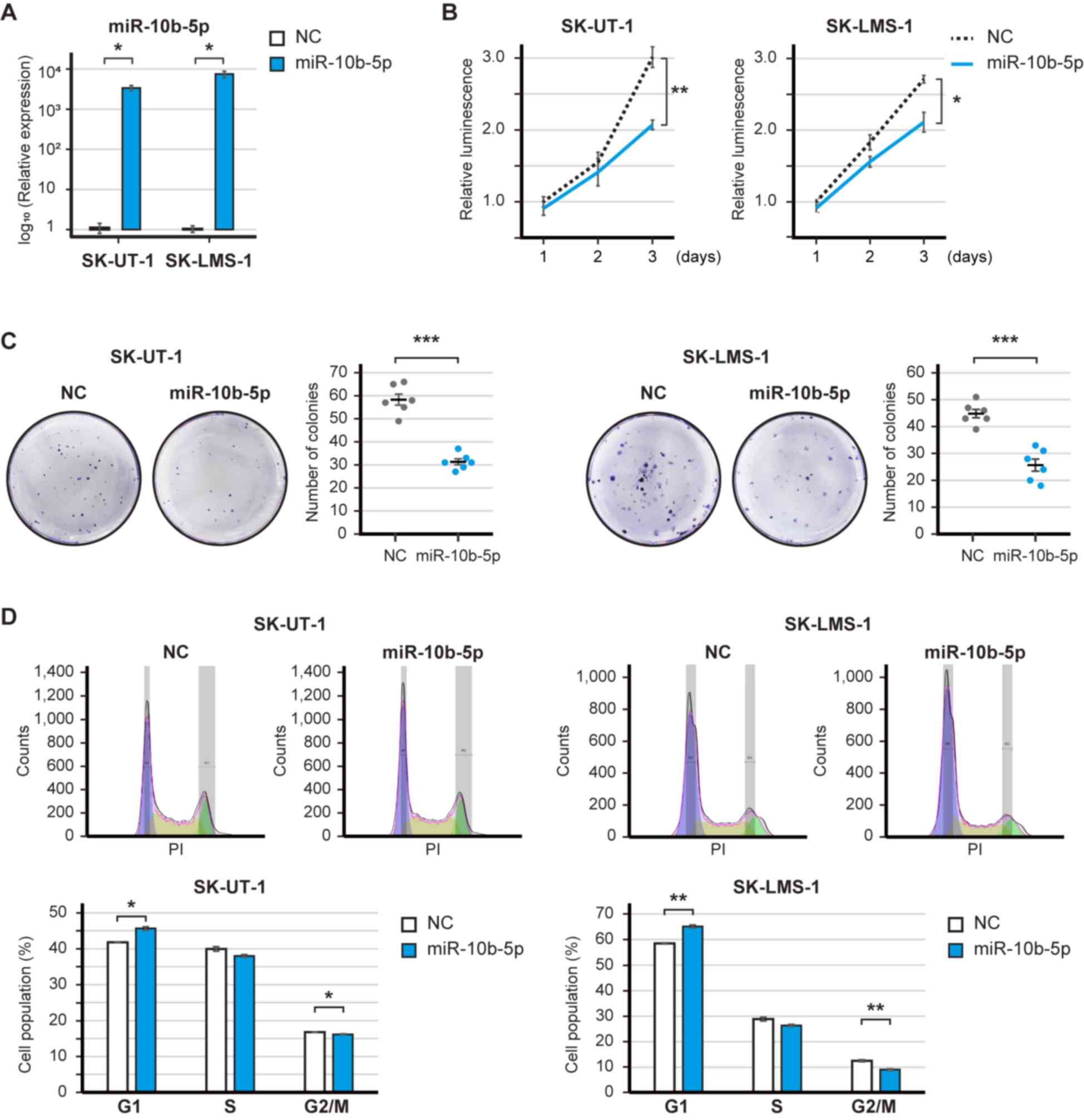
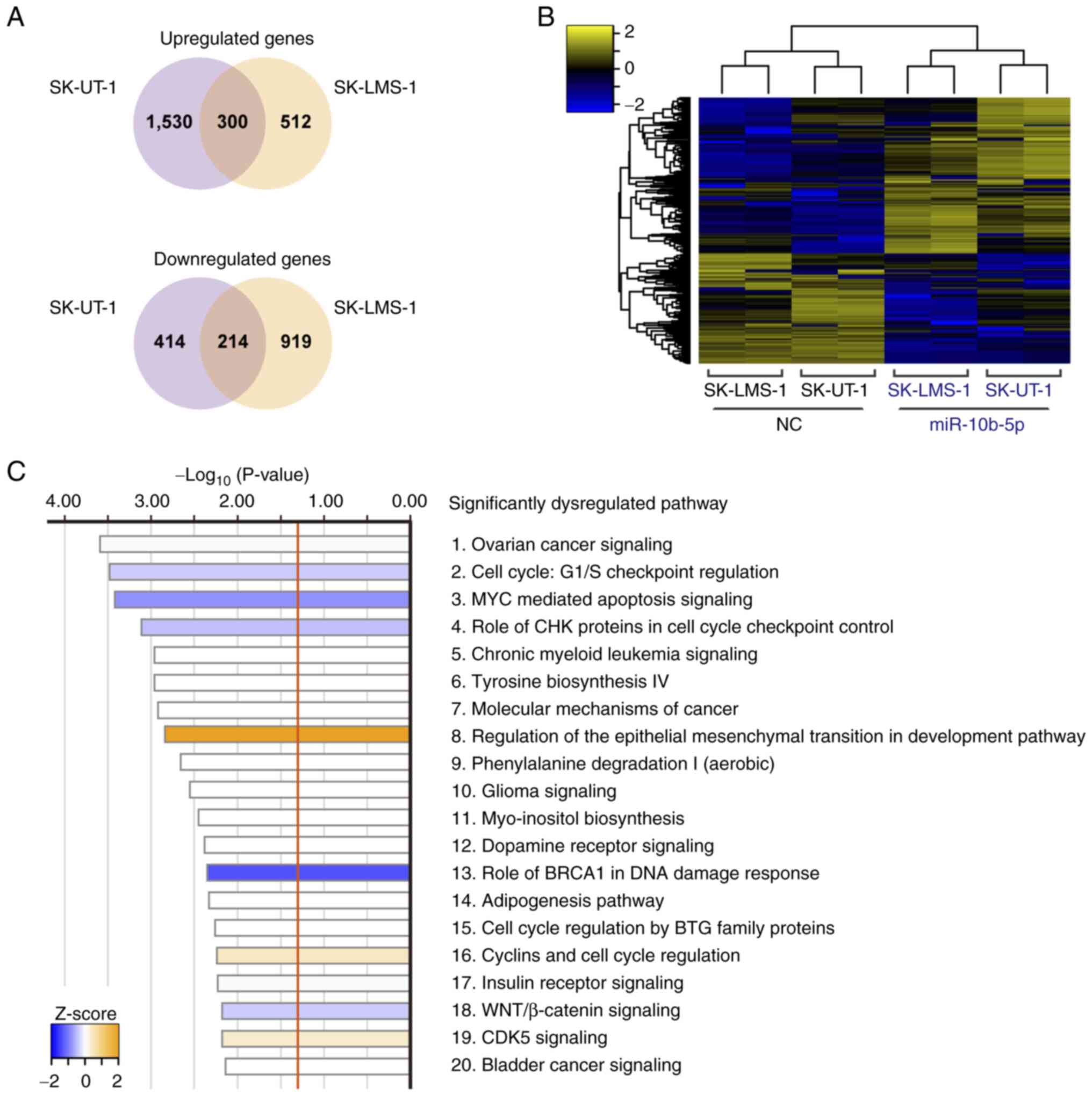
|
1
|
Skorstad M, Kent A and Lieng M: Uterine
leiomyosarcoma-incidence, treatment, and the impact of
morcellation. A nationwide cohort study. Acta Obstet Gynecol Scand.
95:984–990. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
George S, Serrano C, Hensley ML and
Ray-Coquard I: Soft tissue and uterine leiomyosarcoma. J Clin
Oncol. 36:144–150. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Roberts ME, Aynardi JT and Chu CS: Uterine
leiomyosarcoma: A review of the literature and update on management
options. Gynecol Oncol. 151:562–572. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Seagle BL, Sobecki-Rausch J, Strohl AE,
Shilpi A, Grace A and Shahabi S: Prognosis and treatment of uterine
leiomyosarcoma: A national cancer database study. Gynecol Oncol.
145:61–70. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hensley ML, Patel SR, von Mehren M, Ganjoo
K, Jones RL, Staddon A, Rushing D, Milhem M, Monk B, Wang G, et al:
Efficacy and safety of trabectedin or dacarbazine in patients with
advanced uterine leiomyosarcoma after failure of
anthracycline-based chemotherapy: Subgroup analysis of a phase 3,
randomized clinical trial. Gynecol Oncol. 146:531–537. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Blay JY, Schöffski P, Bauer S,
Krarup-Hansen A, Benson C, D'Adamo DR, Jia Y and Maki RG: Eribulin
versus dacarbazine in patients with leiomyosarcoma: Subgroup
analysis from a phase 3, open-label, randomised study. Br J Cancer.
120:1026–1032. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Benson C, Ray-Coquard I, Sleijfer S,
Litière S, Blay JY, Le Cesne A, Papai Z, Judson I, Schöffski P,
Chawla S, et al: Outcome of uterine sarcoma patients treated with
pazopanib: A retrospective analysis based on two European
organisation for research and treatment of cancer (EORTC) soft
tissue and bone sarcoma group (STBSG) clinical trials 62043 and
62072. Gynecol Oncol. 142:89–94. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cuppens T, Moisse M, Depreeuw J, Annibali
D, Colas E, Gil-Moreno A, Huvila J, Carpén O, Zikán M, Matias-Guiu
X, et al: Integrated genome analysis of uterine leiomyosarcoma to
identify novel driver genes and targetable pathways. Int J Cancer.
142:1230–1243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hensley ML, Chavan SS, Solit DB, Murali R,
Soslow R, Chiang S, Jungbluth AA, Bandlamudi C, Srinivasan P, Tap
WD, et al: Genomic landscape of uterine sarcomas defined through
prospective clinical sequencing. Clin Cancer Res. 26:3881–3888.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Astolfi A, Nannini M, Indio V, Schipani A,
Rizzo A, Perrone AM, De Iaco P, Pirini MG, De Leo A, Urbini M, et
al: Genomic database analysis of uterine leiomyosarcoma mutational
profile. Cancers (Basel). 12:21262020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Choi J, Manzano A, Dong W, Bellone S,
Bonazzoli E, Zammataro L, Yao X, Deshpande A, Zaidi S, Guglielmi A,
et al: Integrated mutational landscape analysis of uterine
leiomyosarcomas. Proc Natl Acad Sci USA. 118:e20251821182021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shan W, Akinfenwa PY, Savannah KB,
Kolomeyevskaya N, Laucirica R, Thomas DG, Odunsi K, Creighton CJ,
Lev DC and Anderson ML: A small-molecule inhibitor targeting the
mitotic spindle checkpoint impairs the growth of uterine
leiomyosarcoma. Clin Cancer Res. 18:3352–3365. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yoshida K, Yokoi A, Yamamoto T, Hayashi Y,
Nakayama J, Yokoi T, Yoshida H, Kato T, Kajiyama H and Yamamoto Y:
Aberrant activation of cell cycle-related kinases and the potential
therapeutic impact of PLK1 or CHEK1 inhibition in uterine
leiomyosarcoma. Clin Cancer Res. 28:2147–2159. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kozomara A, Birgaoanu M and
Griffiths-Jones S: miRBase: From microRNA sequences to function.
Nucleic Acids Res. 47(D1): D155–D162. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yoshida K, Yamamoto Y and Ochiya T: miRNA
signaling networks in cancer stem cells. Regen Ther. 17:1–7. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shi G, Perle MA, Mittal K, Chen H, Zou X,
Narita M, Hernando E, Lee P and Wei JJ: Let-7 repression leads to
HMGA2 overexpression in uterine leiomyosarcoma. J Cell Mol Med.
13:3898–3905. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pazzaglia L, Novello C, Conti A, Pollino
S, Picci P and Benassi MS: miR-152 down-regulation is associated
with MET up-regulation in leiomyosarcoma and undifferentiated
pleomorphic sarcoma. Cell Oncol (Dordr). 40:77–88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bray NL, Pimentel H, Melsted P and Pachter
L: Near-optimal probabilistic RNA-seq quantification. Nat
Biotechnol. 34:525–527. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Soneson C, Love MI and Robinson MD:
Differential analyses for RNA-seq: Transcript-level estimates
improve gene-level inferences. F1000Res. 4:15212015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hemming ML, Fan C, Raut CP, Demetri GD,
Armstrong SA, Sicinska E and George S: Oncogenic gene-expression
programs in leiomyosarcoma and characterization of conventional,
inflammatory, and uterogenic subtypes. Mol Cancer Res.
18:1302–1314. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Leitao MM Jr, Hensley ML, Barakat RR,
Aghajanian C, Gardner GJ, Jewell EL, O'Cearbhaill R and Soslow RA:
Immunohistochemical expression of estrogen and progesterone
receptors and outcomes in patients with newly diagnosed uterine
leiomyosarcoma. Gynecol Oncol. 124:558–562. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gonzalez Dos Anjos L, de Almeida BC, Gomes
de Almeida T, Mourão Lavorato Rocha A, De Nardo Maffazioli G,
Soares FA, Werneck da Cunha I, Baracat EC and Carvalho KC: Could
miRNA signatures be useful for predicting uterine sarcoma and
carcinosarcoma prognosis and treatment? Cancers (Basel).
10:3152018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
de Almeida BC, Garcia N, Maffazioli G, dos
Anjos LG, Baracat EC and Carvalho KC: Oncomirs expression profiling
in uterine leiomyosarcoma cells. Int J Mol Sci. 19:522017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang J, Yan Y, Zhang Z and Li Y: Role of
miR-10b-5p in the prognosis of breast cancer. PeerJ. 7:e77282019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Chen D, Li Y, Jin L, Liu J, Su Z, Qi
Z, Shi M, Jiang Z, Ni L, et al: Oncogenic cAMP responsive element
binding protein 1 is overexpressed upon loss of tumor suppressive
miR-10b-5p and miR-363-3p in renal cancer. Oncol Rep. 35:1967–1978.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hu G, Shi Y, Zhao X, Gao D, Qu L, Chen L,
Zhao K, Du J and Xu W: CBFβ/RUNX3-miR10b-TIAM1 molecular axis
inhibits proliferation, migration, and invasion of gastric cancer
cells. Int J Clin Exp Pathol. 12:3185–3196. 2019.PubMed/NCBI
|
|
33
|
Liu F, An X, Zhao X, Zhang N, Chen B, Li Z
and Xu W: MiR-10b-5p inhibits tumorigenesis in gastric cancer
xenograft mice model through down-regulating Tiam1. Exp Cell Res.
407:1128102021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yan T, Wang X, Wei G, Li H, Hao L, Liu Y,
Yu X, Zhu W, Liu P, Zhu Y and Zhou X: Exosomal miR-10b-5p mediates
cell communication of gastric cancer cells and fibroblasts and
facilitates cell proliferation. J Cancer. 12:2140–2150. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lo PK, Zhang Y, Yao Y, Wolfson B, Yu J,
Han SY, Duru N and Zhou Q: Tumor-associated myoepithelial cells
promote the invasive progression of ductal carcinoma in situ
through activation of TGFβ signaling. J Biol Chem. 292:11466–11484.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang B, Zhang Y, Zhang H, Lin F, Tan Q,
Qin Q, Bao W, Liu Y, Xie J and Zeng Q: Long intergenic non-protein
coding RNA 324 prevents breast cancer progression by modulating
miR-10b-5p. Aging (Albany NY). 12:6680–6699. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu C, Jiang W, Hui B, Rong D, Fu K, Dong
C, Tang W and Cao H: The circ_0021977/miR-10b-5p/P21 and P53
regulatory axis suppresses proliferation, migration, and invasion
in colorectal cancer. J Cell Physiol. 235:2273–2285. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li W, Li C, Xiong Q, Tian X and Ru Q:
MicroRNA-10b-5p downregulation inhibits the invasion of glioma
cells via modulating homeobox B3 expression. Exp Ther Med.
17:4577–4585. 2019.PubMed/NCBI
|
|
39
|
Yang Y, Liu X, Zheng J, Xue Y, Liu L, Ma
J, Wang P, Yang C, Wang D, Shao L, et al: Interaction of BACH2 with
FUS promotes malignant progression of glioma cells via the
TSLNC8-miR-10b-5p-WWC3 pathway. Mol Oncol. 14:2936–2959. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li B, Yang C, Zhu Z, Chen H and Qi B:
Hypoxic glioma-derived extracellular vesicles harboring
MicroRNA-10b-5p enhance M2 polarization of macrophages to promote
the development of glioma. CNS Neurosci Ther. 28:1733–1747. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yoshida K, Yokoi A, Kato T, Ochiya T and
Yamamoto Y: The clinical impact of intra- and extracellular miRNAs
in ovarian cancer. Cancer Sci. 111:3435–3444. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shan X, Zhang L, Zhu DX, Zhou X, Zhang H,
Liu QX, Tang JW, Wen W, Wang TS, Zhu W and Liu P: Serum microRNA
expression profiling revealing potential diagnostic biomarkers for
lung adenocarcinoma. Chin Med J (Engl). 133:2532–2542. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cho HJ, Eun JW, Baek GO, Seo CW, Ahn HR,
Kim SS, Cho SW and Cheong JY: Serum exosomal MicroRNA, miR-10b-5p,
as a potential diagnostic biomarker for early-stage hepatocellular
carcinoma. J Clin Med. 9:2812020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ghosh S, Bhowmik S, Majumdar S, Goswami A,
Chakraborty J, Gupta S, Aggarwal S, Ray S, Chatterjee R,
Bhattacharyya S, et al: The exosome encapsulated microRNAs as
circulating diagnostic marker for hepatocellular carcinoma with low
alpha-fetoprotein. Int J Cancer. 147:2934–2947. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yokoi A, Matsuzaki J, Yamamoto Y, Tate K,
Yoneoka Y, Shimizu H, Uehara T, Ishikawa M, Takizawa S, Aoki Y, et
al: Serum microRNA profile enables preoperative diagnosis of
uterine leiomyosarcoma. Cancer Sci. 110:3718–3726. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ravid Y, Formanski M, Smith Y, Reich R and
Davidson B: Uterine leiomyosarcoma and endometrial stromal sarcoma
have unique miRNA signatures. Gynecol Oncol. 140:512–517. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Git A, Dvinge H, Salmon-Divon M, Osborne
M, Kutter C, Hadfield J, Bertone P and Caldas C: Systematic
comparison of microarray profiling, real-time PCR, and
next-generation sequencing technologies for measuring differential
microRNA expression. RNA. 16:991–1006. 2010. View Article : Google Scholar : PubMed/NCBI
|