FAM114A2
suppresses progression of bladder cancer via regulating ∆NP63 by
sponging miR-762. Cell Death Dis. 11:472020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Y, Ge YZ, Xu L and Jia R: Circular RNA
ITCH: A novel tumor suppressor in multiple cancers. Life Sci.
254:1171762020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Verduci L, Strano S, Yarden Y and Blandino
G: The circRNA-microRNA code: Emerging implications for cancer
diagnosis and treatment. Mol Oncol. 13:669–680. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cao MQ, You AB, Zhu XD, Zhang W, Zhang YY,
Zhang SZ, Zhang KW, Cai H, Shi WK, Li XL, et al: miR-182-5p
promotes hepatocellular carcinoma progression by repressing FOXO3a.
J Hematol Oncol. 11:122018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sang Y, Chen B, Song X, Li Y, Liang Y, Han
D, Zhang N, Zhang H, Liu Y, Chen T, et al: circRNA_0025202
regulates tamoxifen sensitivity and tumor progression via
regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther.
27:1638–1652. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xie F, Li Y, Wang M, Huang C, Tao D, Zheng
F, Zhang H, Zeng F, Xiao X and Jiang G: Circular RNA BCRC-3
suppresses bladder cancer proliferation through miR-182-5p/p27
axis. Mol Cancer. 17:1442018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yeh CH, Bellon M and Nicot C: FBXW7: A
critical tumor suppressor of human cancers. Mol Cancer. 17:1152018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yumimoto K and Nakayama KI: Recent insight
into the role of FBXW7 as a tumor suppressor. Semin Cancer Biol.
67:1–15. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–315. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xu Y, Qiu A, Peng F, Tan X, Wang J and
Gong X: Exosomal transfer of circular RNA FBXW7 ameliorates the
chemoresistance to oxaliplatin in colorectal cancer by sponging
miR-18b-5p. Neoplasma. 68:108–118. 2021. View Article : Google Scholar : PubMed/NCBI
|