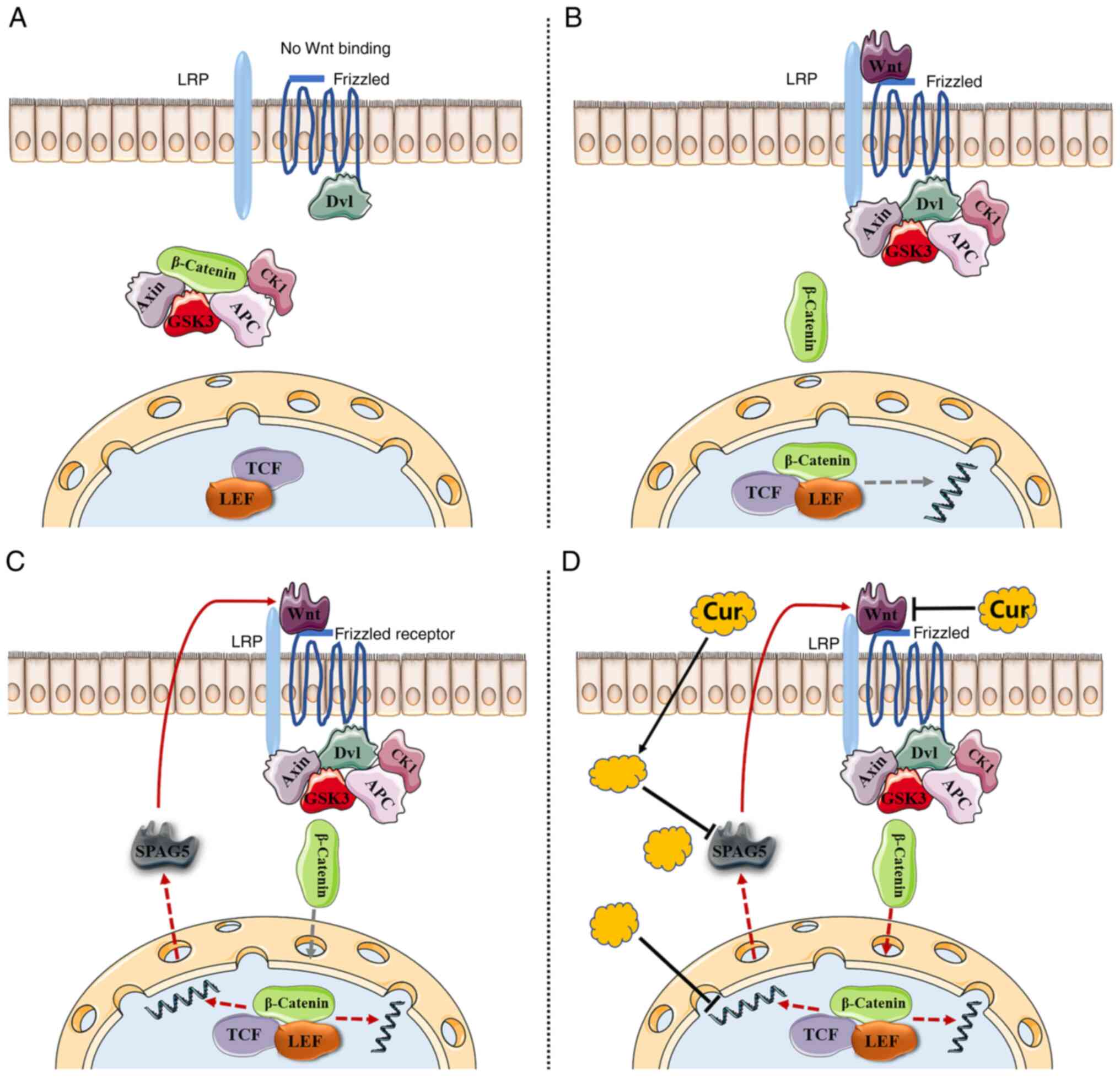
Introduction
Primary liver cancer ranks as the sixth most
prevalent cancer globally and the fifth most common malignant tumor
in China, where it is also the second leading cause of
cancer-related deaths (1,2). Comprising hepatocellular carcinoma
(HCC), intrahepatic cholangiocarcinoma, and combined hepatocellular
cholangiocarcinoma, primary liver cancer is dominated by HCC, which
accounts for 75–85% of cases (1).
The development of HCC has been closely linked with viral
infections, cirrhosis, alcoholism, smoking, aflatoxin, exposure to
harmful chemicals, and genetic factors (2). The disease prevalence significantly
varies across the world, reflecting the differing distribution of
these pathogenic factors, with 72% of cases occurring in Asia and
>50% in China (3). Notably,
sex-based disparities exist in the incidence and mortality rates of
liver cancer. For instance, liver cancer is the second most lethal
form of cancer in men and ranks sixth in women (4). The high mortality rate of liver cancer
is primarily due to late diagnosis and a lack of early treatment,
necessitating the exploration of new diagnostic and therapeutic
approaches. This pursuit, particularly regarding the regulation of
the tumor microenvironment and early drug intervention targets, is
a promising direction for clinical diagnosis and treatment of liver
cancer.
Sperm-associated antigen 5 (SPAG5), a member of the
SPAG family, is a spindle-related protein primarily expressed in
the testis and placenta that regulates spindle assembly and sister
chromatid separation during the M phase of the cell cycle (5). Aberrations in SPAG5 have been
implicated in irregular cell cycle regulation and DNA damage, both
of which are closely associated with tumorigenesis (6,7). An
elevated level of SPAG5 has been documented in numerous tumors
(8–12), fostering an interest in its function
as a cancer testis antigen (CTA) in cancer genesis, especially its
role in promoting cancer via the Wnt/β-catenin pathway. The
Wnt/β-catenin pathway is activated in HCC patients, and the
function of SPAG5 in HCC has been identified as being mediated
through the same pathway (13). In
patients with HCC, SPAG5 has been revealed to downregulate the
expression of SCARA5 via the β-catenin/TCF4 pathway, thereby
exacerbating the progression of the cancer (13). Separate studies by Jiang et
al (14) and Liu et al
(15) explored the influence of
SPAG5 via the Wnt/β-catenin pathway in breast cancer and gastric
cancer, respectively. In addition, Rebouissou et al
(16) reported that ~95% of
patients with HCC exhibited Wnt/β-catenin pathway activation.
However, the biological role and clinical significance of SPAG5 in
HCC remain unclear.
Wnt is a type of secreted glycoprotein that uses
frizzled as its receptor (17,18).
The classical Wnt signaling pathway involves β-catenin accumulation
and subsequent entry into the nucleus to activate target gene
transcription, thus promoting cancer development (19,20).
Accordingly, the Wnt pathway has been considered a suitable
therapeutic target. Despite this, targeted drugs have faced
clinical trial challenges and have not been implemented clinically
due to severe side effects (21).
As a result, research has shifted towards exploring natural
pharmaceutical components capable of inhibiting the Wnt/β-catenin
pathway. For instance, genistein has been revealed to inhibit the
Wnt/β-catenin pathway and regulate the expression of several
Wnt/β-catenin antagonists through epigenetic modifications.
Furthermore, myricetin has been demonstrated to reduce cytoplasmic
and nuclear β-catenin levels, and curcumin has been observed to
limit β-catenin nuclear translocation (22–24).
Curcumin, an active compound extracted from Curcuma
longa, exhibits numerous pharmacological effects such as
antioxidative, anti-inflammatory, free radical scavenging, and
antitumor properties, and is employed in the treatment of
cardiovascular diseases and digestive system diseases (25). The antitumor effect of curcumin has
been widely researched, revealing a close association with the
dysregulation of tumor cell proliferation, apoptosis, and
angiogenesis signaling pathways (26). Curcumin, either in isolation or in
combination with other drugs, impacts cancer-related signaling
pathways (27–28).
There is existing evidence indicating that curcumin
can downregulate the Wnt/β-catenin signaling pathway by inhibiting
Wnt in HCC, a process partially mediated by the activation of
autophagy (29–31). Furthermore, additional research has
identified the inhibitory effect of curcumin on tumors associated
with the Wnt/β-catenin pathway (32). Notably, it has been established that
SPAG5 operates through the Wnt/β-catenin pathway (13). However, to date, no study has
explored the curcumin-SPAG5-Wnt/β-catenin triad.
Based on the analysis of bioinformatics databases
and previous research (33), the
aim of the present study was to combine CTA and SPAG5 with the
cancer-related Wnt/β-catenin signaling pathway to explore the
regulatory effect of SPAG5 on the Wnt/β-catenin pathway.
Confirmation of the hypothesis that curcumin inhibits the
Wnt/β-catenin pathway by acting on SPAG5, was also attempted. The
goal was to identify alternative methods and new targets for
targeted therapeutic drugs aimed at the Wnt pathway, which
currently cannot be used clinically due to their side effects.
Equally important was the exploration of natural drug ingredients
with lower toxicity and side effects for early intervention in HCC,
enhancement of the effectiveness of other treatments, and
alleviation of drug resistance.
Materials and methods
Gene expression analysis
Differential analysis of SPAG5 expression in cancer
tissue compared to adjacent tissue from 374 hepatocellular
carcinoma (HCC) patient tissue samples was carried out using The
Cancer Genome Atlas (TCGA, http://portal.gdc.cancer.gov/). In addition, Kyoto
Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp/kegg/kegg1.html) and Gene Ontology
(GO http://david.ncifcrf.gov/home.jsp) database used for
enrichment analysis. DESeq2 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html)
(34) was used to analyze
differentially expressed genes, with genes deemed differentially
expressed if | log2 FC | ≥1 and P<0.05. Additionally,
differential gene expression analysis associated with SPAG5 was
conducted.
Patients and sample collection
Tumor tissues from patients with HCC patients, along
with corresponding non-tumor tissues, were procured from the Second
People's Hospital of Hunan Province (Changsha, China), between
March 2021 and December 2022. The clinical and demographic data of
participants are outlined in Table
I. The inclusion criteria were as follows: i) Age, ≥30 years;
ii) patients with HCC who were undergoing surgical treatment; and
iii) patients who had previously undergone a physical examination.
The exclusion criteria included mental illness and liver and kidney
dysfunction. Resected specimens were promptly frozen and stored at
−80°C for subsequent analysis. A team of pathologists confirmed the
identification of tumor tissues and adjacent normal tissues. Each
patient provided written informed consent, and the study was
approved (approval no. DK2018002) by the Ethics Committee of the
Second People's Hospital of Hunan Province.
 | Table I.Clinical data of patients with
HCC. |
Table I.
Clinical data of patients with
HCC.
| Sample number | Age | Sex | Clinicopathological
diagnosis |
|---|
| 1 | 62 | Female | Medium
differentiated liver cancer |
| 2 | 50 | Female | Medium-low
differentiated liver cancer |
| 3 | 73 | Male | Medium
differentiated liver cancer |
| 4 | 51 | Male | Medium-high
differentiated liver cancer |
| 5 | 49 | Female | High differentiated
liver cancer |
| 6 | 32 | Male | Medium
differentiated liver cancer |
| 7 | 62 | Male | Medium
differentiated liver cancer |
| 8 | 34 | Male | High differentiated
liver cancer |
| 9 | 50 | Male | High differentiated
liver cancer |
| 10 | 42 | Male | Medium
differentiated liver cancer |
| 11 | 64 | Male | Medium
differentiated liver cancer |
| 12 | 41 | Male | High differentiated
liver cancer |
| 13 | 52 | Male | Medium
differentiated liver cancer |
| 14 | 49 | Male | Medium
differentiated liver cancer |
| 15 | 66 | Male | Medium
differentiated liver cancer |
Cell culture
The human HCC cell lines (Huh7 and HCCLM3) and a
normal human hepatocytes line (MIHA) were procured from the Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). These cells were cultivated in DMEM (Dalian
Meilun Biology Technology Co., Ltd.) supplemented with 10% fetal
bovine serum (FBS; Bioexplorer Life Sciences). The Huh7 cell line,
derived from a Japanese male high-grade HCC, is hepatitis B
virus-negative and is capable of producing cytoplasmic molecules
such as Alb, ATT, and AFP (35).
The HCCLM3 cell line, also known as human highly metastatic
hepatoma cells, was developed via multiple rounds of in vivo
selection in nude mice of the human hepatoma cell line MHCC97-H for
its high lung metastatic potential (36). MIHA denotes normal human
hepatocytes. The cells were incubated at 37°C in an atmosphere
containing 5% CO2.
Plasmid transfection
SPAG5 siRNA plasmid was procured from Shanghai
GeneChem Co., Ltd. The process of plasmid transfection commenced
with seeding of cells in a six-well plate at a density of
2–3×105 and addition of 2 ml of complete medium. The
cells were then placed in a carbon dioxide incubator at 37°C
overnight. Transfection reagents were then prepared. Solution A
consisted of Opti-MEM (125 µl; Gibco; Thermo Fisher Scientific,
Inc.) combined with Lipofectamine®3000 (3.75 µl per
well; Invitrogen; Thermo Fisher Scientific, Inc.). Solution B
comprised Opti-MEM (125 µl), P3000 (5 µl; Invitrogen; Thermo Fisher
Scientific, Inc.), and 2.5 µg of the plasmid to be transfected per
well. Solutions A and B were mixed in equal proportions and left to
rest at room temperature for 15 min. Upon achieving a cell density
of 70–90%, the complete medium was replaced with basal medium. The
combined A + B solution was gradually added to each well, mixed
gently in a cross direction, and then incubated at 37°C in a carbon
dioxide incubator. After 24 h, fluorescence expression was observed
with a fluorescence microscope (Olympus Corporation). The sequence
for SPAG5 siRNA was as follows: 5′-ccAUGCAACUGGAUUAUACAA-3′. The
sequence for the scrambled siRNA was as follows:
5′-UUCUCCGAACGUGUCACGU-3′ (used as the negative control).
Cell Counting Kit-8 (CCK-8) assay
Both Huh7 and HCCLM3 cells were seeded in 96-well
plates (Zhejiang Sorfa Life Science Research Co., Ltd.) at a
density of 5×104 cells/ml in DMEM medium supplemented
with 10% FBS. Following incubation for 24 h at 37°C in an
atmosphere containing 5% CO2, the cells were exposed to
curcumin at varying concentrations (0, 12.5, 50, 100, 125 and 200
µM). After another 24-h incubation period, 10 µl CCK-8 (Dojindo
Laboratories, Inc.) was added to each well and the cells were
incubated for an additional 1 h at 37°C. The absorbance at a
wavelength of 450 nm was measured for each well using a microplate
reader (Thermo Fisher Scientific, Inc.).
Wound-healing assay
Huh7 and HCCLM3 cells were seeded in six-well plates
(Zhejiang Sorfa Life Science Research Co., Ltd.) at a density of
2×105 cells/ml and incubated overnight at 37°C.
Subsequently, when cell confluence reached 80–90%, a wound was
created in the cell layer by manual scraping with a 200-µl pipette
tip. The cells were rinsed with PBS (Dalian Meilun Biology
Technology Co., Ltd.), then exposed to curcumin in serum-free
medium at concentrations of 80 and 100 µM. Images were captured at
0 and 24 h post-wounding to observe the healing process by
fluorescence microscopy.
Immunohistochemistry
Tissues were formalin-fixed (fixed in 4% buffered
formaldehyde at room temperature for 24 h) and embedded in
paraffin. The HCC tissue sections (thickness, 1 µm) underwent
treatment with xylene and fractional ethanol, followed by antigen
retrieval in 0.01 M citric acid buffer. Blocking was accomplished
using 3% hydrogen peroxide (Fuzhou Maixin Biotech Co., Ltd.).
Tissue sections were subsequently subjected to a 30-min incubation
with 5–10% goat serum (cat. no. SL038; Beijing Solarbio Science
& Technology Co., Ltd.) at room temperature, followed by an
overnight incubation at 4°C with anti-SPAG5 monoclonal antibody
(1:100; cat. no. 14726-1-AP; ProteinTech Group, Inc.).
Subsequently, the sections were treated with HRP-conjugated goat
anti-rabbit immunoglobulin G for 30 min at room temperature (1:200;
cat. no. PR30011; ProteinTech Group, Inc). DAB was utilized for
color rendering. Post-staining with hematoxylin (at room
temperature for 3–5 min), images of the tissue sections were
captured using an Olympus light microscope (Olympus Corporation).
Scoring was independently performed by a pathologist (37).
Western blot analysis
Protein extraction of Huh7 and HCCLM3 cell was
accomplished using a lysis buffer (Biosharp Life Sciences),
followed by quantification using the BCA method. Proteins (50 µg)
were separated on 8% SDS-PAGE and transferred onto polyvinylidene
fluoride (PVDF) membranes (0.45 µm; Abiowell). The PVDF membranes
were then blocked with 5% skim milk for 1.5 h at room temperature.
Overnight incubation at 4°C on a shaker followed, with antibodies
against SPAG5 (1:20,000; cat. no. 14726-1-AP; ProteinTech Group,
Inc.), cyclin D1 (1:20,000; cat. no. ab134175; Abcam), and GAPDH
(1:20,000; cat. no. 10494-1-AP; ProteinTech Group, Inc.).
Post-0.05% TBST washing, the membranes underwent incubation with
goat anti-rabbit IgG-HRP (1:20,000; cat. no. PR30011; ProteinTech
Group, Inc.) at 37°C for 1 h. The membrane was visualized using an
enhanced chemiluminescence (ECL) detection system (Biosharp Life
Sciences). The band grayscale value is calculated using ImageJ
(1.53a; National Institutes of Health).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from Huh7 and HCCLM3 cell
using the Ultra-pure total RNA extraction Kit (Simgen Xinjing
Biological). The RNA was subsequently reverse transcribed using the
PrimeScript RT Reagent Kit (Novoprotein Scientific, Inc.) according
to the manufacturer's instructions. For the quantitative polymerase
chain reaction (PCR) analysis, qPCR was conducted using SYBR Premix
Ex Taq (Novoprotein Scientific, Inc.) as per the manufacturer's
instructions. The relative amounts of SPAG5, and GAPDH (internal
control) mRNAs were determined using the Real-Time PCR System. The
thermocycling conditions were as follows: 95°C for 1 min, followed
by 40 cycles at 95°C for 20 sec, and 60°C for 1 min. Relative gene
expression was caculated using the 2−ΔΔCq method
(38). Primers used for the RT-qPCR
assay were as follows: GAPDH forward, 5′-ACAGCCTCAAGATCATCAGC3′ and
reverse, 5′-GGTCATGAGTCCTTCCACGAT-3′; SPAG5 forward,
5′-CATCTCACAGTGGGATAACTAATAAAC-3′ and reverse,
5′-CAGGGATAGGTGAAGCAAGGATA-3′.
Cell apoptosis analysis
The apoptosis rate of the cells was assessed using
Annexin V-APC/PI Apoptosis Kit (cat. no. E-CK-A117; Elabscience
Biotechnology, Inc.) as per the manufacturer's instructions.
Briefly, 5×105 cells were harvested by centrifugation at
300 × g for 5 min at room temperature and rinsed twice with PBS.
The cells were then resuspended in 500 µl binding buffer and
incubated with 5 µl Annexin V-APC and 5 µl PI in the dark at room
temperature for 15 min. Apoptotic events were subsequently detected
using flow cytometry (BD Fortessa; BD Biosciences) Data analysis
was used by FlowJo 10.8.1 (Becton Dickinson and Company).
Statistical analysis
The results, expressed as the mean ± SD, were
derived from at least three independent experiments and analyzed
using GraphPad Prism 8 (GraphPad Software, Inc.). Significant
differences were determined using Student's paired t-test and
two-tailed distribution. P<0.05 was considered to indicate a
statistically significant difference.
Results
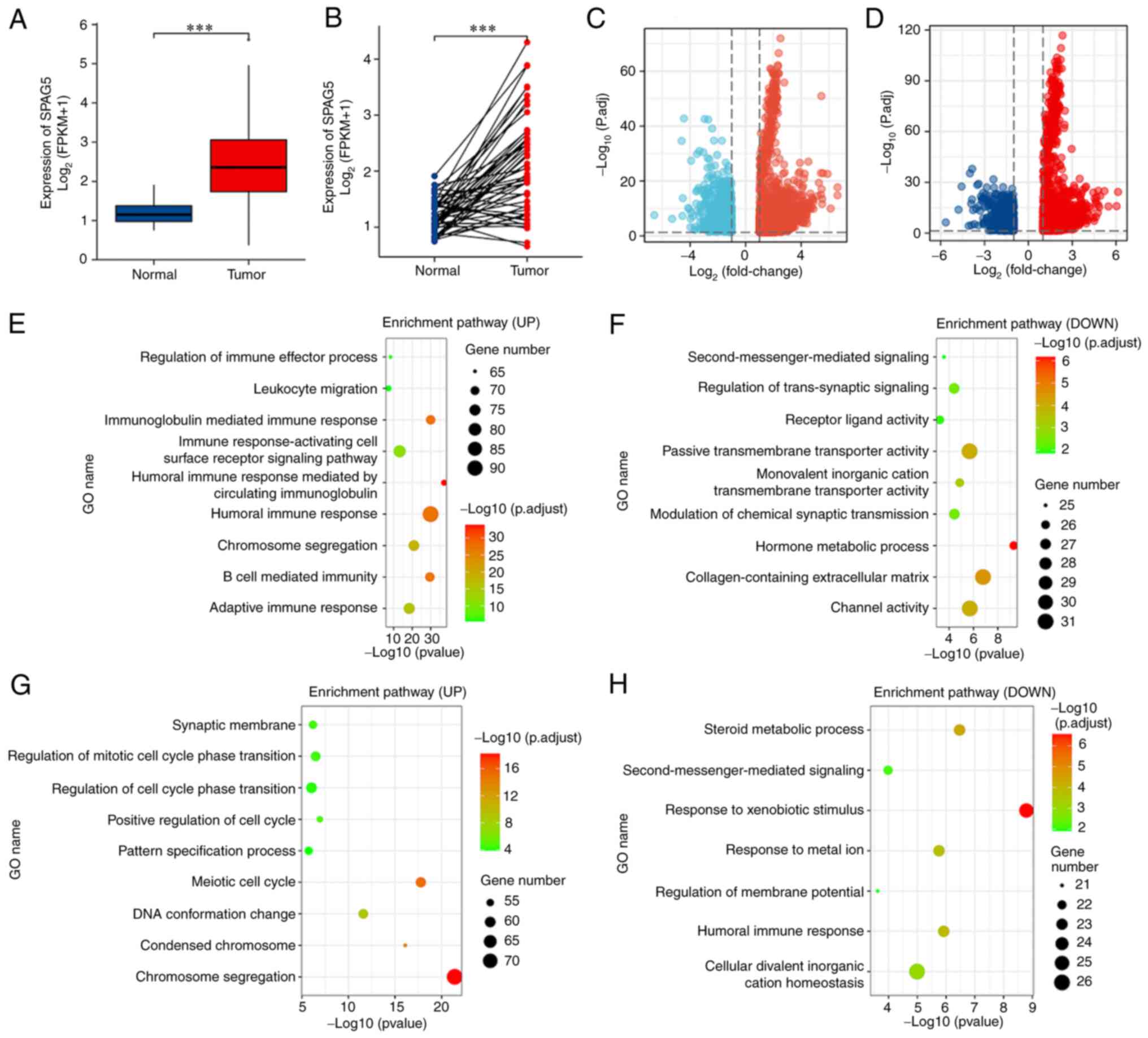
Bioinformatics database analysis of
SPAG5 expression and experimental verification in HCC
The expression level of SPAG5 in the tissue of
patients with HCC was analyzed via TCGA database. Compared with
normal tissues, the tumor tissue of patients with HCC exhibited a
significantly higher expression of SPAG5 (Fig. 1A and B; P<0.001). Functional
enrichment analysis was carried out on transcriptome data from
TCGA. Using the median value of SPAG5 expression as the cutoff
point, the upper 50% was designated as the high expression group
and the lower 50% as the low expression group. This revealed 14,858
differentially expressed genes, including 2,710 upregulated genes
(with top genes being KIF18B, MCM10 and GINS1) and 12,148
downregulated genes (with top genes being SMR3A, MT1B and ANKFN1).
DESeq2 was used to analyze differentially expressed genes, with
genes deemed differentially expressed if | log2 FC | ≥1
and P<0.05. Applying the same methodology but considering the
top 25% as the high expression group and the bottom 25% as the low
expression group, 13,213 genes were found to be differentially
expressed, including 2,677 upregulated genes (top genes being
AL139327, MAGEA4 and LGALS14) and 10,536 downregulated genes (top
genes being SMR3A, BX322559 and TRARG1). As indicated in the
volcano plot, low expression genes are in blue, and high expression
genes are in red (Fig. 1C and D). A
series of enrichment analyses, including KEGG and GO, were
subsequently performed. The results of GO-KEGG enrichment analysis
mainly implicated involvement in the ‘humoral immune response’,
‘immunoglobulin-mediated immune response’, and ‘immune
response-activating cell surface receptor signaling pathway’.
Additionally, these genes were involved in the regulation of cell
division activities, such as ‘chromosome segregation’, ‘regulation
of mitotic cell cycle phase transition’, and ‘positive regulation
of cell cycle’. Notably, these genes also partook in ‘response to
metal ion’, ‘channel activity’, ‘second-messenger-mediated
signaling’, and ‘response to xenobiotic stimulus’ (Fig. 1E-H).
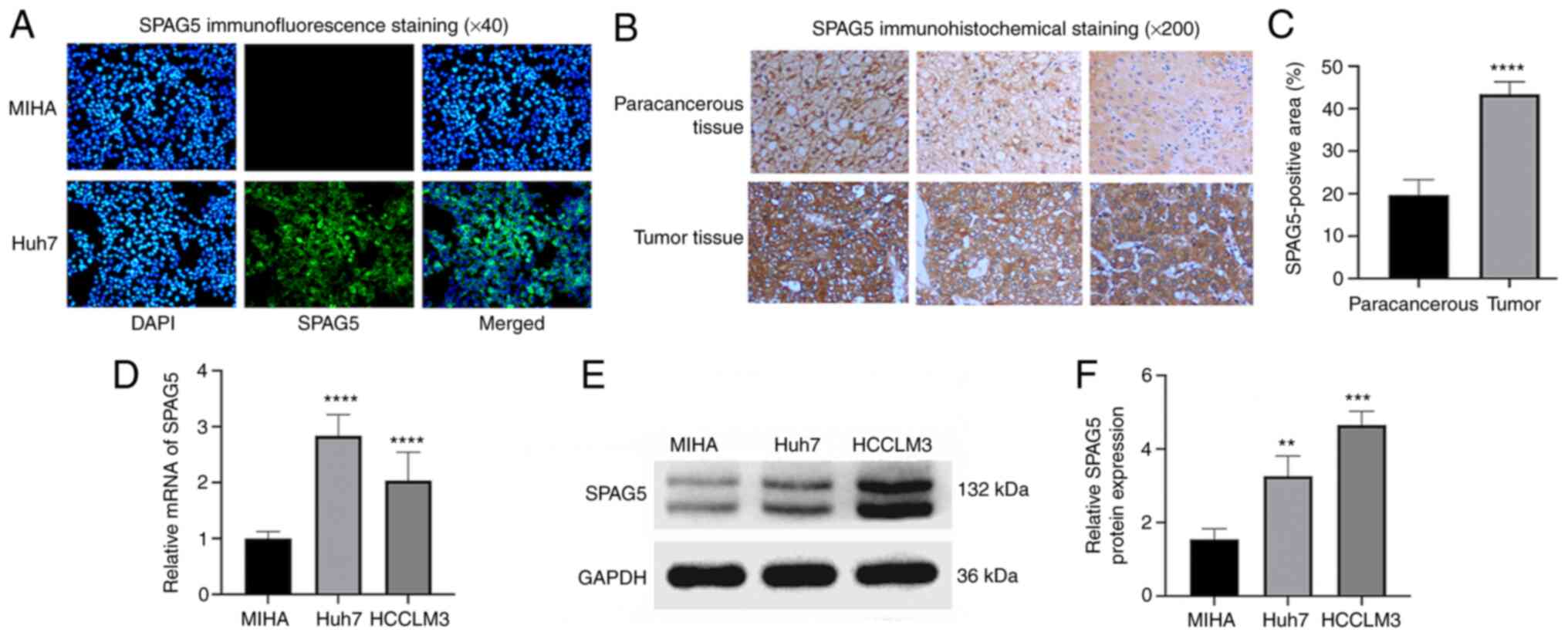
Cellular immunofluorescence analysis results showed
strong fluorescent expression of SPAG5 in Huh7 cells, while normal
human hepatocyte line MIHA displayed no fluorescent expression
(Fig. 2A). The expression of SPAG5
in cancer tissues and adjacent tissues from patients with HCC was
investigated through immunohistochemical staining. This revealed
significantly higher SPAG5 expression in cancer tissues than in
adjacent tissues (P<0.001; Fig. 2B
and C). To determine the expression of SPAG5 in HCC cells,
RT-qPCR experiments were conducted using normal human hepatocyte
line MIHA and two hepatocellular carcinoma cells (Huh7 and HCCLM3).
The mean fold change of SPAG5 mRNA expression was revealed to be
significantly lower in MIHA compared with Huh7 and HCCLM3, with a
2.8-fold increase in Huh7 compared with MIHA cells, and a 2-fold
increase in HCCLM3 compared with MIHA cells (Fig. 2D). Western blot analysis was
conducted to ascertain SPAG5 protein expression in these cell
lines, which also indicated higher SPAG5 expression in HCC cells
compared with MIHA (Fig. 2E and
F).
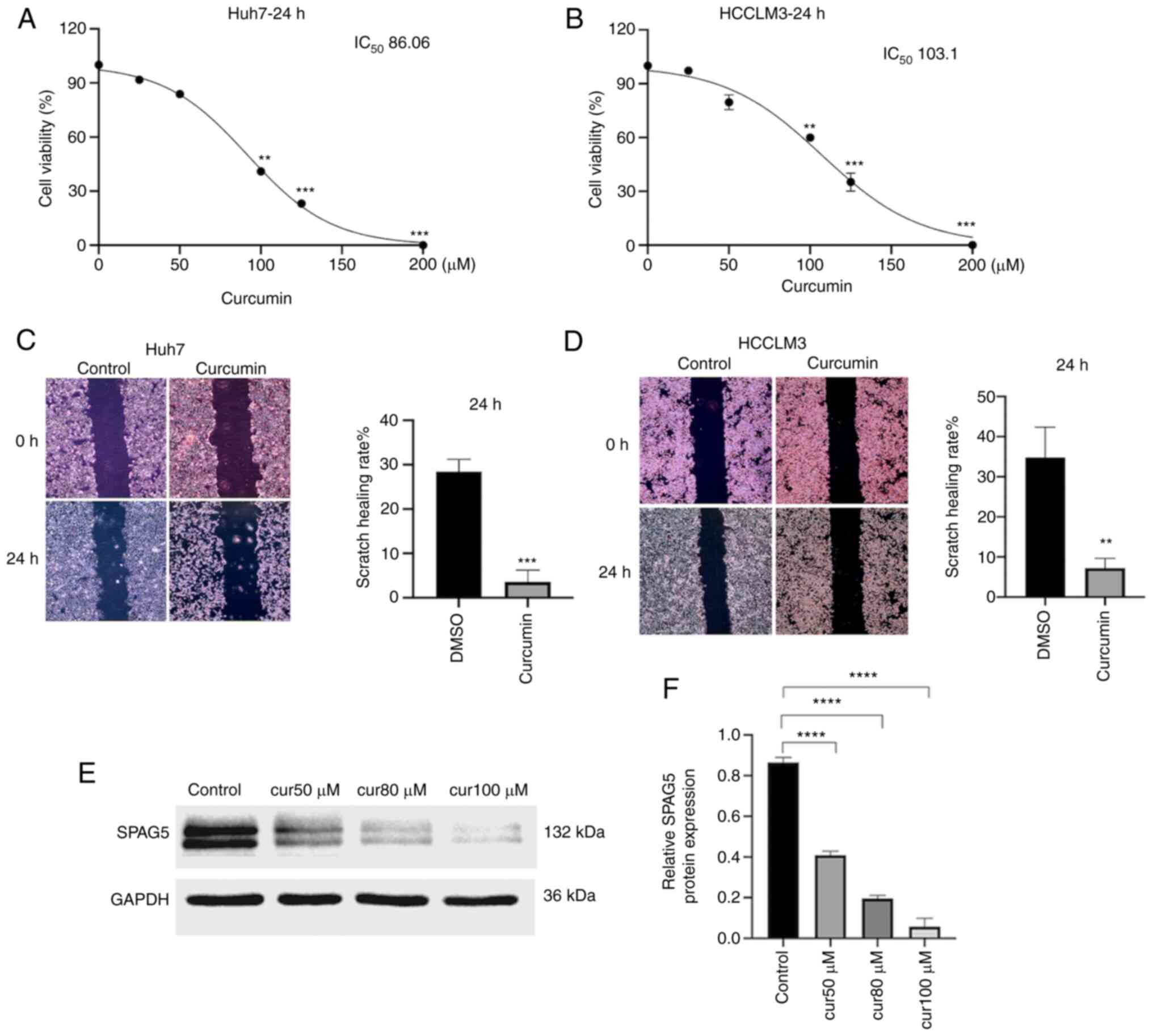
Curcumin inhibits the proliferation,
migration and SPAG5 expression of HCC cells
CCK-8 results confirmed that treatment of Huh7 and
HCCLM3 cells with curcumin for 24 h led to statistically
significant differences in cell viability (P<0.05), with optical
density (OD) decreasing as curcumin concentration increased.
(Fig. 3A and B) The optimal
curcumin concentrations for HCC cells, Huh7 and HCCLM3, were
determined to be 80 and 100 µM respectively, and these were
selected for further experimentation. A scratch test was then
performed, with a control group treated with DMSO and an
experimental group treated with the optimal concentration of
curcumin. The results indicated that after 24 h, the migration
ability of curcumin-treated HCC cells was significantly reduced,
with the difference being statistically significant (P<0.01
Fig. 3C and D). Western blot
results revealed that the expression of SPAG5 protein in Huh7 cells
co-cultured with varying concentrations of curcumin was curcumin
concentration-dependently inhibited (Fig. 3E and F).
Curcumin inhibits SPAG5-induced cyclin
D1 expression, and knockdown of SPAG5 decreases the expression of
β-catenin
The expression of cyclin D1 was examined using
western blot analysis, and the results demonstrated that the
expression of cyclin D1 protein in both cell types was
significantly weakened in the curcumin-treated group (Fig. 4A and B). In Huh7 cells
overexpressing SPAG5, curcumin could significantly inhibit the
expression of SPAG5 and cyclin D1 (Fig.
4C and D). On the other hand, the expression of SPAG5 was
significantly reduced in siSPAG5 HCCLM3 cells, and curcumin could
further reduce the expression of SPAG5 and cyclin D1, (Fig. 4E and F). In the SPAG5-knockdown cell
line, the expression of β-catenin was also significantly decreased
(Fig. 4G and H). Although the
overexpression of SPAG5 promoted proliferation and migration,
curcumin inhibited this promoting effect, but not by inhibiting
cyclin D1.
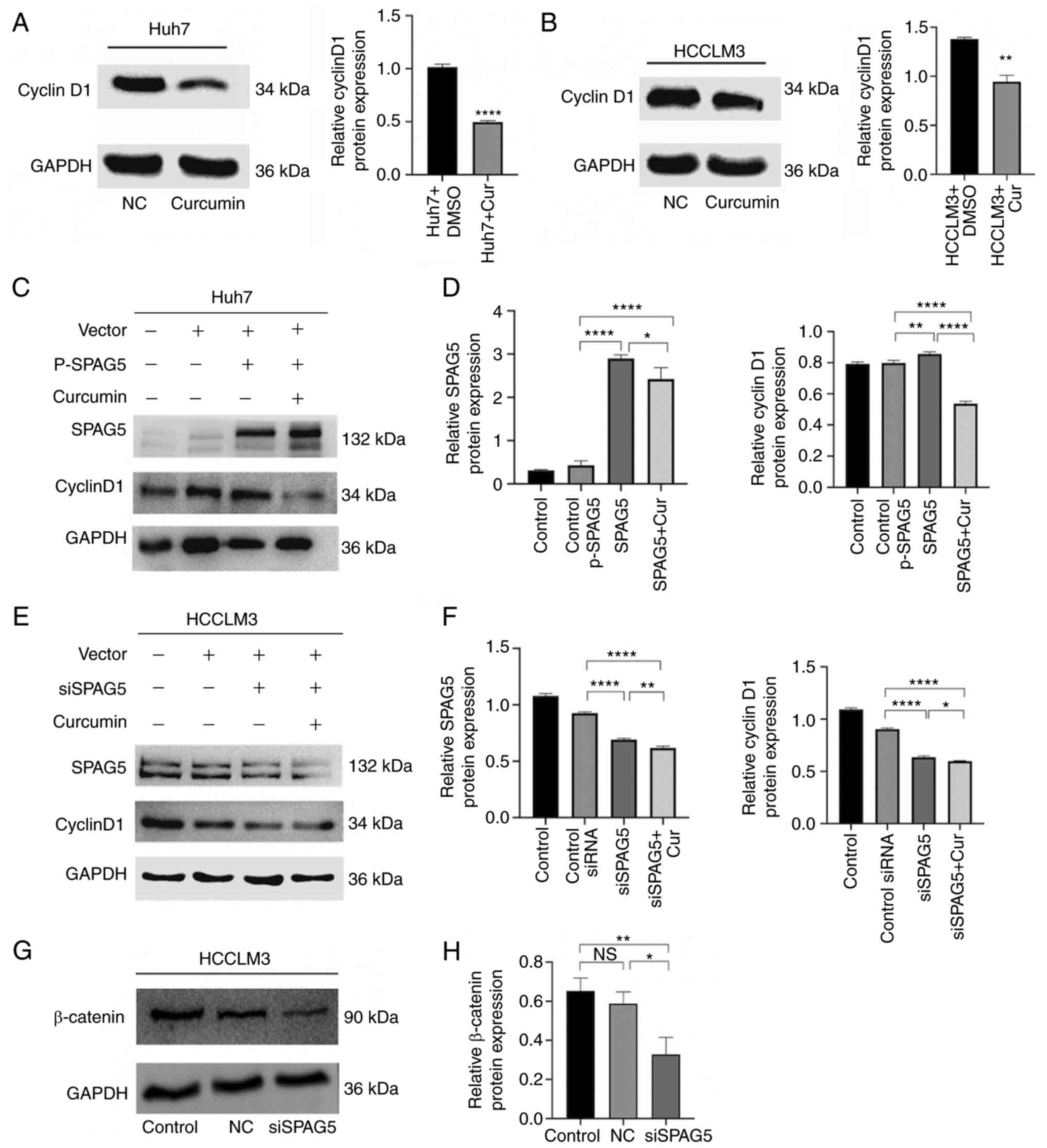 | Figure 4.Inhibitory effect of curcumin on
SPAG5 expression in hepatocellular carcinoma. (A and B) Western
blotting experiments revealed that curcumin can suppress cyclin D1
expression in hepatoma cells. (C and D) Huh7 cells were transfected
with P-SPAG5 plasmid and treated with curcumin, followed by the
analysis of SPAG5 and cyclin D1 expression via western blotting.
Control, Huh7 cells; Control p-SPAG5, cells transfected with a
negative control expression plasmid; SPAG5, cells transfected with
overexpression SPAG5 plasmids; SPAG5 + Cur, cells transfected with
overexpression SPAG5 plasmids and the addition of curcumin. (E and
F) HCCLM3 cells were transfected with siSPAG5 plasmid and treated
with curcumin, leading to the analysis of SPAG5 and cyclin D1
expression via western blotting. Control, HCCLM3 cells; Control
siRNA, cells transfected with a negative control expression
plasmid; siSPAG5, cells transfected with SPAG5-knockdown plasmids;
SPAG5 + Cur, cells transfected with SPAG5-knockdown plasmids and
the addition of curcumin. (G and H) SPAG5-knockdown decreased
β-catenin expression. Control, HCCLM3 cells; NC, cells transfected
with a negative control expression plasmid; siSPAG5, cells
transfected with SPAG5-knockdown plasmids. *P<0.05, **P<0.01
and ****P<0.0001. SPAG5, sperm-associated antigen 5; NS, not
significant. |
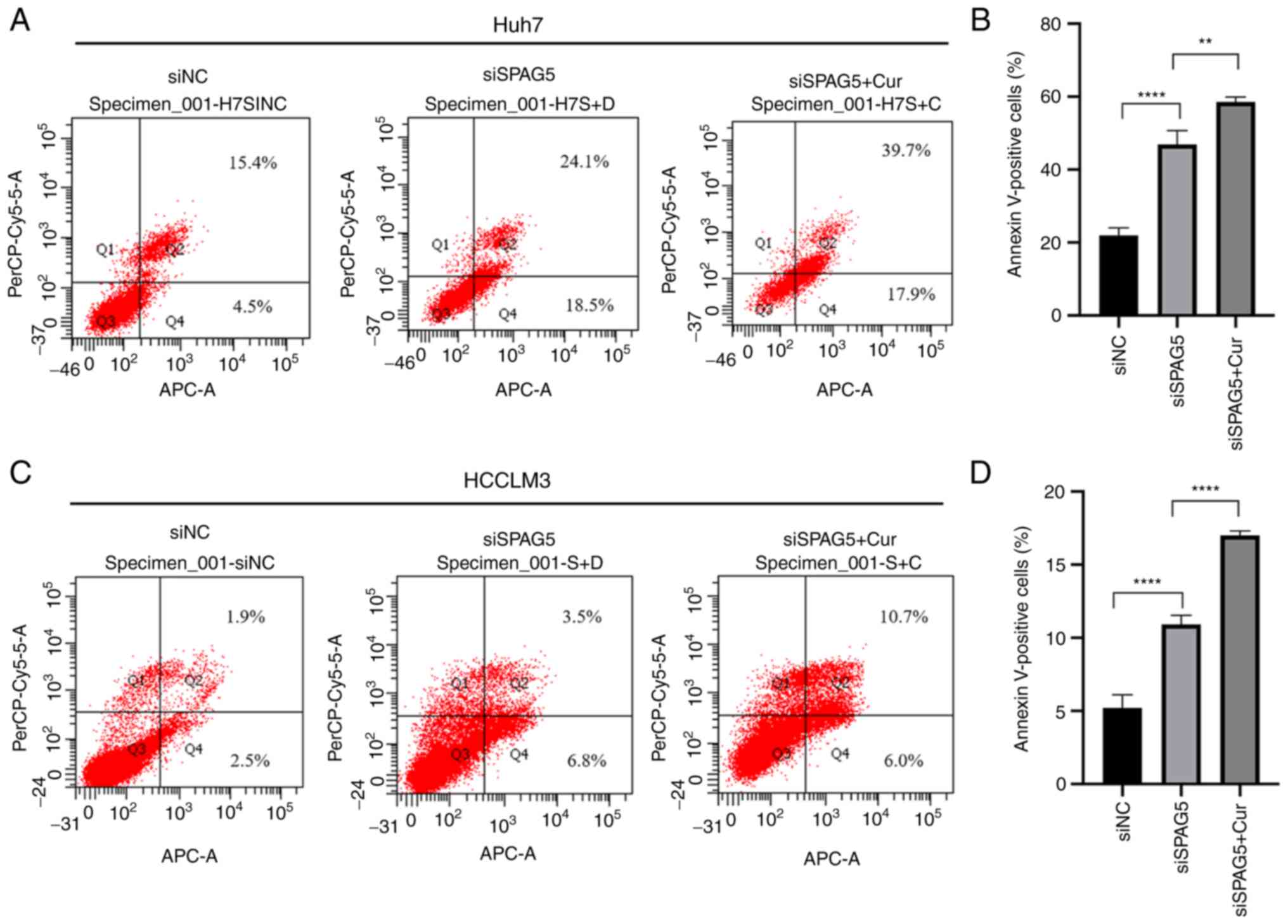
Further investigation was conducted to better
understand the mechanism by which SPAG5 contributes to HCC cell
growth, specifically examining the effects of SPAG5 knockdown on
apoptosis. The findings, as presented in Fig. 5, revealed that the percentage of
apoptotic cells was significantly increased in the SPAG5-knockdown
group compared with the control group. The application of curcumin
resulted in a further increase in the rate of apoptosis.
Collectively, these data indicated that the knockdown of SPAG5
exerted a tumor-suppressive effect in human HCC cells, an effect
which was amplified by the action of curcumin (Fig. 6).
Discussion
The present study investigated the potential
carcinogenic role of SPAG5 in HCC and demonstrated via experimental
evidence that the expression of SPAG5 was significantly elevated in
HCC at both the tissue and cellular levels. Not only was the high
expression of SPAG5 in HCC validated, but also its mechanism and
association with Wnt, and the intervention of curcumin were
revealed. The present research, exploring uncharted territories,
has provided some preliminary conclusions.
Initially, the study revealed that in HCC cell
lines, the most suitable concentration of curcumin, which
effectively inhibited their proliferation and migration, ranged
between 80–100 µM. This inhibition was further confirmed at the
gene level by RT-qPCR and at the protein level by western blot
analysis. Next, the potential effect of curcumin on SPAG5 was
investigated. It was revealed that the expression of SPAG5 in HCC
cell lines exhibited a negative association with curcumin
concentration. Finally, HCC cell lines overexpressing SPAG5 were
co-cultured under the optimal concentration of curcumin, revealing
that curcumin could still significantly inhibit the expression of
SPAG5 and the Wnt/β-catenin pathway-related oncogene cyclin D1.
Moreover, experimental results indicated that curcumin could
directly inhibit the expression of β-catenin in HCC cells. Cyclin
D1 and β-catenin were inhibited when SPAG5 was knocked down. In
addition, when SPAG5 was knocked down, curcumin further inhibited
cyclin D1, which indicated that the inhibition of cyclin D1 by
curcumin was associated with the inhibition of SPAG5.
The present study elucidated the effect of curcumin
on SPAG5 targets in HCC from multiple angles and provided a
theoretical basis for the treatment of clinical liver cancer.
However, whether curcumin influences normal liver cells, was not
addressed during the experimental process of the present study. In
addition, these findings are only preliminary conclusions, as
additional pathway proteins have not yet been used to corroborate
these findings. Whether curcumin directly inhibits Wnt or inhibits
Wnt by inhibiting SPAG5 still necessitates further investigation.
These questions will be the focus of a future follow-up study.
In summary, the findings of the present study
indicated that SPAG5 is a highly significant cancer-promoting
molecule and therapeutic target, especially its association with
the Wnt pathway, which warrants further investigation. This
research could potentially address the issues of drug side effects
targeting Wnt. The inhibition of SPAG5 by curcumin also presents a
valuable approach for the early intervention and adjuvant treatment
of HCC, thus warranting the execution of more clinical trials.
While the effect of curcumin has been experimentally confirmed
again, the correct application of curcumin in clinical practice
requires resolution in the following areas: i) Further elucidation
of the mechanism and target of curcumin; ii) addressing the
bioavailability of curcumin; and iii) conducting clinical
research.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The Science and Technology Program of Traditional Chinese
Medicine of Hunan Province (grant no. 2021033) has provided support
for this study.
Availability of data and materials
The data that support the findings of this study are
available from the corresponding author upon reasonable
request.
Authors' contributions
BR conceived and designed the study. HL performed
the experiments and drafted the manuscript. YQ undertook the
responsibility of data collection and performed data analysis. The
task of gathering and verifying the references was performed by JW,
while YH participated in discussing the results. HL and YQ confirm
the authenticity of all the raw data. All authors have critically
reviewed and edited the manuscript. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved (approval no.
DK2018002) by the Ethics Committee of the Second People's Hospital
of Hunan Province (Changsha, China) and each patient provided
written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
SPAG5
|
sperm-associated antigen 5
|
|
HCC
|
hepatocellular carcinoma
|
|
CTA
|
cancer testis antigen
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
CCK-8
|
Cell Counting Kit-8
|
References
|
1
|
Mejia JC and Pasko J: Primary liver
cancers: Intrahepatic cholangiocarcinoma and hepatocellular
carcinoma. Surg Clin North Am. 100:535–549. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chidambaranathan-Reghupaty S, Fisher PB
and Sarkar D: Hepatocellular carcinoma (HCC): Epidemiology,
etiology and molecular classification. Adv Cancer Res. 149:1–61.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shi JF, Cao M, Wang Y, Bai FZ, Lei L, Peng
J, Feletto E, Canfell K, Qu C and Chen W: Is it possible to halve
the incidence of liver cancer in China by 2050? Int J Cancer.
148:1051–1065. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thein KH, Kleylein-Sohn J, Nigg EA and
Gruneberg U: Astrin is required for the maintenance of sister
chromatid cohesion and centrosome integrity. J Cell Biol.
178:345–354. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Buechler S: Low expression of a few genes
indicates good prognosis in estrogen receptor positive breast
cancer. BMC Cancer. 9:2432009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Välk K, Vooder T, Kolde R, Reintam MA,
Petzold C, Vilo J and Metspalu A: Gene expression profiles of
non-small cell lung cancer: Survival prediction and new biomarkers.
Oncology. 79:283–292. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang YF, Zhang MF, Tian QH, Fu J, Yang X,
Zhang CZ and Yang H: SPAG5 interacts with CEP55 and exerts
oncogenic activities via PI3K/AKT pathway in hepatocellular
carcinoma. Mol Cancer. 17:1172018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang H, Li S, Yang X, Qiao B, Zhang Z and
Xu Y: miR-539 inhibits prostate cancer progression by directly
targeting SPAG5. J Exp Clin Cancer Res. 35:602016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li M, Li A, Zhou S, Lv H and Yang W: SPAG5
upregulation contributes to enhanced c-MYC transcriptional activity
via interaction with c-MYC binding protein in triple-negative
breast cancer. J Hematol Oncol. 12:142019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Abdel-Fatah TMA, Agarwal D, Liu DX,
Russell R, Rueda OM, Liu K, Xu B, Moseley PM, Green AR, Pockley AG,
et al: SPAG5 as a prognostic biomarker and chemotherapy sensitivity
predictor in breast cancer: A retrospective, integrated genomic,
transcriptomic, and protein analysis. Lancet Oncol. 17:1004–1018.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yuan LJ, Li JD, Zhang L, Wang JH, Wan T,
Zhou Y, Tu H, Yun JP, Luo RZ, Jia WH and Zheng M: SPAG5
upregulation predicts poor prognosis in cervical cancer patients
and alters sensitivity to taxol treatment via the mTOR signaling
pathway. Cell Death Dis. 5:e12472014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu H, Hu J, Wei R, Zhou L, Pan H, Zhu H,
Huang M, Luo J and Xu W: SPAG5 promotes hepatocellular carcinoma
progression by downregulating SCARA5 through modifying β-catenin
degradation. J Exp Clin Cancer Res. 37:2292018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang J, Wang J, He X, Ma W, Sun L, Zhou
Q, Li M and Yu S: High expression of SPAG5 sustains the malignant
growth and invasion of breast cancer cells through the activation
of Wnt/β-catenin signalling. Clin Exp Pharmacol Physiol.
46:597–606. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu JY, Zeng QH, Cao PG, Xie D, Yang F, He
LY, Dai YB, Li JJ, Liu XM, Zeng HL, et al: SPAG5 promotes
proliferation and suppresses apoptosis in bladder urothelial
carcinoma by upregulating Wnt3 via activating the AKT/mTOR pathway
and predicts poorer survival. Oncogene. 37:3937–3952. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rebouissou S, Franconi A, Calderaro J,
Letouzé E, Imbeaud S, Pilati C, Nault JC, Couchy G, Laurent A,
Balabaud C, et al: Genotype-phenotype correlation of CTNNB1
mutations reveals different ß-catenin activity associated with
liver tumor progression. Hepatology. 64:2047–2061. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hayat R, Manzoor M and Hussain A: Wnt
signaling pathway: A comprehensive review. Cell Biol Int.
46:863–877. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Clevers H and Nusse R: Wnt/β-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
De A: Wnt/Ca2+ signaling pathway: A brief
overview. Acta Biochim Biophys Sin (Shanghai). 43:745–756. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhan T, Rindtorff N and Boutros M: Wnt
signaling in cancer. Oncogene. 36:1461–1473. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kahn M: Can we safely target the WNT
pathway? Nat Rev Drug Discov. 13:513–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Duchartre Y, Kim YM and Kahn M: The Wnt
signaling pathway in cancer. Crit Rev Oncol Hematol. 99:141–149.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Steinhart Z and Angers S: Wnt signaling in
development and tissue homeostasis. Development. 145:dev1465892018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tafrihi M and Nakhaei Sistani R:
E-Cadherin/β-Catenin Complex: A target for anticancer and
antimetastasis plants/plant-derived compounds. Nutr Cancer.
69:702–722. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gupta SC, Patchva S and Aggarwal BB:
Therapeutic roles of curcumin: Lessons learned from clinical
trials. AAPS J. 15:195–218. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Udagawa T and Wood M: Tumor-stromal cell
interactions and opportunities for therapeutic intervention. Curr
Opin Pharmacol. 10:369–374. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Al-Ejeh F, Kumar R, Wiegmans A, Lakhani
SR, Brown MP and Khanna KK: Harnessing the complexity of DNA-damage
response pathways to improve cancer treatment outcomes. Oncogene.
29:6085–6098. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jin H, Lian N, Zhang F, Chen L, Chen Q, Lu
C, Bian M, Shao J, Wu L and Zheng S: Activation of PPARγ/P53
signaling is required for curcumin to induce hepatic stellate cell
senescence. Cell Death Dis. 7:e21892016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ashrafizadeh M, Ahmadi Z, Mohamamdinejad
R, Yaribeygi H, Serban MC, Orafai HM and Sahebkar A: Curcumin
therapeutic modulation of the wnt signaling pathway. Curr Pharm
Biotechnol. 21:1006–1015. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vallée A, Lecarpentier Y and Vallée JN:
Curcumin: A therapeutic strategy in cancers by inhibiting the
canonical WNT/β-catenin pathway. J Exp Clin Cancer Res. 38:3232019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Patel SS, Acharya A, Ray RS, Agrawal R,
Raghuwanshi R and Jain P: Cellular and molecular mechanisms of
curcumin in prevention and treatment of disease. Crit Rev Food Sci
Nutr. 60:887–939. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ghasemi F, Shafiee M, Banikazemi Z,
Pourhanifeh MH, Khanbabaei H, Shamshirian A, Amiri Moghadam S,
ArefNezhad R, Sahebkar A, Avan A and Mirzaei H: Curcumin inhibits
NF-kB and Wnt/β-catenin pathways in cervical cancer cells. Pathol
Res Pract. 215:1525562019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen W, Chen X, Li S and Ren B:
Expression, immune infiltration and clinical significance of SPAG5
in hepatocellular carcinoma: A gene expression-based study. J Gene
Med. 22:e31552020. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou M, Zhao K, Yao Y, Yuan Y, Pei R, Wang
Y, Chen J, Hu X, Zhou Y, Chen X and Wu C: Productive HBV infection
of well-differentiated, hNTCP-expressing human hepatoma-derived
(Huh7) cells. Virol Sin. 32:465–475. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li B, Zhang Y, Wu W, Du G, Cai L, Shi H
and Chen S: Neovascularization of hepatocellular carcinoma in a
nude mouse orthotopic liver cancer model: A morphological study
using X-ray in-line phase-contrast imaging. BMC Cancer. 17:732017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ram S, Vizcarra P, Whalen P, Deng S,
Painter CL, Jackson-Fisher A, Pirie-Shepherd S, Xia X and Powell
EL: Pixelwise H-score: A novel digital image analysis-based metric
to quantify membrane biomarker expression from immunohistochemistry
images. PLoS One. 16:e02456382021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|