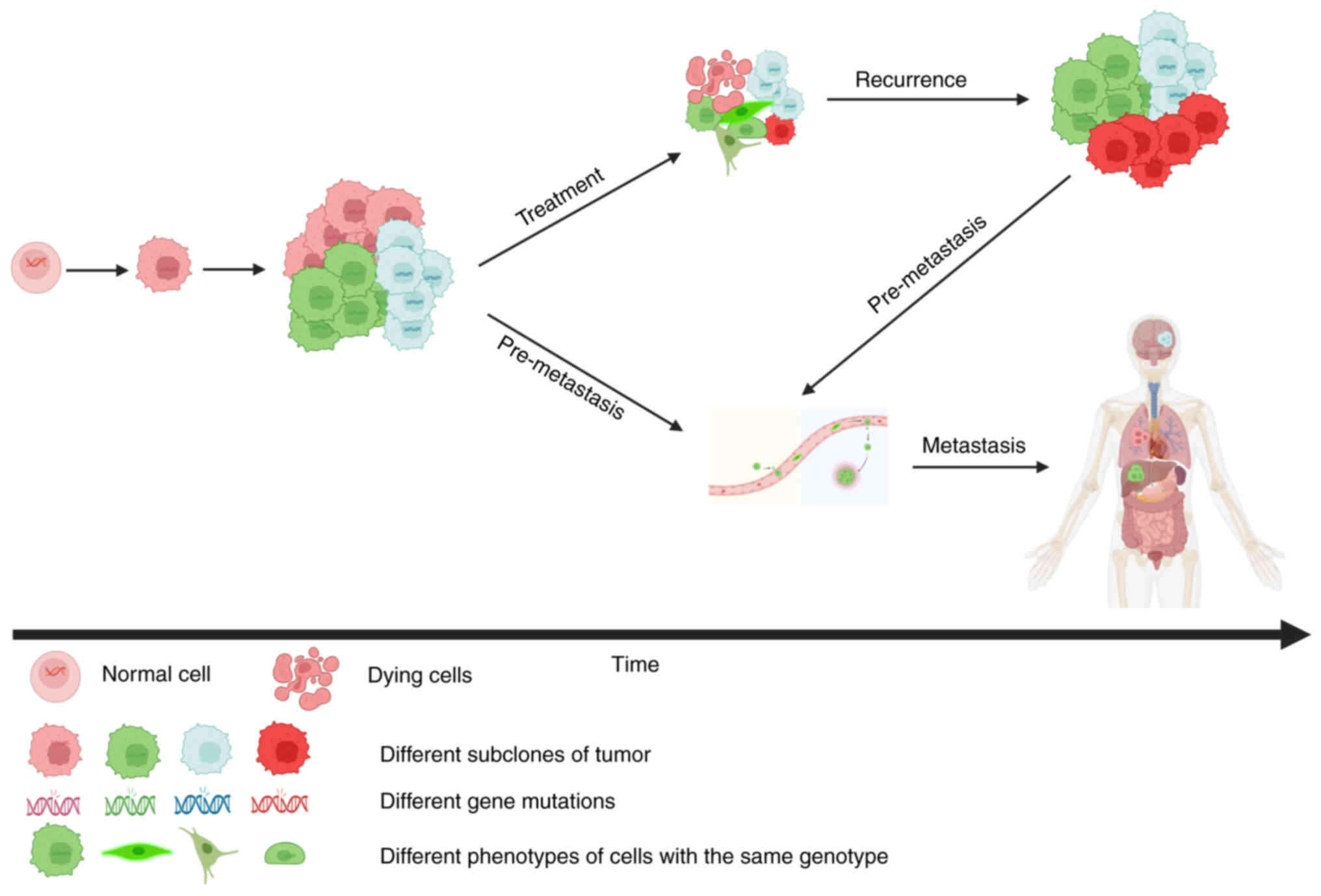
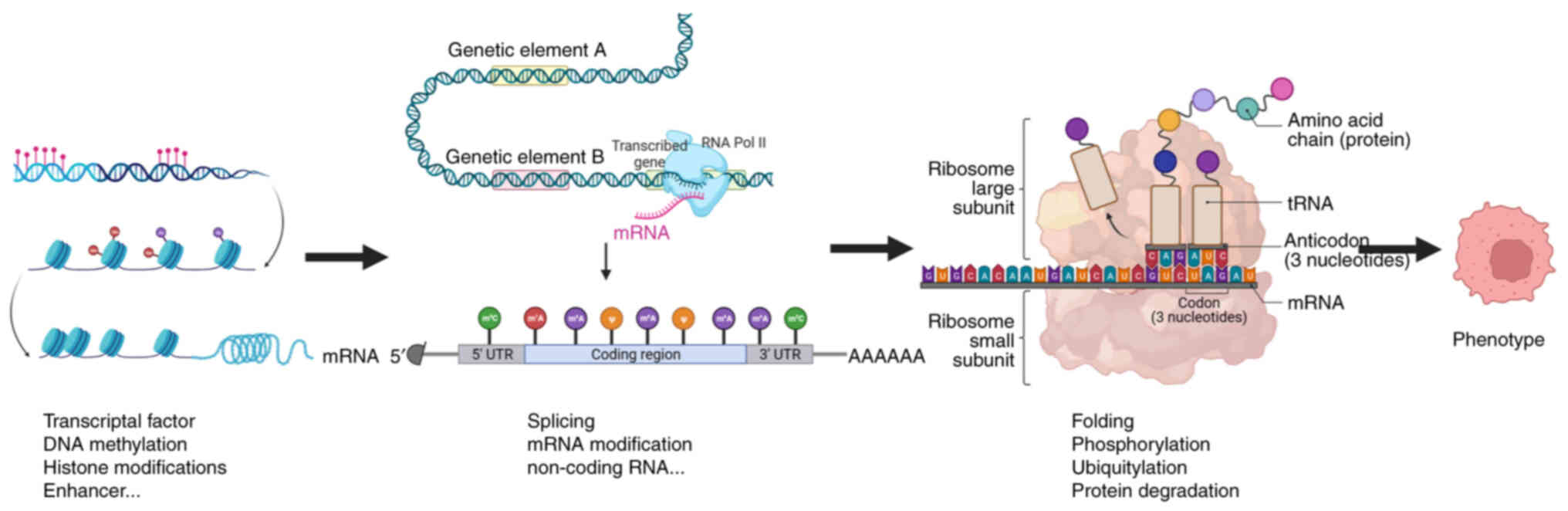
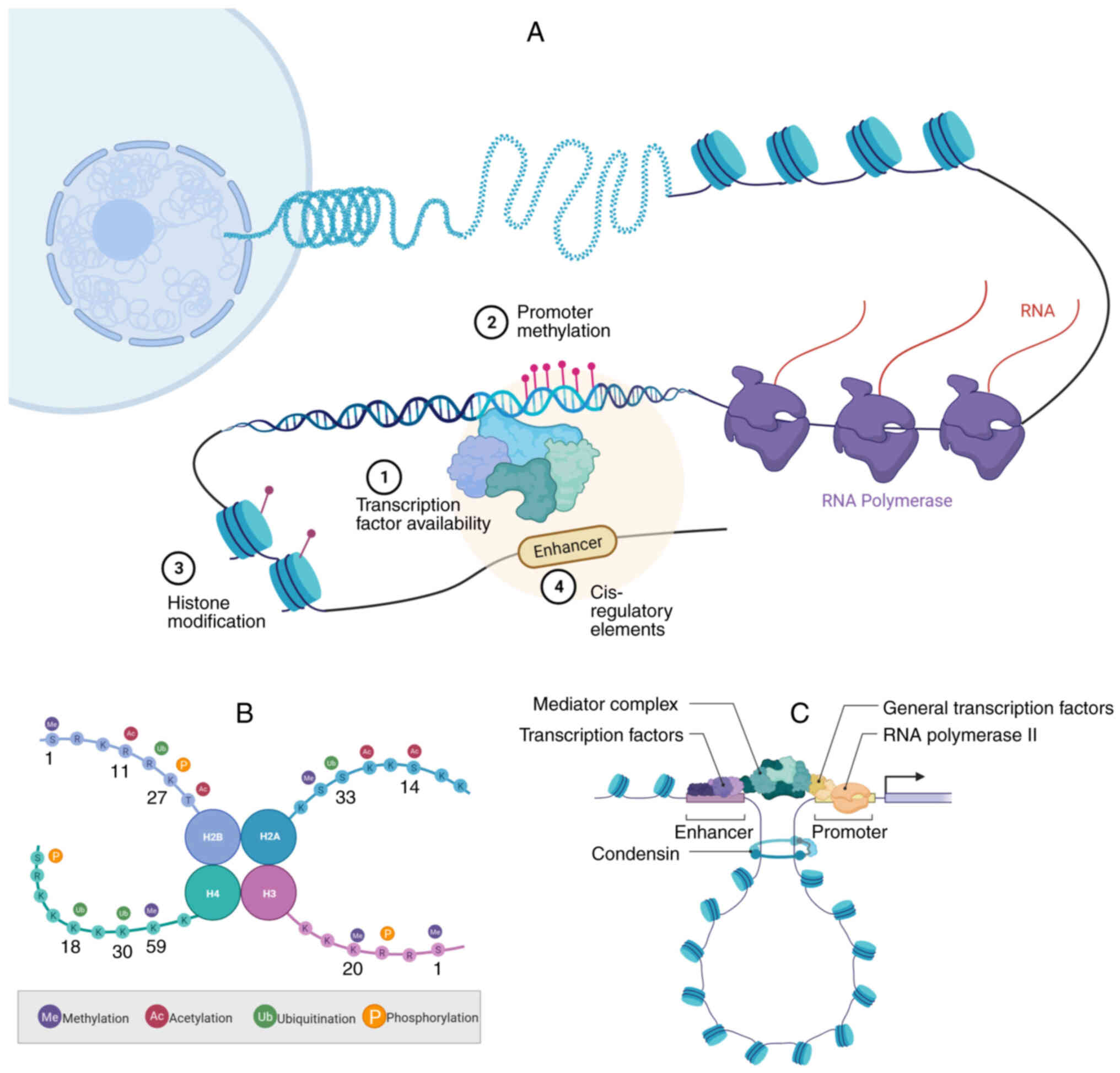
|
1
|
Lyden D, Ghajar CM, Correia AL,
Aguirre-Ghiso JA, Cai S, Rescigno M, Zhang P, Hu G, Fendt SM, Boire
A, et al: Metastasis. Cancer Cell. 40:787–791. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang D, Jones MG, Naranjo S, Rideout WM
III, Min KHJ, Ho R, Wu W, Replogle JM, Page JL, Quinn JJ, et al:
Lineage tracing reveals the phylodynamics, plasticity, and paths of
tumor evolution. Cell. 185:1905–1923.e25. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vendramin R, Litchfield K and Swanton C:
Cancer evolution: Darwin and beyond. EMBO J. 40:e1083892021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saha S, Pradhan N B N, Mahadevappa R,
Minocha S and Kumar S: Cancer plasticity: Investigating the causes
for this agility. Semin Cancer Biol. 88:138–156. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Barkley D, Moncada R, Pour M, Liberman DA,
Dryg I, Werba G, Wang W, Baron M, Rao A, Xia B, et al: Cancer cell
states recur across tumor types and form specific interactions with
the tumor microenvironment. Nat Genet. 54:1192–1201. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Lih TM, Dhanasekaran SM, Mannan R,
Chen L, Cieslik M, Wu Y, Lu RJ, Clark DJ, Kolodziejczak I, et al:
Histopathologic and proteogenomic heterogeneity reveals features of
clear cell renal cell carcinoma aggressiveness. Cancer Cell.
41:139–163.e17. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Drapkin BJ and Minna JD: Studying lineage
plasticity one cell at a time. Cancer Cell. 38:150–152. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lambert AW and Weinberg RA: Linking EMT
programmes to normal and neoplastic epithelial stem cells. Nat Rev
Cancer. 21:325–338. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Aggarwal V, Montoya CA, Donnenberg VS and
Sant S: Interplay between tumor microenvironment and partial EMT as
the driver of tumor progression. iScience. 24:1021132021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hanahan D: Hallmarks of cancer: New
dimensions. Cancer Discov. 12:31–46. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chang HY and Qi LS: Reversing the Central
Dogma: RNA-guided control of DNA in epigenetics and genome editing.
Mol Cell. 83:442–451. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Buccitelli C and Selbach M: mRNAs,
proteins and the emerging principles of gene expression control.
Nat Rev Genet. 21:630–644. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fabbri L, Chakraborty A, Robert C and
Vagner S: The plasticity of mRNA translation during cancer
progression and therapy resistance. Nat Rev Cancer. 21:558–577.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee LJ, Papadopoli D, Jewer M, Del Rincon
S, Topisirovic I, Lawrence MG and Postovit LM: Cancer plasticity:
The role of mRNA translation. Trends Cancer. 7:134–145. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pope SD and Medzhitov R: Emerging
principles of gene expression programs and their regulation. Mol
Cell. 71:389–397. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao LY, Song J, Liu Y, Song CX and Yi C:
Mapping the epigenetic modifications of DNA and RNA. Protein Cell.
11:792–808. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Joung J, Ma S, Tay T, Geiger-Schuller KR,
Kirchgatterer PC, Verdine VK, Guo B, Arias-Garcia MA, Allen WE,
Singh A, et al: A transcription factor atlas of directed
differentiation. Cell. 186:209–229.e26. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu Y, Zhuo S, Zhou Y, Ma L, Sun Z, Wu X,
Wang XW, Gao B and Yang Y: Yap-Sox9 signaling determines hepatocyte
plasticity and lineage-specific hepatocarcinogenesis. J Hepatol.
76:652–664. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Park S, Mossmann D, Chen Q, Wang X, Dazert
E, Colombi M, Schmidt A, Ryback B, Ng CKY, Terracciano LM, et al:
Transcription factors TEAD2 and E2A globally repress acetyl-CoA
synthesis to promote tumorigenesis. Mol Cell. 82:4246–4261.e11.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tan SH and Barker N: Stemming colorectal
cancer growth and metastasis: HOXA5 forces cancer stem cells to
differentiate. Cancer Cell. 28:683–685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Perekatt AO, Shah PP, Cheung S, Jariwala
N, Wu A, Gandhi V, Kumar N, Feng Q, Patel N, Chen L, et al: SMAD4
Suppresses WNT-Driven dedifferentiation and oncogenesis in the
differentiated gut epithelium. Cancer Res. 78:4878–4890. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu R, Zhou Z, Zhao D and Chen C: The
induction of KLF5 transcription factor by progesterone contributes
to progesterone-induced breast cancer cell proliferation and
dedifferentiation. Mol Endocrinol. 25:1137–1144. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thier B, Zhao F, Stupia S, Bruggemann A,
Koch J, Schulze N, Horn S, Coch C, Hartmann G, Sucker A, et al:
Innate immune receptor signaling induces transient melanoma
dedifferentiation while preserving immunogenicity. J Immunother
Cancer. 10:e0038632022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kopanja D, Chand V, O'Brien E,
Mukhopadhyay NK, Zappia MP, Islam A, Frolov MV, Merrill BJ and
Raychaudhuri P: Transcriptional repression by FoxM1 suppresses
tumor differentiation and promotes metastasis of breast cancer.
Cancer Res. 82:2458–2471. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kalisz M, Bernardo E, Beucher A, Maestro
MA, Del Pozo N, Millan I, Haeberle L, Schlensog M, Safi SA, Knoefel
WT, et al: HNF1A recruits KDM6A to activate differentiated acinar
cell programs that suppress pancreatic cancer. EMBO J.
39:e1028082020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nilsson T, Waraky A, Ostlund A, Li S,
Staffas A, Asp J, Fogelstrand L, Abrahamsson J and Palmqvist L: An
induced pluripotent stem cell t(7;12)(q36;p13) acute myeloid
leukemia model shows high expression of MNX1 and a block in
differentiation of the erythroid and megakaryocytic lineages. Int J
Cancer. 151:770–782. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Simeoni F, Romero-Camarero I, Camera F,
Amaral FMR, Sinclair OJ, Papachristou EK, Spencer GJ, Lie ALM,
Lacaud G, Wiseman DH, et al: Enhancer recruitment of transcription
repressors RUNX1 and TLE3 by mis-expressed FOXC1 blocks
differentiation in acute myeloid leukemia. Cell Rep. 36:1097252021.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao L, Zhang P, Galbo PM, Zhou X, Aryal
S, Qiu S, Zhang H, Zhou Y, Li C, Zheng D, et al: Transcription
factor MEF2D is required for the maintenance of MLL-rearranged
acute myeloid leukemia. Blood Adv. 5:4727–4740. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baggiolini A, Callahan SJ, Montal E, Weiss
JM, Trieu T, Tagore MM, Tischfield SE, Walsh RM, Suresh S, Fan Y,
et al: Developmental chromatin programs determine oncogenic
competence in melanoma. Science. 373:eabc10482021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xie W, Jiang Q, Wu X, Wang L, Gao B, Sun
Z, Zhang X, Bu L, Lin Y, Huang Q, et al: IKBKE phosphorylates and
stabilizes Snail to promote breast cancer invasion and metastasis.
Cell Death Differ. 29:1528–1540. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang J, Mani SA, Donaher JL, Ramaswamy S,
Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and
Weinberg RA: Twist, a master regulator of morphogenesis, plays an
essential role in tumor metastasis. Cell. 117:927–939. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu M, Zhang Y, Yang J, Zhan H, Zhou Z,
Jiang Y, Shi X, Fan X, Zhang J, Luo W, et al: Zinc-Dependent
regulation of ZEB1 and YAP1 coactivation promotes
Epithelial-Mesenchymal transition plasticity and metastasis in
pancreatic cancer. Gastroenterology. 160:1771–1783.e1. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chan JM, Zaidi S, Love JR, Zhao JL, Setty
M, Wadosky KM, Gopalan A, Choo ZN, Persad S, Choi J, et al: Lineage
plasticity in prostate cancer depends on JAK/STAT inflammatory
signaling. Science. 377:1180–1191. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tang S, Xue Y, Qin Z, Fang Z, Sun Y, Yuan
C, Pan Y, Zhao Y, Tong X, Zhang J, et al: Counteracting
lineage-specific transcription factor network finely tunes lung
adeno-to-squamous transdifferentiation through remodeling tumor
immune microenvironment. Natl Sci Rev. 10:nwad0282023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shiode Y, Kodama T, Shigeno S, Murai K,
Tanaka S, Newberg JY, Kondo J, Kobayashi S, Yamada R, Hikita H, et
al: TNF receptor-related factor 3 inactivation promotes the
development of intrahepatic cholangiocarcinoma through
NF-κB-inducing kinase-mediated hepatocyte transdifferentiation.
Hepatology. 77:395–410. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Choi SI, Yoon C, Park MR, Lee D, Kook MC,
Lin JX, Kang JH, Ashktorab H, Smoot DT, Yoon SS, et al: CDX1
Expression induced by CagA-Expressing helicobacter pylori promotes
gastric tumorigenesis. Mol Cancer Res. 17:2169–2183. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guo L, Lee YT, Zhou Y and Huang Y:
Targeting epigenetic regulatory machinery to overcome cancer
therapy resistance. Semin Cancer Biol. 83:487–502. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Greenberg MVC and Bourc'his D: The diverse
roles of DNA methylation in mammalian development and disease. Nat
Rev Mol Cell Biol. 20:590–607. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nishiyama A and Nakanishi M: Navigating
the DNA methylation landscape of cancer. Trends Genet.
37:1012–1027. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liang W, Zhao Y, Huang W, Gao Y, Xu W, Tao
J, Yang M, Li L, Ping W, Shen H, et al: Non-invasive diagnosis of
early-stage lung cancer using high-throughput targeted DNA
methylation sequencing of circulating tumor DNA (ctDNA).
Theranostics. 9:2056–2070. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Maire CL, Fuh MM, Kaulich K, Fita KD,
Stevic I, Heiland DH, Welsh JA, Jones JC, Gorgens A, Ricklefs T, et
al: Genome-wide methylation profiling of glioblastoma cell-derived
extracellular vesicle DNA allows tumor classification. Neuro Oncol.
23:1087–1099. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xu Z, Sandler DP and Taylor JA: Blood DNA
Methylation and breast cancer: A prospective Case-Cohort analysis
in the sister study. J Natl Cancer Inst. 112:87–94. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wainwright EN and Scaffidi P: Epigenetics
and cancer stem cells: Unleashing, hijacking, and restricting
cellular plasticity. Trends Cancer. 3:372–386. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Davalos V, Lovell CD, Von Itter R,
Dolgalev I, Agrawal P, Baptiste G, Kahler DJ, Sokolova E, Moran S,
Pique L, et al: An epigenetic switch controls an alternative NR2F2
isoform that unleashes a metastatic program in melanoma. Nat
Commun. 14:18672023. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mancini M, Grasso M, Muccillo L, Babbio F,
Precazzini F, Castiglioni I, Zanetti V, Rizzo F, Pistore C, De
Marino MG, et al: DNMT3A epigenetically regulates key microRNAs
involved in epithelial-to-mesenchymal transition in prostate
cancer. Carcinogenesis. 42:1449–1460. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu S, Cheng K, Zhang H, Kong R, Wang S,
Mao C and Liu S: Methylation status of the nanog promoter
determines the switch between cancer cells and cancer stem cells.
Adv Sci (Weinh). 7:19030352020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Morinishi L, Kochanowski K, Levine RL, Wu
LF and Altschuler SJ: Loss of TET2 Affects proliferation and drug
sensitivity through altered dynamics of Cell-State transitions.
Cell Syst. 11:86–94.e5. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li S, Wei T and Panchenko AR: Histone
variant H2A.Z modulates nucleosome dynamics to promote DNA
accessibility. Nat Commun. 14:7692023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bouyahya A, Mechchate H, Oumeslakht L,
Zeouk I, Aboulaghras S, Balahbib A, Zengin G, Kamal MA, Gallo M,
Montesano D and El Omari N: The role of epigenetic modifications in
human cancers and the use of natural compounds as epidrugs:
Mechanistic pathways and pharmacodynamic actions. Biomolecules.
12:3672022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang Y, Zhang Q, Zhang Y and Han J: The
role of histone modification in DNA Replication-Coupled nucleosome
assembly and cancer. Int J Mol Sci. 24:49392023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhao S, Allis CD and Wang GG: The language
of chromatin modification in human cancers. Nat Rev Cancer.
21:413–430. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xu Y and Zhu Q: Histone modifications
represent a key epigenetic feature of Epithelial-to-Mesenchyme
transition in pancreatic cancer. Int J Mol Sci. 24:48202023.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
He Y, Ji Z, Gong Y, Fan L, Xu P, Chen X,
Miao J, Zhang K, Zhang W, Ma P, et al: Numb/Parkin-directed
mitochondrial fitness governs cancer cell fate via metabolic
regulation of histone lactylation. Cell Rep. 42:1120332023.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yuan S, Natesan R, Sanchez-Rivera FJ, Li
J, Bhanu NV, Yamazoe T, Lin JH, Merrell AJ, Sela Y, Thomas SK, et
al: Global regulation of the histone mark H3K36me2 underlies
epithelial plasticity and metastatic progression. Cancer Discov.
10:854–871. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Carrer A, Trefely S, Zhao S, Campbell SL,
Norgard RJ, Schultz KC, Sidoli S, Parris JLD, Affronti HC, Sivanand
S, et al: Acetyl-CoA metabolism supports multistep pancreatic
tumorigenesis. Cancer Discov. 9:416–435. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liau BB, Sievers C, Donohue LK, Gillespie
SM, Flavahan WA, Miller TE, Venteicher AS, Hebert CH, Carey CD,
Rodig SJ, et al: Adaptive chromatin remodeling drives glioblastoma
stem cell plasticity and drug tolerance. Cell Stem Cell.
20:233–246.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yao J, Chen J, Li LY and Wu M: Epigenetic
plasticity of enhancers in cancer. Transcription. 11:26–36. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li GH, Qu Q, Qi TT, Teng XQ, Zhu HH, Wang
JJ, Lu Q and Qu J: Super-enhancers: A new frontier for epigenetic
modifiers in cancer chemoresistance. J Exp Clin Cancer Res.
40:1742021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chen H and Liang H: A High-Resolution map
of human enhancer RNA loci characterizes super-enhancer activities
in cancer. Cancer Cell. 38:701–715.e5. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mirzadeh Azad F and Atlasi Y: Deregulation
of transcriptional enhancers in cancer. Cancers (Basel).
13:35322021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bi M, Zhang Z, Jiang YZ, Xue P, Wang H,
Lai Z, Fu X, De Angelis C, Gong Y, Gao Z, et al: Enhancer
reprogramming driven by high-order assemblies of transcription
factors promotes phenotypic plasticity and breast cancer endocrine
resistance. Nat Cell Biol. 22:701–715. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Han J, Meng J, Chen S, Wang X, Yin S,
Zhang Q, Liu H, Qin R, Li Z, Zhong W, et al: YY1 complex promotes
quaking expression via Super-Enhancer binding during EMT of
hepatocellular carcinoma. Cancer Res. 79:1451–1464. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Manning KS and Cooper TA: The roles of RNA
processing in translating genotype to phenotype. Nat Rev Mol Cell
Biol. 18:102–114. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Barbieri I and Kouzarides T: Role of RNA
modifications in cancer. Nat Rev Cancer. 20:303–322. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Shi Y: Mechanistic insights into precursor
messenger RNA splicing by the spliceosome. Nat Rev Mol Cell Biol.
18:655–670. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Bonnal SC, Lopez-Oreja I and Valcarcel J:
Roles and mechanisms of alternative splicing in cancer-implications
for care. Nat Rev Clin Oncol. 17:457–474. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kahles A, Lehmann KV, Toussaint NC, Huser
M, Stark SG, Sachsenberg T, Stegle O, Kohlbacher O, Sander C; ancer
Genome Atlas Research Network, ; Rätsch G: Comprehensive Analysis
of Alternative Splicing Across Tumors from 8,705 Patients. Cancer
Cell. 34:211–224.e6. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Bradley RK and Anczukow O: RNA splicing
dysregulation and the hallmarks of cancer. Nat Rev Cancer.
23:135–155. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Other-Gee Pohl S and Myant KB: Alternative
RNA splicing in tumour heterogeneity, plasticity and therapy. Dis
Model Mech. 15:dmm0492332022. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Labrecque MP, Brown LG, Coleman IM, Lakely
B, Brady NJ, Lee JK, Nguyen HM, Li D, Hanratty B, Haffner MC, et
al: RNA splicing factors SRRM3 and SRRM4 distinguish molecular
phenotypes of castration-resistant neuroendocrine prostate cancer.
Cancer Res. 81:4736–4750. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Li J, Choi PS, Chaffer CL, Labella K,
Hwang JH, Giacomelli AO, Kim JW, Ilic N, Doench JG, Ly SH, et al:
An alternative splicing switch in FLNB promotes the mesenchymal
cell state in human breast cancer. Elife. 7:e371842018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Xu T, Verhagen M, Joosten R, Sun W,
Sacchetti A, Munoz Sagredo L, Orian-Rousseau V and Fodde R:
Alternative splicing downstream of EMT enhances phenotypic
plasticity and malignant behavior in colon cancer. Elife.
11:e820062022. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Roundtree IA, Evans ME, Pan T and He C:
Dynamic RNA modifications in gene expression regulation. Cell.
169:1187–1200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Frye M, Harada BT, Behm M and He C: RNA
modifications modulate gene expression during development. Science.
361:1346–1349. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Penning A, Jeschke J and Fuks F: Why novel
mRNA modifications are so challenging and what we can do about it.
Nat Rev Mol Cell Biol. 23:385–386. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Xu Y, Song M, Hong Z, Chen W, Zhang Q,
Zhou J, Yang C, He Z, Yu J, Peng X, et al: The N6-methyladenosine
METTL3 regulates tumorigenesis and glycolysis by mediating
m6A methylation of the tumor suppressor LATS1 in breast
cancer. J Exp Clin Cancer Res. 42:102023. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Tao M, Li X, He L, Rong X, Wang H, Pan J,
Lu Z, Zhang X and Peng Y: Decreased RNA m6A methylation
enhances the process of the epithelial mesenchymal transition and
vasculogenic mimicry in glioblastoma. Am J Cancer Res. 12:893–906.
2022.PubMed/NCBI
|
|
78
|
Lin X, Chai G, Wu Y, Li J, Chen F, Liu J,
Luo G, Tauler J, Du J, Lin S, et al: RNA m6A methylation
regulates the epithelial mesenchymal transition of cancer cells and
translation of Snail. Nat Commun. 10:20652019. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Yue B, Song C, Yang L, Cui R, Cheng X,
Zhang Z and Zhao G: METTL3-mediated N6-methyladenosine modification
is critical for epithelial-mesenchymal transition and metastasis of
gastric cancer. Mol Cancer. 18:1422019. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Cheng C, Wu Y, Xiao T, Xue J, Sun J, Xia
H, Ma H, Lu L, Li J, Shi A, et al: METTL3-mediated m6A
modification of ZBTB4 mRNA is involved in the smoking-induced EMT
in cancer of the lung. Mol Ther Nucleic Acids. 23:487–500. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Xia P, Zhang H, Xu K, Jiang X, Gao M, Wang
G, Liu Y, Yao Y, Chen X, Ma W, et al: MYC-targeted WDR4 promotes
proliferation, metastasis, and sorafenib resistance by inducing
CCNB1 translation in hepatocellular carcinoma. Cell Death Dis.
12:6912021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Slack FJ and Chinnaiyan AM: The role of
non-coding RNAs in oncology. Cell. 179:1033–1055. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhang C and Peng G: Non-coding RNAs: An
emerging player in DNA damage response. Mutat Res Rev Mutat Res.
763:202–211. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yu W, Liang S and Zhang C: Aberrant miRNAs
regulate the biological hallmarks of glioblastoma. Neuromolecular
Med. 20:452–474. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhang C, Zhou Y, Gao Y, Zhu Z, Zeng X,
Liang W, Sun S, Chen X and Wang H: Radiated glioblastoma
cell-derived exosomal circ_0012381 induce M2 polarization of
microglia to promote the growth of glioblastoma by CCL2/CCR2 axis.
J Transl Med. 20:3882022. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Goodall GJ and Wickramasinghe VO: RNA in
cancer. Nat Rev Cancer. 21:22–36. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Li X, Li L and Wu J: The members of the
miR-148/152 family inhibit cancer stem cell-like properties in
gastric cancer via negative regulation of ITGA5. J Transl Med.
21:1052023. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Xu M, Zhang J, Lu X, Liu F, Shi S and Deng
X: MiR-199a-5p-Regulated SMARCA4 promotes oral squamous cell
carcinoma tumorigenesis. Int J Mol Sci. 24:47562023. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Ma Y, Zhu Y, Shang L, Qiu Y, Shen N, Wang
J, Adam T, Wei W, Song Q, Li J, et al: LncRNA XIST regulates breast
cancer stem cells by activating proinflammatory IL-6/STAT3
signaling. Oncogene. 42:1419–1437. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Fan C, Wang Q, Kuipers TB, Cats D, Iyengar
PV, Hagenaars SC, Mesker WE, Devilee P, Tollenaar RAEM, Mei H and
Ten Dijke P: LncRNA LITATS1 suppresses TGF-beta-induced EMT and
cancer cell plasticity by potentiating TbetaRI degradation. EMBO J.
42:e1128062023. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Xiong H, Liu B, Liu XY, Xia ZK, Lu M, Hu
CH and Liu P: circ_rac GTPase-activating protein 1 facilitates
stemness and metastasis of non-small cell lung cancer via
polypyrimidine tract-binding protein 1 recruitment to promote
sirtuin-3-mediated replication timing regulatory factor 1
deacetylation. Lab Invest. 103:1000102023. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Wang J, Zheng L, Hu C, Kong D, Zhou Z, Wu
B, Wu S, Fei F and Shen Y: CircZFR promotes pancreatic cancer
progression through a novel circRNA-miRNA-mRNA pathway and
stabilizing epithelial-mesenchymal transition protein. Cell Signal.
107:1106612023. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Mirzaei S, Gholami MH, Hushmandi K,
Hashemi F, Zabolian A, Canadas I, Zarrabi A, Nabavi N, Aref AR,
Crea F, et al: The long and short non-coding RNAs modulating EZH2
signaling in cancer. J Hematol Oncol. 15:182022. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
McCabe EM and Rasmussen TP: lncRNA
involvement in cancer stem cell function and epithelial-mesenchymal
transitions. Semin Cancer Biol. 75:38–48. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Pan G, Liu Y, Shang L, Zhou F and Yang S:
EMT-associated microRNAs and their roles in cancer stemness and
drug resistance. Cancer Commun (Lond). 41:199–217. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Kristensen LS, Jakobsen T, Hager H and
Kjems J: The emerging roles of circRNAs in cancer and oncology. Nat
Rev Clin Oncol. 19:188–206. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Gerstberger S, Jiang Q and Ganesh K:
Metastasis. Cell. 186:1564–1579. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Pastushenko I and Blanpain C: EMT
Transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Zhang N, Ng AS, Cai S, Li Q, Yang L and
Kerr D: Novel therapeutic strategies: Targeting
epithelial-mesenchymal transition in colorectal cancer. Lancet
Oncol. 22:e358–e368. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Liu Y, Qi X, Donnelly L,
Elghobashi-Meinhardt N, Long T, Zhou RW, Sun Y, Wang B and Li X:
Mechanisms and inhibition of Porcupine-mediated Wnt acylation.
Nature. 607:816–822. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zarzosa P, Garcia-Gilabert L, Hladun R,
Guillen G, Gallo-Oller G, Pons G, Sansa-Girona J, Segura MF,
Sanchez de Toledo J, Moreno L, et al: Targeting the hedgehog
pathway in rhabdomyosarcoma. Cancers (Basel). 15:7272023.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Bondarev AD, Attwood MM, Jonsson J,
Chubarev VN, Tarasov VV and Schioth HB: Recent developments of HDAC
inhibitors: Emerging indications and novel molecules. Br J Clin
Pharmacol. 87:4577–4597. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Tian C, Liu Y, Xue L, Zhang D, Zhang X, Su
J, Chen J, Li X, Wang L and Jiao S: Sorafenib inhibits ovarian
cancer cell proliferation and mobility and induces radiosensitivity
by targeting the tumor cell epithelial-mesenchymal transition. Open
Life Sci. 17:616–625. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Chan CY, Hong SC, Chang CM, Chen YH, Liao
PC and Huang CY: Oral squamous cell carcinoma cells with acquired
resistance to erlotinib are sensitive to Anti-Cancer effect of
quercetin via pyruvate kinase M2 (PKM2). Cells. 12:1792023.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Ali S, Rehman MU, Yatoo AM, Arafah A, Khan
A, Rashid S, Majid S, Ali A and Ali MN: TGF-β signaling pathway:
Therapeutic targeting and potential for anti-cancer immunity. Eur J
Pharmacol. 947:1756782023. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Dhanyamraju PK, Schell TD, Amin S and
Robertson GP: Drug-Tolerant persister cells in cancer therapy
resistance. Cancer Res. 82:2503–2514. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
De Conti G, Dias MH and Bernards R:
Fighting drug resistance through the targeting of Drug-Tolerant
persister cells. Cancers (Basel). 13:11182021. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Kim HD, Yoo C, Ryu MH and Kang YK: A
randomised phase 2 study of continuous or intermittent dosing
schedule of imatinib re-challenge in patients with tyrosine kinase
inhibitor-refractory gastrointestinal stromal tumours. Br J Cancer.
129:275–282. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
East MP and Johnson GL: Adaptive chromatin
remodeling and transcriptional changes of the functional kinome in
tumor cells in response to targeted kinase inhibition. J Biol Chem.
298:1015252022. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Deng S, Wang C, Wang Y, Xu Y, Li X,
Johnson NA, Mukherji A, Lo UG, Xu L, Gonzalez J, et al: Ectopic
JAK-STAT activation enables the transition to a stem-like and
multilineage state conferring AR-targeted therapy resistance. Nat
Cancer. 3:1071–1087. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Matsushima K, Yang D and Oppenheim JJ:
Interleukin-8: An evolving chemokine. Cytokine. 153:1558282022.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Qin Q, Li X, Liang X, Zeng L, Wang J, Sun
L and Zhong D: Targeting the EMT transcription factor Snail
overcomes resistance to osimertinib in EGFR-mutant non-small cell
lung cancer. Thorac Cancer. 12:1708–1715. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Shi ZD, Pang K, Wu ZX, Dong Y, Hao L, Qin
JX, Wang W, Chen ZS and Han CH: Tumor cell plasticity in targeted
therapy-induced resistance: mechanisms and new strategies. Signal
Transduct Target Ther. 8:1132023. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Garcia-Mayea Y, Mir C, Masson F, Paciucci
R and ME LL: Insights into new mechanisms and models of cancer stem
cell multidrug resistance. Semin Cancer Biol. 60:166–180. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Wang J, Yu H, Dong W, Zhang C, Hu M, Ma W,
Jiang X, Li H, Yang P and Xiang D: N6-methyladenosine-mediated
up-regulation of FZD10 regulates liver cancer stem cells'
properties and lenvatinib resistance through WNT/β-Catenin and
hippo signaling pathways. Gastroenterology. 164:990–1005. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Hao L, Chen H, Wang L, Zhou H, Zhang Z,
Han J, Hou J, Zhu Y, Zhang H and Wang Q: Transformation or tumor
heterogeneity: Mutations in EGFR, SOX2, TP53, and RB1 persist in
the histological rapid conversion from lung adenocarcinoma to
small-cell lung cancer. Thorac Cancer. 14:1036–1041. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Quintanal-Villalonga A, Chan JM, Yu HA,
Pe'er D, Sawyers CL, Sen T and Rudin CM: Lineage plasticity in
cancer: A shared pathway of therapeutic resistance. Nat Rev Clin
Oncol. 17:360–371. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Marcoux N, Gettinger SN, O'Kane G, Arbour
KC, Neal JW, Husain H, Evans TL, Brahmer JR, Muzikansky A, Bonomi
PD, et al: EGFR-Mutant adenocarcinomas that transform to small-cell
lung cancer and other neuroendocrine carcinomas: Clinical outcomes.
J Clin Oncol. 37:278–285. 2019. View Article : Google Scholar : PubMed/NCBI
|