Introduction
Circular RNAs (circRNAs) are a class of closed
circular RNA molecules composed of upstream splicing receptors and
downstream splicing donors. Owing to the lack of a 5′-cap and a
3′-polyadenylated [poly(A)] tail, circRNAs are more stable compared
with linear RNA, and these molecules are not easily degraded by
nucleic acid exonucleases. Originally, circRNAs were hypothesized
to arise as the products of splicing errors or as intermediate
products that escaped from the ‘lasso’ structure of introns, but
given the development of high-throughput sequencing technologies
and bioinformatics-based tools, circRNAs are no longer considered
to be a class of RNA molecules that lack any role in humans, and
their highly stable structure and remarkable tissue specificity
support this notion. Conversely, an increasing number of circRNAs
have been found to regulate tumor proliferation, migration,
invasion, apoptosis, differentiation, metabolism, and angiogenesis
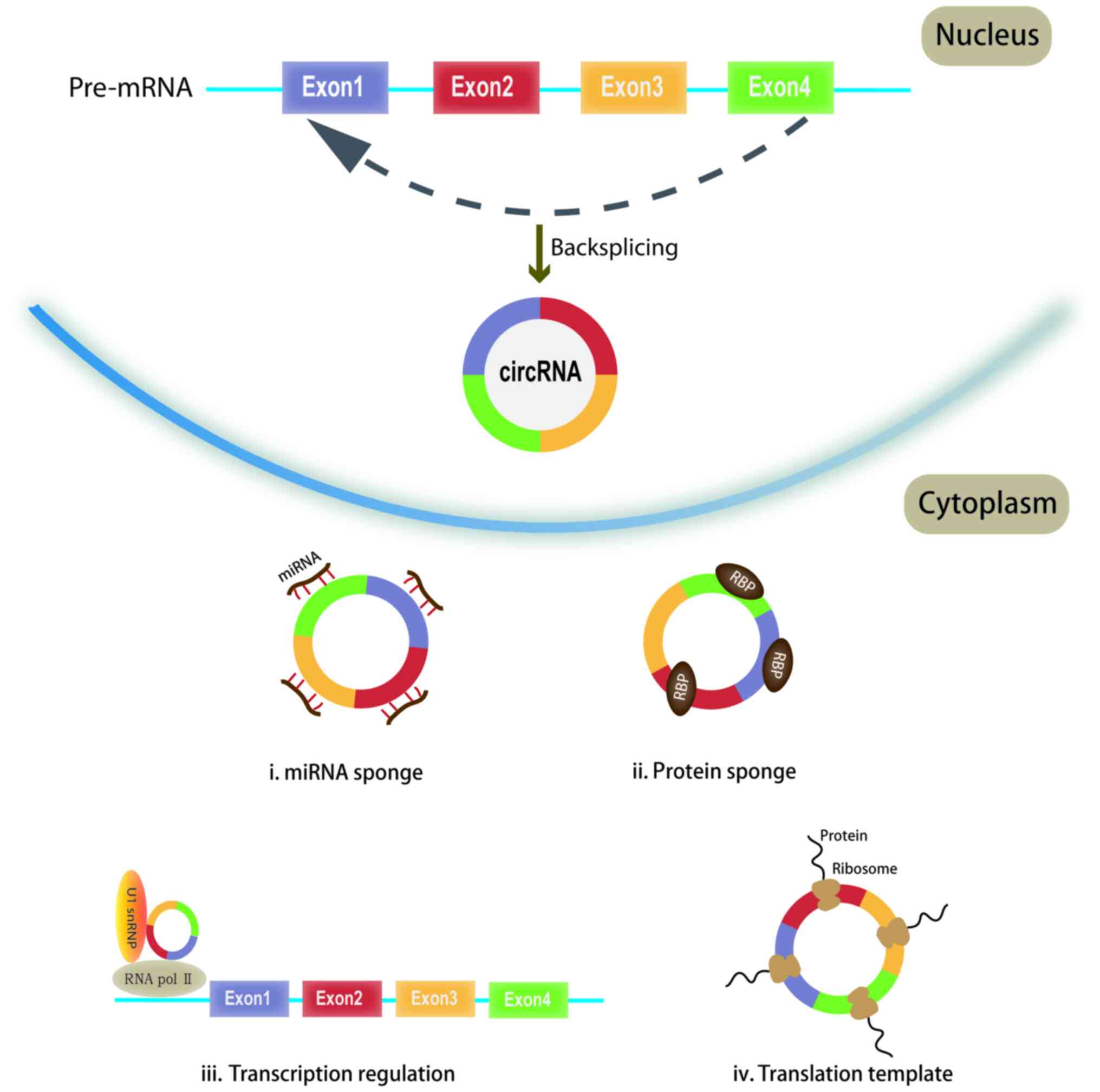
(1,2) by acting as ‘sponges’ of microRNAs
(miRNAs/miRs), interacting with RNA-binding proteins, regulating
parent gene expression, and regulating alternative splicing in a
number of molecular methods (Fig.
1). circRNAs regulate tumor progression through different
mechanisms, and clinical trials targeting circRNA to treat cancer
have shown promising therapeutic effects, showing that they play
significant roles in the pathogenesis of cancer and have potential
as biomarkers of cancer (3). For
example, circHGS regulates the progression of bladder cancer by
acting as a sponge for miR-513a-5p (4); when an intron-derived circRNA such as
ci-ankrd52 was knocked down, the expression of its corresponding
parent gene was also found to be significantly decreased,
indicating that circRNAs may exert cis-effects on the
expression of their parent genes (5); circPLIN2 promotes the progression of
clear cell renal cell carcinoma by binding to insulin-like growth
factor-2 mRNA-binding protein 2 (IGF2BP) (6); and circMbl is produced by the second
exon of the splicing factor MBL gene, and has a role in gene
regulation through competing with linear splicing (7). Based on the conventional opinion, 5′
and 3′ untranslated regions (UTR) are essential elements for
translation initiation in eukaryotic cells. Due to the absence of
5′ and 3′ ends, circRNAs have previously been uniformly classified
as non-coding RNAs that cannot be translated (8). However, given the recent advances that
have been made in this area of research, it has been shown that
certain circRNAs are translated into functional proteins, which
subsequently regulate the transcriptional extension of various
oncogenes, and fulfill specific roles in promoting or suppressing
cancer. Their protein-encoding products are considered to be
potentially reliable prognostic biomarkers for certain types of
tumors. For example, smo-193aa was shown to be highly expressed in
glioblastoma and may be a crucial biomarker for glioblastoma
development (9). In addition,
breast cancer formation and development have been demonstrated to
be regulated by SEMA4B-211aa and EIF6-224aa (10,11).
Studies have also found that protein-encoded synthetic circRNAs can
be used as therapeutics and vaccines, showing higher translational
efficiency and longer stability than linear mRNA-based vaccines
(12). Therefore, it has been
convincingly shown that circRNA-encoded proteins exert a
significant impact on cancer, and their translational function has
become a research focus. The present review will subsequently focus
on the progress that has been made in terms of understanding
circRNA-encoded protein functions.
Translational mechanisms of circRNAs
Protein synthesis (also termed ‘translation’) is the
name given to the process via which the genetic information stored
in DNA is decoded by various catalysts that enable amino acids to
be precisely located in proteins. According to the central dogma of
genetics, eukaryotic mRNAs are generally translated into proteins
through initiation, extension, and termination processes. The
process of translation relies on the 5′-end m7GpppN'
(m7G) cap and the 3′-end poly-A tail (13). Typically, mRNA translation relies on
the 5′-cap to recognize the eukaryotic initiation factor 4F complex
(eIF4F), and 43S ribosomes are recruited to direct protein
synthesis. circRNA lacks a 5′-cap and a 3′-poly(A) tail, which
makes it impossible to translate via this mechanism. However, mass
spectrometry studies have shown that certain circRNAs can be
efficiently translated into detectable peptides. Researchers have
hypothesized that the presence of an internal ribosome entry site
(IRES) allows it to directly recruit initiation factors and
ribosomes to the circRNA independently of a cap, and subsequently,
it was shown that cap-independent translation could be performed
via an m6A-mediated mechanism (14).
Therefore, circRNA translation requires two basic
elements, namely the translation initiation element and the open
reading frame (ORF). Owing to the covalent ring structure of
circRNAs, special translation initiation elements are required
(15). At present, the
translational methods of circRNAs primarily include IRES-mediated
translation and non-IRES-mediated translation. Non-IRES-mediated
translation can be further divided into
N6-methyladenosine (m6A)
modification-initiated translation, rolling circle amplification
(RCA) translation, and translation mediated by UTRs or other
promoter elements (Fig. 2).
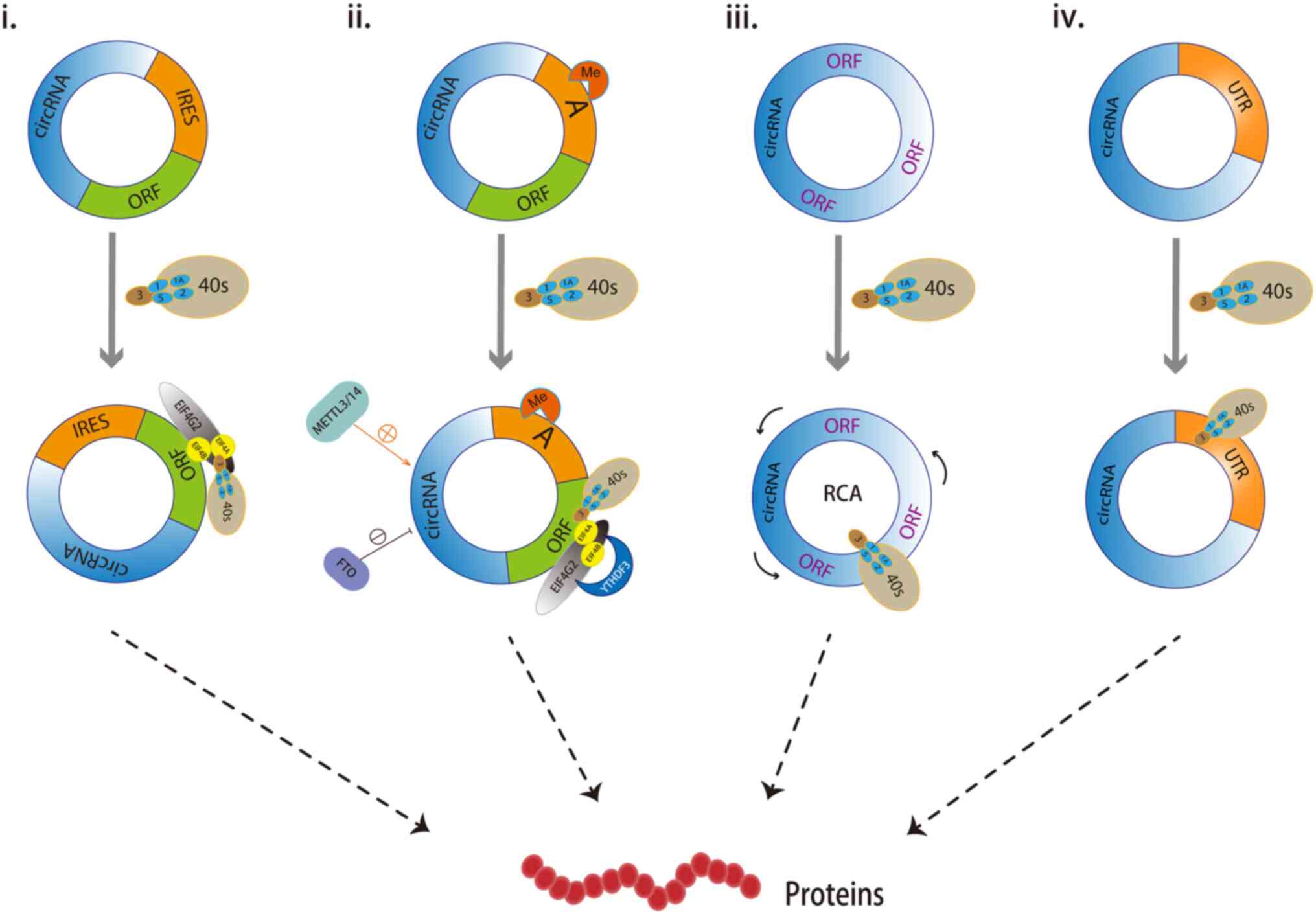 | Figure 2.Common translational mechanisms of
circRNAs. i) IRES-mediated circRNAs translation. The circRNAs are
recognized by eIF4G2 via IRES, which recruit ribosomes for
translation into proteins; ii) m6A-mediated circRNA
translation. circRNAs with m6A modification are recognized by the
YTHDF3 protein, which then binds to eukaryotic translation
initiation factor eIF4G2, which recruits translation initiation
factors eIF4A and eIF4B for translation into proteins. METTL3/14
increases circRNA translation, while FTO reduces it. iii) Rolling
circle amplification translation. Due to the absence of a stop
codon, circRNAs have an infinite ORF, which can be translated into
a circle like RCA after binding with the ribosome to produce
protein expression. iv) Translation mediated by UTR elements. When
certain circRNAs are reverse clipped by pre-mRNA, the clipped site
is located in the UTR, resulting in the generated circRNAs having
part of the same UTR sequence as the linear homologous transcripts,
and the UTRs can recruit ribosomes and drive circRNA translation
proteins. circRNA, circular RNA; IRES, internal ribosome entry
site; RCA, rolling circle amplification; UTR, untranslated region;
ORF, open reading frame. |
IRES-mediated circRNA translation
The IRES is a genetic element ~150-250 bp in length,
and this RNA sequence folds into a structure similar to the
starting tRNA, which is recognized by eIG4 γ2 (eIF4G2) and recruits
ribosomes, thereby initiating protein translation. Initially,
several IRES elements were identified in poliovirus and
encephalomyocarditis virus, and subsequently, IRESs were also found
in a variety of viruses from different viral families. IRES
sequences were found to exist not only in viruses, but also in
mammals, plants, and yeast, although certain differences in the
functional mechanisms of different IRESs were noted (16,17).
Subsequently, IRES-mediated eukaryotic translation was found to act
as an emergency ‘maintenance’ mechanism to ensure that the body's
protein requirements are met in times of stress (18). As early as 1995, it was found that
an IRES sequence was also present upstream of the circRNA start
codon, and in a previous study, a circRNA containing IRES element
was transferred into rabbit reticulocyte lysates to obtain the
predicted protein product (19).
Recent studies have also demonstrated that endogenous circRNAs
mediated by IRESs can be translated to produce proteins (20). For example, an IRES sequence of
circ-EIF6 was identified by bioinformatics analysis, and
subsequently, the putative full-length IRES sequence, as well as
differentially truncated mutants, were cloned into the pGL3-Basic
vector; the IRES activity was then assayed using a dual luciferase
assay (11). The results showed
that the putative IRES sequence had a strong ability to initiate
protein translation, whereas the mutant IRES sequence could not
initiate protein translation. Subsequently, red fluorescent protein
(RFP) and green fluorescent protein (GFP) were cloned into the
double cis-trans reporter gene construct on both sides of the
putative IRES full-length sequence and the mutant sequence,
respectively, to verify again whether the IRES full-length sequence
could induce ribosome entry and initiate translation. The results
obtained showed that both RFP and GFP were detected in cells
transfected with the putative IRES plasmid under normal conditions,
whereas the putative IRES plasmid only induced GFP expression when
eIF4E was inhibited. Furthermore, the mutated IRES also induced
significantly lower levels of GFP expression compared with the
full-length IRES, suggesting that the full-length IRES sequence
induced ribosome entry and initiated translation. In conclusion,
IRES-mediated circRNA translation has been shown to have extended
the diverse range of cellular life processes and is a novel tool of
significant research value.
m6A-mediated circRNA
translation
The m6A modification is the most abundant
base modification in eukaryotes, with a shared sequence motif of
RRACH. It is present primarily on adenine bases in the RRACH
sequence, and its associated biological roles are mediated by
‘writer’ (methyltransferase), ‘eraser’ (demethylase), and ‘reader’
(recognition) enzymes (21). Writer
enzymes are methyltransferases, including methyltransferase-like 3
(METTL3), methyltransferase-like 14 (METTL14), Wilms tumor 1
associated protein (WTAP), and Vir-like m6A
methyltransferase-associated (VIRMA); by contrast, AlkB homolog 5
(ALKBH5) and fat mass and obesity-associated protein (FTO) are
demethylases (erasers) that reverse methylation. m6A
modifications are recognized by m6A-binding proteins,
and it has been shown that m6A-binding proteins
(readers) possess YTH domain proteins, including YTH domain family
proteins 1–3 (YTHDF1-3), YTH domain-containing-1 and −2 (YTHDC-1
and −2) and the heterogeneous nuclear ribonucleoprotein (HNRNP)
family, including HNRNPA2B1 and HNRNPC (22). Recently, it was shown that circRNAs
containing m6A residues can be translated by
non-cap-dependent structures. RNA immunoprecipitation (RIP) assays
with m6A antibodies, followed by enrichment of circRNAs
and RNA sequencing (RNA-seq), has allowed the identification of all
potential circRNAs that contain m6A sites (23). For example, Yang et al
(24) identified m6A
sites on several circRNA sequences, subsequently constructed
mutants containing circRNA m6A motifs, and found that
circRNAs in the negative control group without an IRES could
initiate translation; moreover, they identified that these circRNAs
all contained an m6A-modified RRACH motif near the start
codon when analyzing the sequences, which also successfully
confirmed that m6A modification could drive the
translation of circRNAs. m6A-modified circRNAs are
recognized by the YTHDF3 protein, which then binds to eIF4G2.
eIF4G2 recruits the translation initiation factors eIF4A and eIF4B
to form the translation initiation complex eIF4, which subsequently
initiates the translation of circRNA. The m6A
demethylase FTO decreases the rate of circRNA translation, whereas
the methyltransferase METTL3/14 increases the translation rate
(8). Zhao et al (25) found that circE7 in carcinogenic
human papillomavirus (HPV) possesses multiple potential
m6A modification sites and is able to translate the E7
protein, thereby promoting the proliferation of human papilloma
cells. In addition, Duan et al (26) showed that circMAP3K4 encodes
circMAP3K4-455aa, which protected hepatocellular carcinoma (HCC)
cells from cisplatin exposure driven by m6A
modifications. circ-ZNF609 has been shown to be translated both in
an IRES-dependent manner and through m6A
modification-dependent, cap-non-dependent translation (27,28).
This suggests that there are multiple mechanisms through which the
translation of circRNAs may be driven. In conclusion,
m6A-modification-binding proteins can interact with
transcription initiation factors to drive the translation of
circRNAs.
RCA translation
In 1995, Chen and Sarnow (19) first proposed the process of RCA
translation of circRNAs. RCA translation is a fast, sensitive, and
constant-temperature technique of single-strand DNA amplification,
which is capable of infinite single-strand amplification of
circular DNA molecules. Certain circRNAs have infinite open reading
frames (iORFs) due to the absence of stop codons, which allows them
to be translated in a similar manner to RCAs in a rolling circle to
produce high-molecular-weight proteins. For example, Das et
al (29) found that both
circASPH_219 and cicASPH_264 variants have iORFs that lack any stop
codons; subsequently, they hypothesized and identified that
circASPH splice variants without stop codons could be translated
into macromolecular proteins with repetitive sequences through RCA
translation. Gao et al (30)
also found that circular E-cadherin (circ-E-Cad) encoded the
protein C-E-Cad, producing a 254-amino-acid protein in
glioblastoma. Initially, Perriman and Ares demonstrated that
circRNAs could be translated in vivo in E. coli (31,32).
Subsequently, Abe et al (33) constructed iORF circRNAs in E. coli
and found that they were translated in a manner similar to that of
the RCA reaction of polymerase, and that the circRNA molecules
produced 100 times more product compared with their homologous
linear mRNAs over a given period of time. In a further study by Abe
et al (34), they sought to
confirm whether circRNAs could be translated into proteins in
cells. They confirmed that circRNAs synthesized in vitro
could be translated in living human cells without internal
initiation of specific elements by transferring circRNAs containing
an iORF into rabbit reticulocyte lysates. Subsequently, Liu et
al (35) expanded further on
our understanding of the translation mechanism of RCA. First, they
constructed 3×Flag-tagged-circ-EGFR vectors, transfected them into
293T cells, and detected the presence of rolling translated
epidermal growth factor receptor (rtEGFR) in vivo using
either specific antibody immunoblotting or mass spectrometry
methods to demonstrate the presence of ‘rolling translation’ in
vivo. Next, they found that programmed-1 ribosomal
frameshifting (−1PRF), mutation-induced out-of-frame stop codons
could terminate infinite rolling ring translation, suggesting that
the circRNAs of ‘rolling translation’ could also be terminated.
Finally, they also identified the biological functions of certain
RCA products through cell function experiments. Taken together,
these studies show that the majority of the newly identified
proteins translated by RCA were formerly unknown, and their
underlying biological mechanisms need to be further studied.
Translation mediated by 3′-UTRs or
other primers
Under certain conditions of stress, cap-dependent
mRNA translation is usually suppressed, whereas the levels of
cap-independent circRNA translation products are increased under
conditions such as heat shock, hypoxia, and starvation (36). This suggests that circRNA-encoded
proteins exert a role in stress responses. In addition, 3′-UTRs
recruit ribosomes and drive circRNA-translated proteins. Pamudurti
et al (37) demonstrated
that circRNAs were associated with the translation of ribosomes.
These researchers found that the 3′-UTR of a set of circRNAs in the
head of Drosophila bound to the ribosome and detected the
subsequently generated protein products. To exclude the translation
interference of homologous linear mRNAs, the overexpression of
eIF4E binding protein (4EBP) effectively inhibited the translation
of linear mRNAs, and the protein products of their circRNAs were
not affected, indicating that circRNAs can be non-dependently
translated by 3′-UTR-driven caps.
Bioinformatics tools for the analysis of
circRNA-encoded proteins
Tools for predicting circRNA-encoded
proteins
CircRNAs require an ORF and translation initiation
elements for their translation. The translational capacity of
circRNAs has been predicted using bioinformatics tools (Table I). At present, the most commonly
used prediction tools consist of a set of methods with different
objectives; the first to be discussed is ORF prediction. For this
method, using circPrimer software, the user inputs the full-length
sequence of the circRNA into the ORF finder to retrieve the
possible ORFs and derive the corresponding amino acid sequence.
Through inputting a specific circRNA, users can visualize its
structure and design their own primers to verify the circRNA
according to its full-length sequence, determine the presence of
codable sequences according to its full-length sequence, and
predict the possible ORFs and IRES. The second method that may be
used (TransCirc database) is for the purposes of predicting the
circRNA translational ability. Coding potential calculator 2 (CPC2)
and coding-potential assessment tool (CPAT) are both able to
quickly identify coding and non-coding texts from a large number of
candidate texts, and to quickly and accurately assess the coding
potential of RNA transcripts: The closer the assessment score is to
1, the more likely it is that the protein will be translated
(38,39). The TransCirc database can integrate
all types of evidence associated with translation, and the
retrieval results are designed to intuitively present the
information associated with the translation products, whilst also
rating them (40). The RiboCIRC
database is a comprehensive database for predicting cyclic RNA
translation, containing predictions (and the validation) of cORF,
IRES, m6A, and mass spectrometry, also providing
cross-species conservation evaluation of translatable circRNAs and
systematic annotation of putative cyclic RNA-encoded peptides.
Although this database does not include as much coding evidence as
the TransCirc database, its visualization, primer design, and
peptide structure analysis features are more stable and reliable
than those of the TransCirc database (41). The third type of
bioinformatics-based method is circRNA translation initiation mode
prediction. circRNAs are generally modified by an IRES or
m6A prior to the initiation of translation, and
therefore it is necessary to predict whether circRNA sequences
contain IRES or m6A motifs. The circRNADb database is
the first database to summarize human circRNAs that encode
proteins. This database contains both the IRES and the ORF
information corresponding to the circRNAs, and the basic features
of the predicted peptides. Alternatively, the IRESite database
contains IRESs that have been experimentally validated. Through
entering the sequence of a circRNA, a user can predict all the
potential IRESs on the circRNAs; scores are assigned, and their
corresponding positions in the sequence are listed (42,43).
IRESfinder is a bioinformatics tool based on logit models, which
features 19 k-mer parameters with 80% precision and 73% accuracy;
this tool has a higher predictive efficiency compared with the
IRESite (44). The majority of the
aforementioned IRES prediction methods are based on traditional
machine-learning algorithms, which are still limited as far as
linear RNA IRES predictions are concerned, and at present, there
are no prediction methods specifically suited for circRNA IRESs. To
address this, Zhou et al (45) of Zhejiang University used DeepCIP,
which, through a multi-modal deep learning approach, has been
developed as a tool dedicated to the prediction of circRNA IRESs:
This tool has enabled the study of the encoding potential of
circRNAs and to more effectively capture the characteristics of
circRNA IRESs. The SRAMP software is able to identify mammalian
m6A sites with single-nucleotide resolution; users input
mammalian circRNA sequences in order to predict the m6A
motif. M6APred-EL software has been developed to predict the
m6A genomic sequences of circRNAs in Saccharomyces
cerevisiae and has a higher degree of accuracy compared with the
other methods in terms of identification of m6A sites
(46). Alternatively, DeepM6ASeq
software based on miCLIP-Seq experimental data has been employed to
detect m6A sites with single-base resolution and to
obtain the biological characteristics that surround the
m6A sites, enabling the visualization of the
m6A sites. DeepM6ASeq has been shown to possess better
predictive performance compared with the other machine learning
methods (47). The fourth and final
method to be discussed is the prediction of protein product
function. The Pfam 32.0 database can be used to predict protein
function: The user enters the ORF sequence of the circRNA into the
Pfam 32.0 database, which subsequently can be used to search for
the potential protein domain based on the ORF sequence, enabling
the function of the protein product to be predicted (48).
 | Table I.Biological information tool for
circRNA encoded proteins. |
Table I.
Biological information tool for
circRNA encoded proteins.
Identification and verification of the
protein-coding abilities of circRNAs
First, it is important to determine whether the
circRNA is associated with ribosomes. To meet this aim, ribosome
profiling is performed; the RNA that is not protected by a ribosome
is degraded using RNase R, and the mRNA fragments that are
protected by a ribosome are subsequently separated using sucrose
density gradient centrifugation. Sequencing of these fragments, and
screening for circRNAs bound to ribosomes, were subsequently
performed (49). Second, the
activity of the IRES or the m6A motif should be
investigated. The luciferase activity of each plasmid compared with
the luciferase activity in sea kidney is measured by constructing
different luciferase reporter gene tandem plasmids. For IRES,
luciferase assays revealed that the activity of the full-length
IRES-induced Luc/Rluc plasmid was the highest. Regarding
m6A motifs, GFP protein production, and activity could
be detected by transfecting 293T cells with a short fragment
containing a different copy of the m6A motif in a
position prior to the circRNA reporter start codon (24). Subsequently, the ORF translational
ability of circRNA should be verified. A vector containing the
circRNA ORF and Flag tags was constructed, and the vector was
transferred into an in vitro transcription-translation
system. Flag antibody was subsequently used to detect the formation
of the Flag fusion protein to verify whether the ORF could be
encoded in vitro. A circRNA overexpression vector containing
a Flag label was then constructed and transfected into the cells.
Subsequently, a Flag label antibody was used to detect whether a
protein could be generated with the same molecular weight as that
determined for the protein by western blotting, to verify the
translational ability of the circRNA ORF in cells. The fourth
consideration is to probe with design-specific antibodies based on
the ORF protein product sequence. The reliability of protein
translation is detected by western blot analysis, and the product
sequence is subsequently verified by immunoprecipitation and mass
spectrometry to confirm the consistency of the predicted protein
sequence. Fifth, it is important to verify the interaction between
the circRNA and its translation initiation factors. In a previously
published study, the interaction of circRNAs with eIF4G2 and
IRES-transacting factors (ITAFs) associated with IRES-mediated
translation (50) or YTH-domain
proteins associated with m6A-mediated translation was
co-verified by RIP and RNA pull-down assays (51). Finally, researchers should verify
the effects of specific stimuli or proteins on circRNA translation.
To meet this aim, the cells are subjected to conditions of stress
such as heat shock, hypoxia, or starvation to detect whether these
influence the characteristics of the circRNA-encoded proteins.
Construction of antibodies against
circRNA encoding proteins
First, a circRNA overexpression plasmid containing a
FLAG tag should be constructed by inserting a FLAG tag prior to the
ORF termination codon of the circRNA, which is then transfected
into 293T cells. One approach that has been used was to transfect
the 293T cells, extract the total protein, and then use liquid
chromatic-tandem mass spectrometry (LC-MS) to identify the specific
peptide sequence of this new protein. Another method was to purify
the new protein from 293T cells transfected with Co-IP and Flag
antibodies, which was subsequently detected using SDS-PAGE. The new
proteins were then collected for LC-MS/MS analysis to identify
their specific peptide sequences. The resultant peptide sequences
should match up with the predicted sequence exactly. Subsequently,
by analyzing the specificity, hydrophilicity, and immunogenicity of
the translation peptide, a unique fragment of the new protein is
selected as the antigenic region for the preparation of the
specific antibody, and the antibody is constructed to specifically
target the putative circRNA translation protein (11,52).
Role of circRNA-encoded proteins in
cancer
In recent years, there have been numerous reports
published on newly identified circRNA-encoded proteins, and several
studies have demonstrated that proteins or peptides encoded by
circRNAs are able to contribute to the promotion or inhibition of
cancer through a variety of different modes of action (Table II). The discovery of the proteins
encoded by circRNAs has significantly expanded our understanding of
the biological functions of circRNAs and provided novel
perspectives for cancer therapy.
 | Table II.Expression and mechanism of
circRNA-encoded proteins/peptides in cancer. |
Table II.
Expression and mechanism of
circRNA-encoded proteins/peptides in cancer.
| First author,
year | Cancer | circRNA | Encoding protein or
polypeptide | Role in cancer | Change in
expression | (Refs.) |
|---|
| Peng et al,
2021 | Gastric cancer | circAXIN1 | AXIN1-295aa | Oncogene | Upregulation | (54) |
| Jiang et al,
2021 | Gastric cancer | circMAPK1 | MAPK1-109aa | Tumor suppressor
gene | Downregulation | (57) |
| Zhang et al,
2021 | Gastric cancer | circDIDO1 | DIDO1-529aa | Tumor suppressor
gene | Downregulation | (52) |
| Li et al,
2023 | Gastric cancer | circ-E-Cad | C-E-Cad | Oncogene | Upregulation | (58) |
| Geng et al,
2021 | Gastric cancer | circCOL6A3_030 |
COL6A3_030_198aa | Oncogene | Upregulation | (59) |
| Zheng et al,
2019 | Colon cancer | circPPP1R12A | PPP1R12A-73aa | Oncogene | Upregulation | (61) |
| Pan et al,
2020 | Colon cancer | circFNDC3B | FNDC3B-218aa | Tumor suppressor
gene | Downregulation | (62) |
| Liang et al,
2021 | Colon cancer | circPLCE1 | circPLCE1-411 | Tumor suppressor
gene | Downregulation | (63) |
| Wang et al,
2021 | Colon cancer | circMAPK14 | MAPK14-175aa | Tumor suppressor
gene | Downregulation | (64) |
| Zhang et al,
2021 | Colon cancer |
hsa_circ_0006401 |
hsa_circ_0006401-198aa | Oncogene | Upregulation | (65) |
| Liang et al,
2019 | Liver cancer | circβ-catenin |
β-catenin-370aa | Oncogene | Upregulation | (20) |
| Li et al,
2021 | Liver cancer | circARHGAP35 |
ARHGAP35-1289aa | Oncogene | Upregulation | (68) |
| Li et al,
2022 | Liver cancer | circMRPS35 |
circMRPS35-168aa | Oncogene | Upregulation | (70) |
| Duan et al,
2022 | Liver cancer | circMAP3K4 |
circMAP3K4-455aa | Oncogene | Upregulation | (26) |
| Li et al,
2022 | Liver cancer | circGGNBP2 | cGGNBP2-184aa | Oncogene | Upregulation | (71) |
| Song et al,
2023 | Liver cancer | circZKSCAN1 | circZKSaa | Tumor suppressor
gene | Downregulation | (73) |
| Wang et al,
2021 | Lung cancer | circASK1 | ASK1-272aa | Tumor suppressor
gene | Downregulation | (75) |
| Zhao et al,
2022 | Lung cancer | circPPP1R12A |
circPPP73R1A-73aa | Oncogene | Upregulation | (76) |
| Yang et al,
2018 | Glioblastoma | circFBXW7 | FBXW7-185aa | Tumor suppressor
gene | Downregulation | (80) |
| Xia et al,
2019 | Glioblastoma | circ-AKT3 | AKT3-174aa | Tumor suppressor
gene | Downregulation | (81) |
| Zhang et al,
2018 | Glioblastoma | circ-SHPRH | SHPRH-146aa | Tumor suppressor
gene | Downregulation | (82) |
| Zhang et al,
2018 | Glioblastoma | circPINTexon2 | PINT87aa | Tumor suppressor
gene | Downregulation | (83) |
| Wu et al,
2021 | Glioblastoma | circSMO | SMO-193aa | Oncogene | Upregulation | (9) |
| Gao et al,
2021 | Glioblastoma | circ-E-Cad | C-E-Cad | Oncogene | Upregulation | (30) |
| Liu et al,
2021 | Glioblastoma | circ-EGFR | rtEGFR | Oncogene | Upregulation | (35) |
| Saunders et
al, 2023 | Glioblastoma | circ-HGF | C-HGF | Oncogene | Upregulation | (88) |
| Ye et al,
2019 | Breast cancer | circFBXW7 | FBXW7-185aa | Tumor suppressor
gene | Downregulation | (90) |
| Li et al,
2020 | Breast cancer | circ-HER2 | HER2-103 | Oncogene | Upregulation | (91) |
| Li et al,
2022 | Breast cancer | circ-EIF6 | EIF6-224aa | Oncogene | Upregulation | (11) |
| Zhao et al,
2019 | Cervical
cancer | circE7 | E7 | Oncogene | Upregulation | (25) |
| Gu et al,
2018 | Bladder cancer | circGprc5a |
circGprc5a-peptide | Oncogene | Upregulation | (93) |
| Tang et al,
2021 | Multiple
myeloma | circBUB1B |
circBUB1B_544aa | Oncogene | Upregulation | (95) |
| Tang et al,
2022 | Multiple
myeloma | circHNRNPU |
circHNRNPU_603aa | Oncogene | Upregulation | (96) |
| Li et al,
2021 | Endometrial
cancer |
has-circ-0000437 | CORO1C-47aa | Tumor suppressor
gene | Downregulation | (97) |
CircRNA-encoded proteins and gastric
cancer (GC)
GC is one of the most common malignant tumors in the
world and is always associated with high morbidity and mortality
rates (53). Several research
groups have recently discovered that circRNAs encode novel proteins
in GC, and that these are crucial for the occurrence and
progression of GC. According to a study by Yin et al
(54), circAXIN1 encodes the novel
protein AXIN1-295aa, which competes with the tumor suppressor
adenomatous polyposis coli (APC) to stimulate the Wnt/β-catenin
signaling pathway, thereby enhancing the progression of GC. The
MAPK pathway is a signal transduction pathway that is
well-established to be involved in cell proliferation,
inflammation, and apoptosis (55,56).
Jiang et al (57) used
circRNADb to both predict and confirm that circMAPK1 encoded a
novel protein, MAPK1-109aa, which exerts an inhibitory role in GC.
In addition, this novel protein was able to inhibit MAPK1
phosphorylation through competitive binding with MEK1, thereby
inhibiting the MAPK pathway, which demonstrated that MAPK1-109aa
has a role in tumor inhibition. This suggests that circMAPK1 may
serve as a therapeutic target for GC. Subsequently, Zhang et
al (52) also employed
circRNADb to predict and confirm the inhibitory effects of
circDIDO1 in GC by identifying a novel protein containing 529 amino
acids, DIDO1-529aa. Through mass spectrometry, immunofluorescence,
immunoprecipitation, and western blotting, the encoded DIDO1-529aa
protein was found to interact with polyADP-ribose polymerase 1
(PARP1) and inhibit its activity. In addition, circDIDO1 was shown
to promote the ubiquitination and degradation of peroxiredoxin 2
(PRDX2), thereby inhibiting GC cell growth and invasiveness.
circDIDO1 is hypothesized to be a potential prognostic biomarker
and therapeutic target for GC. C-E-Cad, the protein encoded by
circ-E-Cad, has been shown to exert a notable influence on the
progression of glioblastoma, although its specific role in GC has
yet to be fully elucidated (30).
Recently, Li et al (58)
demonstrated that circ-E-Cad-encoded C-E-Cad also exerts specific
effects on GC and promotes the proliferation and migration of GC
cells. In addition, Geng et al (59) found that circCOL6A3_030 regulates GC
metastasis by encoding the protein circCOL6A3_030_198aa, which also
provides a putative therapeutic direction for the diagnosis and
prognosis of GC.
circRNA-encoded proteins and colon
cancer
Colon cancer (CC) is the third most common type of
cancer worldwide, and the fourth most common cause of
cancer-associated death. A variety of factors, including alcohol,
obesity, and genetic and epigenetic shifts, contribute to the
progression of CC (60). Xiao et
al (61) found that
circPPP1R12A is upregulated in CC and carries an ORF that encodes a
functional protein, namely circPPP1R12A-73aa. Relevant functional
experiments confirmed that circPPP1R12A-73aa was conducive to the
rapid proliferation of CC cells, and that this protein was involved
in the regulation of the Hippo-Yes-associated protein (YAP)
signaling pathway. Pan et al (62) showed that circFNDC3B may encode a
novel protein containing 218 amino acids, circFNDC3b-218aa, which
was shown to inhibit CC proliferation, invasion, and migration both
in vitro and in vivo. Moreover, it further regulates
the Snail/fructose-1,6-bisphosphatase
(FBP1)/epithelial-to-mesenchymal transition (EMT) axis, thus
possessing a tumor-associated role. A study by Liang et al
(63) confirmed that circPLCE-411
encoded by circPLCE1 is involved in the regulation of
NF-κB-mediated signal transduction, and that this may serve
as a novel and promising therapeutic target and prognostic marker
for CC. Wang et al (64)
found that the circMAPK14-encoded protein circMAPK14-175aa was able
to competitively bind with the kinase MKK14 to decrease the nuclear
translocation of MAPK6, thereby promoting the ubiquitin-mediated
degradation of Forkhead Box C1 (FOXC1), signifying its potential as
a biomarker for the therapeutic treatment of colorectal cancer
(CRC). In addition, Zhang et al (65) predicted that the circRNA
hsa_circ_0006401 encoded a 198-amino-acid protein. An expression
vector with circular transcript capability was constructed and
transfected into 293T cells, whereupon it was found that the 198-aa
protein was indeed encoded by hsa_circ_0006401, which has a role in
CRC via promoting the proliferation and metastasis of CRC both
in vitro and in vivo.
circRNA-encoded proteins and liver
cancer
Liver cancer is one of the most common malignant
tumors in China. No obvious symptoms arise during the early stage
of liver cancer, and the majority of patients are diagnosed with
liver cancer at an advanced stage; as a consequence, the survival
rate is low (66,67). Therefore, the search for valuable
biomarkers and therapeutic targets is urgently required. Liang
et al (20) found that
circβ-catenin encoded β-catenin-370aa, which promoted the growth of
hepatocellular cancer cells. β-catenin-370aa encoded by
circβ-catenin may act as a decoy for glycogen synthase kinase-3β
(GSK3β) to escape GSK-3β-induced β-catenin degradation;
consequently, full-length β-catenin is stabilized and the Wnt
pathway is activated. circRNAs can regulate the atypical functions
of HCC cells through the Wnt/β-catenin pathway, which provides a
novel strategy for studying the mechanism of HCC formation. Li
et al (68) found that
circARHGAP35 initiated protein translation through m6A
modifications in liver cancer, and the circARHGAP35 protein
interacted with the transcription factor TFI–I in the nucleus to
promote the progression of cancer cells. Studies have shown that
non-coding RNAs also play an important role in resistance to
different cancer therapies by rewiring basic signaling pathways
(69). Cisplatin chemotherapy
resistance has been detected in the majority of patients with liver
cancer receiving long-term chemotherapy, which is a major clinical
challenge in liver cancer chemotherapy. A previous study found that
circMRPS35 promoted the malignant progression of liver cancer and
cisplatin resistance, and its encoded protein circMRPS35-168aa was
induced by chemotherapeutic drugs to promote cisplatin resistance
in liver cancer. This finding provides a novel direction for the
management of drug-resistant liver cancer (70). Duan et al (26) found that circMAP3K4 is an
upregulated circRNA with coding potential in HCC. Driven by
m6A modifications, circMAP3K4 encodes the protein
circMAP3K4-455aa, which was shown to promote the growth of liver
cancer. The association between inflammation and tumors has also
been a focus of research in recent years. A recent study identified
that GGNBP2-184aa, encoded by circGNNBP2, when induced by
interleukin-6, was able to activate the JAK-STAT signaling pathway,
and promote the growth and proliferation of liver cancer cells
(71). This study also provided a
novel avenue to explore targeted therapies in the case of a liver
cancer diagnosis. Sorafenib is the first-line agent in the
treatment of liver cancer, as it can improve the survival rate of
patients with advanced HCC (72).
Song et al (73) showed that
circZKSCAN1, and the protein it encodes, inhibit HCC progression
and sensitize HCC cells to sorafenib. This finding demonstrated
that the encoded protein (ZKSCAN1aa) is a potential therapeutic
target and biomarker for liver cancer.
circRNA-encoded proteins and lung
cancer
Lung cancer is the second most common malignancy in
the world after breast cancer. Approximately 85% of patients with
lung cancer have non-small cell lung cancer (NSCLC), of which lung
adenocarcinoma (LUAD) is the most commonly observed histological
subtype (74). Wang et al
(75) showed circASK1, which
encodes the recently identified protein ASK1-272aa, in LUAD was
markedly downregulated, and the sensitivity of LUAD cells to
gefitinib in gefitinib-resistant cells was enhanced. The protein
ASK1-272aa is essential for ASK1/JNK/p38 signaling activation and
mediates the chemosensitivity induction of circASK1 in LUAD. This
finding highlights a novel therapeutic target for addressing
gefitinib resistance in patients with LUAD. Subsequently,
circPPP1R12A was shown to encode a newly identified protein,
circPPP1R12A-73aa, which has been previously reported to play a
catalytic role in colon cancer (61). Subsequently, Zhao et al
(76) found that the expression of
circPPP1R12A was also significantly increased in small-cell lung
cancer, and its associated protein circPPP73R1A-73aa could promote
the proliferation of small-cell lung cancer through the AKT
pathway. Therefore, this study also provided useful insights and
suggestions for the clinical treatment of NSCLC.
circRNA-encoding proteins and
glioblastoma
Glioblastoma is the most common primary malignant
tumor of the adult central nervous system, accounting for ~80% of
all malignant brain tumors (77).
An increasing number of studies have found that circRNAs are
differentially expressed in glioblastoma and participate in the
occurrence and development of glioblastoma by acting as miRNA
sponges and encoding proteins (78,79).
In glioblastoma, circRNAs have been identified that may encode
proteins of peptides, and these may fulfill certain roles in the
occurrence and development of glioblastoma. Several studies
confirmed that circRNA translation proteins regulate the occurrence
and development of brain glioma (80–83).
Yang et al (80) found that
circFBXW7 encodes a novel 21 kDa protein, FBXW7-185aa, and that
FBXW7-185aa inhibits the formation of malignant glioblastoma
through antagonizing USP28-induced c-Myc stability by reducing the
half-life of c-Myc. Furthermore, Xia et al (81) found that circ-AKT3 is expressed at a
low level in glioblastoma, and its encoded protein, AKT3-174aa,
also exerts an inhibitory role in glioblastoma. circ-SHPRH is a
novel tumor-associated circRNA that has been shown to be associated
with a variety of tumors, including HCC (84), GC (85), bile duct carcinoma (86), and pancreatic ductal adenocarcinoma
(87), amongst others. Zhang et
al (82) showed that
circ-SHPRH, which encodes a 146-amino-acid protein (SHPRH-146aa),
has an inhibitory role in tumorigenicity and that SHPRH-146aa
protects full-length SHPRH from ubiquitin-proteasome degradation.
Therefore, both circ-SHPRH and SHPRH-146aa may be used as potential
biomarkers in glioblastoma. These researchers subsequently
discovered a 53-amino-acid peptide encoded by the circRNA long
intergenic non-protein-coding RNA p53-induced transcript
(LINC-PINT) that was induced by long intergene non-protein-coding
RNA p87, and inhibited glioblastoma cell proliferation both in
vitro and in vivo. PINT87aa, which is encoded by the
circular form but not linear LINC-PINT, can regulate the RNA
elongation of multiple oncogenes and exert tumor inhibitory effects
(83). In addition, activation of
the Hedgehog signaling pathway also has a role in glioblastoma, but
the sensitization mechanism has yet to be fully elucidated. Wu
et al (9) screened circSMO
with abundant Hedgehog gene expression in the database and found
that it encoded a novel protein, SMO-193aa, which was later
confirmed by in vitro experiments to be associated with
Hedgehog signaling and to promote the formation of glioblastoma.
Gao et al (30) found that
the tumor suppressor gene E-Cadherin forms a circRNA (circ-E-Cad),
and that the translated protein (C-E-Cad) is able to promote
cancer. In addition, this research group found that circ-EGFR has
an infinite cycle of ORFs, and that this particular translational
product has the ability to promote the initiation of tumorigenesis
in brain tumors (35). Their study
also demonstrated that targeting rtEGFR leads to an improvement in
the efficiency of EGFR-targeted therapy for glioblastoma. Just
recently, Saunder et al (88) team also recently found that circHGF
encoding translates the HGF protein variant, which promotes
glioblastoma growth by stimulating c-MET signaling, and they boldly
speculated that targeting C-HGF may have therapeutic potential for
the management of GBM. Taken together, these findings on the
translational function of circRNAs have provided novel ideas and
insights for the treatment of glioma.
circRNA-encoded proteins and
triple-negative breast cancer (TNBC)
Breast cancer has overtaken lung cancer as the most
common type of cancer worldwide, according to new figures released
by the World Health Organization's International Agency for
Research on Cancer (89).
Approximately 15–20% of patients with breast cancer are diagnosed
with TNBC, which is a subtype of breast cancer defined by the lack
of expression of the estrogen receptor (ER), progesterone receptor
(PR), and human epidermal growth factor receptor type 2 (Her-2).
circFBXW7 has previously been reported as a tumor suppressor in
glioma, which is translated into a protein product consisting of
185 amino acids (FBXW7-185aa) (80). Ye et al (90) also identified this circRNA in TNBC,
wherein it exhibited low expression levels. Subsequently, they also
explored the potential function of FBXW7-185aa in TNBC. According
to the results of pre-existing studies, FBXW7-185aa has anti-cancer
activity and can be used as a therapeutic target of TNBC and is a
biomarker for its prognosis. Li et al (91) screened and verified the function of
circ-HER2 in TNBC, and found that circ-HER2 encoded a protein,
HER2-103, composed of 103 amino acids. Both in vitro and
in vivo experiments confirmed that circ-HER and HER2-103
promote the cell proliferation, invasion and tumorigenicity of
TNBC. In addition, Li et al (11) identified a highly expressed
circ-EIF6 in TNBC through sequencing experiments and determined
that circ-EIF6 had good coding ability according to polysome
analysis with sucrose density gradient centrifugation. It was
confirmed that circ-EIF6 encoded the protein EIF6-224aa and could
promote the proliferation and invasion of TNBC cells. EIF6-224aa
also promotes TNBC progression by activating the MYH9/Wnt/β-catenin
pathway. In conclusion, the newly identified protein EIF6-224aa has
added to our understanding of the underlying mechanisms of TNBC,
and it is expected to serve as a potential prognostic and
therapeutic target for patients with TNBC.
circRNA-encoded proteins and cervical
cancer
Cervical cancer is one of the most common types of
malignant tumors of the female reproductive system, and the second
most common type of cancer in women worldwide. Recent studies have
shown that circE7 in cervical cancer drives translation to produce
E7 oncoprotein through m6A modifications. circE7 is able
to inhibit cancer cell growth by reducing the level of the
oncogenic E7 protein in cervical cancer cells. Since high-risk HPV
detection has been established in cervical cancer screening, it
remains to be determined whether circE7-translated protein can be
used as a sensitive marker for high-risk HPV populations, and
whether its abundance is correlated with cervical cancer prognosis
(25).
circRNA-encoded proteins and bladder
cancer
Bladder cancer is one of the most lethal cancers in
the world. In recent years, numerous circRNAs have been found to be
involved in a large range of biological processes in bladder
cancer. These circRNAs are primarily involved in regulating the
occurrence and development of bladder cancer by acting as a miRNA
sponge (92). To date, several
studies have discovered circRNAs with protein-coding functions in
bladder cancer. Gu et al (93) found that circGprc5a was upregulated
in bladder tumors and in bladder cancer stem cells. Subsequently,
circGprc5a was found to have strong coding potential, as detected
by western blotting, and the circRNA acted in a peptide-dependent
manner. It was also found that a circGprc5a-peptide/Gprc5a
signaling axis could be used for targeting both the bladder and
bladder cancer stem cells. This finding provides novel directions
and concepts for the treatment and prognosis of bladder cancer.
circRNA-encoded proteins and multiple
myeloma
Multiple myeloma (MM) is a malignant proliferative
disease of plasma cells, often accompanied by monoclonal
immunoglobulin or light chain (M protein) hyperplasia (94). Despite the clinical use of targeted
drugs to improve the prognosis of patients with MM, due to drug
resistance, patients are prone to relapse or refractory MM
following treatment with one or more drugs, and under these
circumstances, the disease becomes life-threatening and incurable.
It was found that BUB1 mitotic checkpoint serine/threonine kinase B
(BUB1B) was upregulated in patients with MM and it could promote
the proliferation of MM cells and induce drug resistance. circBUB1B
encodes a novel protein, circBUB1B_544aa, which is a noteworthy
protein in that it contains a BUB1B kinase catalytic center, and it
can be secreted into the bone marrow microenvironment. Both in
vitro and in vivo studies have shown that
circBUB1B_544aa also promotes cell proliferation and drug
resistance of MM, which were attributed to the induction of
activation of chromosomal instability (CIN) and centrosomal protein
170 (CEP170). These findings have demonstrated that both BUB1B and
circBUB1B-544aa are promising potential therapeutic targets for MM
(95). Subsequently, it was also
found that circHNRNPU secreted by MM cells encodes a protein named
circHNRNPU_603aa, and overexpression of circHNRNPU_603aa could
promote the proliferation and cloning of MM cells both in
vitro and in vivo. Moreover, circHNRNPU_603aa has the
potential mechanism of regulating SKP2 exon skipping, inhibiting
c-Myc ubiquitin and stabilizing c-Myc expression. The results
obtained suggested that circHNRNPU_603aa may serve as a potential
biomarker and therapeutic target for MM (96).
circRNA-encoding proteins and
endometrial carcinoma (EC)
EC is the name given to a group of epithelial
malignant tumors that occur in the endometrium, also known as
uterine body cancer, and it is one of the three most common
malignant tumors of the female genital tract. Ling et al
(97) found through RNA sequencing
that the levels of hsa-circ-0000437 were significantly reduced in
EC, and that it contained a short ORF encoding the functional
peptide of CORO1C-47aa. It was subsequently found that CORO1C-47aa
could negatively regulate tumor development and inhibit tumor
angiogenesis. The mechanism of action of CORO1C-47aa in EC cells
was then explored, and it was found that CORO1C-47aa and aryl
hydrocarbon receptor nuclear translocator (ARNT) competitively
bound to and inhibited the co-activation of the vascular
endothelial growth factor (VEGF) promoter, TACC3, thereby
inhibiting VEGF expression. Taken together, the results obtained
showed that the peptide CORO1C-47aa may serve as a potential target
for developing effective anti-angiogenesis therapies against
EC.
Concluding remarks
With the development and application of
high-throughput sequencing and bioinformatics tools, an increasing
number of circRNAs have been shown to have the potential to encode
proteins. In the present review, we have explored the translation
mechanisms associated with circRNAs, the bioinformatics tools that
encode proteins, and the mechanism of action of circRNA-encoded
proteins or peptides in cancer. The majority of these circRNAs have
a role in inhibiting or promoting cancer through different signal
transduction pathways. However, research on the function and
mechanism of circRNA-encoded peptides/proteins essentially remains
in its infancy, and there remain several questions to be explored
and solved. For example: i) Whether there are other translational
methods that are employed by circRNAs; ii) whether the
bioinformatics tools still need to be further improved (many
current technologies have certain limitations, and the accuracy of
the prediction methods still needs to be improved); iii) whether
relevant experimental techniques still need to be improved (for
example, ribosome profiling and deep sequencing are not mature
enough, and mass spectrometry cannot be used to detect small
peptides <10 kDa in size; as a result, certain circRNA
translation products cannot be detected, which presents obstacles
in terms of research on the translational abilities of the
circRNAs); iv) studies on these encoded peptides/proteins, up to
this point, have primarily focused on cells and animals, and more
human specimens, such as serum and plasma, should be considered in
the future; v) the majority of the circRNAs and their translated
proteins are only detected at the molecular level, and establishing
their networks with other disease-causing genes in tumors needs to
be further analyzed in terms of improving the strategies for
clinical diagnosis and treatment; and vi) a standard nomenclature
for circRNA-encoded proteins is lacking at present. The naming of
the circRNAs themselves also lacks a uniform standard, since
multiple circRNAs can be generated at the same gene position.
Simply using the naming method of adding ‘circ’ as a prefix
proceeded by the gene name cannot accurately define a given
circRNA. Chen Lingling and his team from the Chinese Academy of
Sciences, together with international experts, proposed a unified
naming method for circRNAs. Chen et al (98) proposed to add corresponding exon
serial number information to the ‘circ’ prefix and gene name to
distinguish circRNAs with the same gene location but different exon
composition. For instance, other variable splicing forms existing
inside circRNA, it is recommended to add specific characters such
as retention intron (RI), short (S), or long (L) to retain intron
or exon information generated by internal variable splicing. For
circRNAs that contain exons that are not present in existing
annotations, it is recommended to use new exon (NE) as a
placeholder to distinguish them. For circRNAs derived from fusion
genes, experts suggest unification with the latest international
fusion gene naming method, marking two fusion genes with ‘::’, and
adding the serial number information of the exon contained. The
implementation of this naming convention can effectively
distinguish circRNAs that originate from the same gene location but
have different exon compositions. This nomenclature is based on the
international standard for gene and transcript nomenclature. Of
course, in addition to the need for a uniform naming convention,
the provision of matching gene (and transcript) annotation
information is also important to standardize circular RNA naming.
In the future, it is desirable to add some additional annotation
information, including the location of the circular RNA genome (and
its version), the specific sequence, and a clear structural
diagram. At present, the naming method for circRNA-encoded proteins
is still based on the length of the ORFs with coding potential
obtained from database predictions and subsequent experimental
verification. The newly identified proteins encoded by circRNAs can
be efficiently identified and discriminated if the names of
circRNAs are presented in a standard way. The investigation of
circRNA-encoded proteins has undoubtedly begun a new era in the
study of circRNAs. We hypothesize that circRNAs, and the proteins
that they encode, are set to become highly intriguing clinical
diagnostic targets and cancer biomarkers in the near future, as
further studies on the translational function of circRNAs are
published.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by the Key Medical
Research Projects of Jiangsu Provincial Health Commission (grant
no. ZD2022008) and Jiangsu Transfusion Association ‘InTec
PRODUCTS.INC’ project (grant no. HXKT20221035).
Availability of data and materials
Not applicable.
Authors' contributions
HC and XZ provided direction and guidance throughout
the preparation of this manuscript. WY collected and analyzed
studies and was a major contributor in writing and editing the
manuscript. HC, XZ, and WY reviewed and revised the manuscript. All
authors read and approved the final manuscript. Data authentication
is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T
and Shu Y: CircRNAs in cancer metabolism: A review. J Hematol
Oncol. 12:902019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Beilerli A, Gareev I, Beylerli O, Yang G,
Pavlov V, Aliev G and Ahmad A: Circular RNAs as biomarkers and
therapeutic targets in cancer. Semin Cancer Biol. 83:242–252. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yan H and Bu P: Non-coding RNA in cancer.
Essays Biochem. 65:625–639. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu Y, Zuo L, Xiong H, Li S, Chen R and
Liu H: CircHGS enhances the progression of bladder cancer by
regulating the miR-513a-5p/VEGFC axis and activating the AKT/mTOR
signaling pathway. Cell Cycle. 22:919–938. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao B, Huang C, Pan J, Hu H, Liu X, Zhang
K, Zhou F, Shi X, Wu J, Yu B, et al: CircPLIN2 promotes clear cell
renal cell carcinoma progression by binding IGF2BP proteins and
miR-199a-3p. Cell Death Dis. 13:10302022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: CircRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
cancer. Mol Cancer. 19:302020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu X, Xiao S, Zhang M, Yang L, Zhong J, Li
B, Li F, Xia X, Li X, Zhou H, et al: A novel protein encoded by
circular SMO RNA is essential for Hedgehog signaling activation and
glioblastoma tumorigenicity. Genome Biol. 22:332021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Jian W, Luo Q and Fang L:
CircSEMA4B inhibits the progression of breast cancer by encoding a
novel protein SEMA4B-211aa and regulating AKT phosphorylation. Cell
Death Dis. 13:7942022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Wang Z, Su P, Liang Y, Li Z, Zhang
H, Song X, Han D, Wang X, Liu Y, et al: Circ-EIF6 encodes
EIF6-224aa to promote TNBC progression via stabilizing MYH9 and
activating the Wnt/beta-catenin pathway. Mol Ther. 30:415–430.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu X, Zhang Y, Zhou S, Dain L, Mei L and
Zhu G: Circular RNA: An emerging frontier in RNA therapeutic
targets, RNA therapeutics, and mRNA vaccines. J Control Release.
348:84–94. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kozak M: Initiation of translation in
prokaryotes and eukaryotes. Gene. 234:187–208. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Prats AC, David F, Diallo LH, Roussel E,
Tatin F, Garmy-Susini B and Lacazette E: Circular RNA, the Key for
translation. Int J Mol Sc. 21:85912020. View Article : Google Scholar
|
|
15
|
Dong HJ, Zhang R, Kuang Y and Wang XJ:
Selective regulation in ribosome biogenesis and protein production
for efficient viral translation. Arch Microbiol. 203:1021–1032.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fan X, Yang Y, Chen C and Wang Z:
Pervasive translation of circular RNAs driven by short IRES-like
elements. Nat Commun. 13:37512022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jang SK, Kräusslich HG, Nicklin MJ, Duke
GM, Palmenberg AC and Wimmer E: A segment of the 5′nontranslated
region of encephalomyocarditis virus RNA directs internal entry of
ribosomes during in vitro translation. J Virol. 62:2636–43. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shatsky IN, Dmitriev SE, Terenin IM and
Andreev DE: Cap- and IRES-independent scanning mechanism of
translation initiation as an alternative to the concept of cellular
IRESs. Mol Cells. 30:285–93. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen CY and Sarnow P: Initiation of
protein synthesis by the eukaryotic translational apparatus on
circular RNAs. Science. 268:415–7. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liang WC, Wong CW, Liang PP, Shi M, Cao Y,
Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM, et al: Translation of the
circular RNA circβ-catenin promotes liver cancer cell growth
through activation of the Wnt pathway. Genome Biol. 20:842019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Meyer KD and Jaffrey SR: Rethinking
m6A readers, writers, and erasers. Annu Rev Cell Dev
Biol. 33:319–342. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qin S, Zhang Q, Xu Y, Ma S, Wang T, Huang
Y and Ju S: M6A-modified circRNAs: Detections,
mechanisms, and prospects in cancers. Mol Med. 28:792022.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jiang X, Liu B, Nie Z, Duan L, Xiong Q,
Jin Z, Yang C and Chen Y: The role of m6A modification
in the biological functions and diseases. Signal Transduct Target
Ther. 6:742021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res. 27:626–641.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao J, Lee EE, Kim J, Yang R, Chamseddin
B, Ni C, Gusho E, Xie Y, Chiang CM, Buszczak M, et al: Transforming
activity of an oncoprotein-encoding circular RNA from human
papillomavirus. Nat Commun. 10:23002019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Duan JL, Chen W, Xie JJ, Zhang ML, Nie RC,
Liang H, Mei J, Han K, Xiang ZC, Wang FW, et al: A novel peptide
encoded by N6-methyladenosine modified circMAP3K4 prevents
apoptosis in hepatocellular carcinoma. Mol Cancer. 21:932022.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang L, Yu P, Wang J, Xu G, Wang T, Feng
J, Bei Y, Xu J, Wang H, Das S, et al: Downregulation of circ-ZNF609
promotes heart repair by modulating RNA N6-methyladenosine-modified
Yap expression. Research (Wash DC). 2022:98259162022.PubMed/NCBI
|
|
29
|
Das A, Sinha T, Mishra SS, Das D and Panda
AC: Identification of potential proteins translated from circular
RNA splice variants. Eur J Cell Biol. 102:1512862023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gao X, Xia X, Li F, Zhang M, Zhou H, Wu X,
Zhong J, Zhao Z, Zhao K, Liu D, et al: Circular RNA-encoded
oncogenic E-cadherin variant promotes glioblastoma tumorigenicity
through activation of EGFR-STAT3 signaling. Nat Cell Biol.
23:278–291. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Perriman R and Ares M Jr: Circular mRNA
can direct translation of extremely long repeating-sequence
proteins in vivo. RNA. 4:1047–54. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Perriman R: Circular mRNA encoding for
monomeric and polymeric green fluorescent protein. Methods Mol
Biol. 183:69–85. 2002.PubMed/NCBI
|
|
33
|
Abe N, Hiroshima M, Maruyama H, Nakashima
Y, Nakano Y, Matsuda A, Sako Y, Ito Y and Abe H: Rolling circle
amplification in a prokaryotic translation system using small
circular RNA. Angew Chem Int Ed Engl. 52:7004–8. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Abe N, Matsumoto K, Nishihara M, Nakano Y,
Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, et al:
Rolling circle translation of circular RNA in living human cells.
Sci Rep. 5:164352015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu Y, Li Z, Zhang M, Zhou H, Wu X, Zhong
J, Xiao F, Huang N, Yang X, Zeng R, et al: Rolling-translated EGFR
variants sustain EGFR signaling and promote glioblastoma
tumorigenicity. Neuro Oncol. 23:743–756. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tatomer DC and Wilusz JE: An unchartered
journey for ribosomes: Circumnavigating circular RNAs to produce
proteins. Mol Cell. 66:1–2. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kang YJ, Yang DC, Kong L, Hou M, Meng YQ,
Wei L and Gao G: CPC2: A fast and accurate coding potential
calculator based on sequence intrinsic features. Nucleic Acids Res.
45:W12–W16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang L, Park HJ, Dasari S, Wang S, Kocher
JP and Li W: CPAT: Coding-potential assessment tool using an
alignment-free logistic regression model. Nucleic Acids Res.
41:e742013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang W, Ling Y, Zhang S, Xia Q, Cao R,
Fan X, Fang Z, Wang Z and Zhang G: TransCirc: An interactive
database for translatable circular RNAs based on multi-omics
evidence. Nucleic Acids Res. 49:D236–D242. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li H, Xie M, Wang Y, Yang L, Xie Z and
Wang H: RiboCIRC: A comprehensive database of translatable
circRNAs. Genome Biol. 22:792021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mokrejs M, Masek T, Vopálensky V, Hlubucek
P, Delbos P and Pospísek M: IRESite-a tool for the examination of
viral and cellular internal ribosome entry sites. Nucleic Acids
Res. 38:D131–6. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mokrejs M, Vopálenský V, Kolenaty O, Masek
T, Feketová Z, Sekyrová P, Skaloudová B, Kríz V and Pospísek M:
IRESite: The database of experimentally verified IRES structures.
www.iresite.orgNucleic Acids Res. 34:D125–D130. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhao J, Wu J, Xu T, Yang Q, He J and Song
X: IRESfinder: Identifying RNA internal ribosome entry site in
eukaryotic cell using framed k-mer features. J Genet Genomics.
45:403–406. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhou Y, Wu J, Yao S, Xu Y, Zhao W, Tong Y
and Zhou Y: DeepCIP: A multimodal deep learning method for the
prediction of internal ribosome entry sites of circRNAs. bioRxiv.
2022.2010.2003.510726. 2022.
|
|
46
|
Wei L, Chen H and Su R: M6APred-EL: A
sequence-based predictor for identifying N6-methyladenosine sites
using ensemble learning. Mol Ther Nucleic Acids. 12:635–644. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang Y and Hamada M: DeepM6ASeq:
Prediction and characterization of m6A-containing
sequences using deep learning. BMC Bioinformatics. 19:5242018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
El-Gebali S, Mistry J, Bateman A, Eddy SR,
Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart
A, et al: The Pfam protein families database in 2019. Nucleic Acids
Res. 47:D427–D432. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ingolia NT, Hussmann JA and Weissman JS:
Ribosome profiling: Global views of translation. Cold Spring Harb
Perspect Biol. 11:a0326982019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mei F: Research progress of viral IRES
structure and IRES mediated protein translation. Life Science.
407–418. 2021.
|
|
51
|
Marín-Béjar O and Huarte M: RNA pulldown
protocol for in vitro detection and identification of
RNA-associated proteins. Methods Mol Biol. 1206:87–95. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang Y, Jiang J, Zhang J, Shen H, Wang M,
Guo Z, Zang X, Shi H, Gao J, Cai H, et al: CircDIDO1 inhibits
gastric cancer progression by encoding a novel DIDO1-529aa protein
and regulating PRDX2 protein stability. Mol Cancer. 20:1012021.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
López MJ, Carbajal J, Alfaro AL, Saravia
LG, Zanabria D, Araujo JM, Quispe L, Zevallos A, Buleje JL, Cho CE,
et al: Characteristics of gastric cancer around the world. Crit Rev
Oncol Hematol. 181:1038412023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Peng Y, Xu Y, Zhang X, Deng S, Yuan Y, Luo
X, Hossain MT, Zhu X, Du K, Hu F, et al: A novel protein
AXIN1-295aa encoded by circAXIN1 activates the Wnt/β-catenin
signaling pathway to promote gastric cancer progression. Mol
Cancer. 20:1582021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Asl ER, Amini M, Najafi S, Mansoori B,
Mokhtarzadeh A, Mohammadi A, Lotfinejad P, Bagheri M, Shirjang S,
Lotfi Z, et al: Interplay between MAPK/ERK signaling pathway and
MicroRNAs: A crucial mechanism regulating cancer cell metabolism
and tumor progression. Life Sci. 278:1194992021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Fang JY and Richardson BC: The MAPK
signalling pathways and colorectal cancer. Lancet Oncol. 6:322–7.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jiang T, Xia Y, Lv J, Li B, Li Y, Wang S,
Xuan Z, Xie L, Qiu S, He Z, et al: A novel protein encoded by
circMAPK1 inhibits progression of gastric cancer by suppressing
activation of MAPK signaling. Mol Cancer. 20:662021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li F, Tang H, Zhao S, Gao X, Yang L and Xu
J: Circ-E-Cad encodes a protein that promotes the proliferation and
migration of gastric cancer via the TGF-β/Smad/C-E-Cad/PI3K/AKT
pathway. Mol Carcinog. 62:360–368. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Geng X, Wang J, Zhang C, Zhou X, Jing J
and Pan W: Circular RNA circCOL6A3_030 is involved in the
metastasis of gastric cancer by encoding polypeptide.
Bioengineered. 12:8202–8216. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
He YC, Hao ZN, Li Z and Gao DW:
Nanomedicine-based multimodal therapies: Recent progress and
perspectives in colon cancer. World J Gastroenterol. 29:670–681.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z,
Xu B, Wu C, Zhou Q, Hu W, Wu C, et al: A novel protein encoded by a
circular RNA circPPP1R12A promotes tumor pathogenesis and
metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer.
18:472019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Pan Z, Cai J, Lin J, Zhou H, Peng J, Liang
J, Xia L, Yin Q, Zou B, Zheng J, et al: A novel protein encoded by
circFNDC3B inhibits tumor progression and EMT through regulating
Snail in colon cancer. Mol Cancer. 19:712020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liang ZX, Liu HS, Xiong L, Yang X, Wang
FW, Zeng ZW, He XW, Wu XR and Lan P: A novel NF-κB regulator
encoded by circPLCE1 inhibits colorectal carcinoma progression by
promoting RPS3 ubiquitin-dependent degradation. Mol Cancer.
20:1032021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang L, Zhou J, Zhang C, Chen R, Sun Q,
Yang P, Peng C, Tan Y, Jin C, Wang T, et al: A novel tumour
suppressor protein encoded by circMAPK14 inhibits progression and
metastasis of colorectal cancer by competitively binding to MKK6.
Clin Transl Med. 11:e6132021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhang C, Zhou X, Geng X, Zhang Y, Wang J,
Wang Y, Jing J, Zhou X and Pan W: Circular RNA hsa_circ_0006401
promotes proliferation and metastasis in colorectal carcinoma. Cell
Death Dis. 12:4432021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shi JF, Cao M, Wang Y, Bai FZ, Lei L, Peng
J, Feletto E, Canfell K, Qu C and Chen W: Is it possible to halve
the incidence of liver cancer in China by 2050? Int J Cancer.
148:1051–1065. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang W and Wei C: Advances in the early
diagnosis of hepatocellular carcinoma. Genes Dis. 7:308–319. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li Y, Chen B, Zhao J, Li Q, Chen S, Guo T,
Li Y, Lai H, Chen Z, Meng Z, et al: HNRNPL circularizes ARHGAP35 to
produce an oncogenic protein. Adv Sci (Weinh). 8:20017012021.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Chen B, Dragomir MP, Yang C, Li Q, Horst D
and Calin GA: Targeting non-coding RNAs to overcome cancer therapy
resistance. Signal Transduct Target Ther. 7:1212022. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Li P, Song R, Yin F, Liu M, Liu H, Ma S,
Jia X, Lu X, Zhong Y, Yu L, et al: circMRPS35 promotes malignant
progression and cisplatin resistance in hepatocellular carcinoma.
Mol Ther. 30:431–447. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Li H, Lan T, Liu H, Liu C, Dai J, Xu L,
Cai Y, Hou G, Xie K, Liao M, et al: IL-6-induced cGGNBP2 encodes a
protein to promote cell growth and metastasis in intrahepatic
cholangiocarcinoma. Hepatology. 75:1402–1419. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Rimassa L, Personeni N, Czauderna C,
Foerster F and Galle P: Systemic treatment of HCC in special
populations. J Hepatol. 74:931–943. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Song R, Ma S, Xu J, Ren X, Guo P, Liu H,
Li P, Yin F, Liu M, Wang Q, et al: A novel polypeptide encoded by
the circular RNA ZKSCAN1 suppresses HCC via degradation of mTOR.
Mol Cancer. 22:162023. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Quintanal-Villalonga A, Molina-Pinelo S,
Cirauqui C, Ojeda-Márquez L, Marrugal Á, Suarez R, Conde E,
Ponce-Aix S, Enguita AB, Carnero A, et al: FGFR1 cooperates with
EGFR in lung cancer oncogenesis, and their combined inhibition
shows improved efficacy. J Thorac Oncol. 14:641–655. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wang T, Liu Z, She Y, Deng J, Zhong Y,
Zhao M, Li S, Xie D, Sun X, Hu X, et al: A novel protein encoded by
circASK1 ameliorates gefitinib resistance in lung adenocarcinoma by
competitively activating ASK1-dependent apoptosis. Cancer Lett.
520:321–331. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhao W, Xue Y, Zhang Y, Zhu Y, Chen Z and
Zhao X: A peptide translated from circPPP1R12A promotes the
malignancy of non-small cell lung cancer cells through AKT
signaling pathway. J Clin Lab Anal. 36:e246442022. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Chen A, Zhong L, Ju K, Lu T, Lv J and Cao
H: Plasmatic circRNA predicting the occurrence of human
glioblastoma. Cancer Manag Res. 12:2917–2923. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Nie JH, Li TX, Zhang XQ and Liu J: Roles
of non-coding RNAs in normal human brain development, brain tumor,
and neuropsychiatric disorders. Noncoding RNA. 5:362019.PubMed/NCBI
|
|
79
|
Goenka A, Tiek DM, Song X, Iglesia RP, Lu
M, Hu B and Cheng SY: The role of non-coding RNAs in glioma.
Biomedicines. 10:20312022. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–15. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Xia X, Li X, Li F, Wu X, Zhang M, Zhou H,
Huang N, Yang X, Xiao F, Liu D, et al: A novel tumor suppressor
protein encoded by circular AKT3 RNA inhibits glioblastoma
tumorigenicity by competing with active phosphoinositide- dependent
Kinase-1. Mol Cancer. 18:1312019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang M, Huang N, Yang X, Luo J, Yan S,
Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al: A novel protein
encoded by the circular form of the SHPRH gene suppresses glioma
tumorigenesis. Oncogene. 37:1805–1814. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei
P, Liu H, Xu J, Xiao F, Zhou H, et al: A peptide encoded by
circular form of LINC-PINT suppresses oncogenic transcriptional
elongation in glioblastoma. Nat Commun. 9:44752018. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Su Y, Xu C, Liu Y, Hu Y and Wu H: Circular
RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression
via multiple miRNAs sponge. Aging (Albany NY). 11:3362–3375. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Li WH, Song YC, Zhang H, Zhou ZJ, Xie X,
Zeng QN, Guo K, Wang T, Xia P and Chang DM: Decreased expression of
Hsa_circ_00001649 in gastric cancer and its clinical significance.
Dis Markers. 2017:45876982017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Xu Y, Yao Y, Zhong X, Leng K, Qin W, Qu L,
Cui Y and Jiang X: Downregulated circular RNA hsa_circ_0001649
regulates proliferation, migration and invasion in
cholangiocarcinoma cells. Biochem Biophys Res Commun. 496:455–461.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Jiang Y, Wang T, Yan L and Qu L: A novel
prognostic biomarker for pancreatic ductal adenocarcinoma:
Hsa_circ_0001649. Gene. 675:88–93. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Saunders JT, Kumar S, Benavides-Serrato A,
Holmes B, Benavides KE, Bashir MT, Nishimura RN and Gera J:
Translation of circHGF RNA encodes an HGF protein variant promoting
glioblastoma growth through stimulation of c-MET. J Neurooncol.
163:207–218. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Arnold M, Morgan E, Rumgay H, Mafra A,
Singh D, Laversanne M, Vignat J, Gralow JR, Cardoso F, Siesling S,
et al: Current and future burden of breast cancer: Global
statistics for 2020 and 2040. Breast. 66:15–23. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou
X, Xie X and Tang H: CircFBXW7 inhibits malignant progression by
sponging miR-197-3p and encoding a 185-aa protein in
triple-negative breast cancer. Mol Ther Nucleic Acids. 18:88–98.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Li J, Ma M, Yang X, Zhang M, Luo J, Zhou
H, Huang N, Xiao F, Lai B, Lv W, et al: Circular HER2 RNA positive
triple negative breast cancer is sensitive to Pertuzumab. Mol
Cancer. 19:1422020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Li Y, Li G, Guo X, Yao H, Wang G and Li C:
Non-coding RNA in bladder cancer. Cancer Lett. 485:38–44. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Gu C, Zhou N, Wang Z, Li G, Kou Y, Yu S,
Feng Y, Chen L, Yang J and Tian F: CircGprc5a promoted bladder
oncogenesis and metastasis through Gprc5a-targeting peptide. Mol
Ther Nucleic Acids. 13:633–641. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Cowan AJ, Green DJ, Kwok M, Lee S, Coffey
DG, Holmberg LA, Tuazon S, Gopal AK and Libby EN: Diagnosis and
management of multiple myeloma: A review. JAMA. 327:464–477. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Tang X, Guo M, Ding P, Deng Z, Ke M, Yuan
Y, Zhou Y, Lin Z, Li M, Gu C, et al: BUB1B and circBUB1B_544aa
aggravate multiple myeloma malignancy through evoking chromosomal
instability. Signal Transduct Target Ther. 6:3612021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Tang X, Deng Z, Ding P, Qiang W, Lu Y, Gao
S, Hu Y, Yang Y, Du J and Gu C: A novel protein encoded by
circHNRNPU promotes multiple myeloma progression by regulating the
bone marrow microenvironment and alternative splicing. J Exp Clin
Cancer Res. 41:852022. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Li F, Cai Y, Deng S, Yang L, Liu N, Chang
X, Jing L, Zhou Y and Li H: A peptide CORO1C-47aa encoded by the
circular noncoding RNA circ-0000437 functions as a negative
regulator in endometrium tumor angiogenesis. J Biol Chem.
297:1011822021. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Chen LL, Bindereif A, Bozzoni I, Chang HY,
Matera AG, Gorospe M, Hansen TB, Kjems J, Ma XK, Pek JW, et al: A
guide to naming eukaryotic circular RNAs. Nat Cell Biol. 25:1–5.
2023. View Article : Google Scholar : PubMed/NCBI
|