Introduction
Lung cancer (LC) is the most frequently diagnosed
cancer, is the most common cause of cancer-related death worldwide
and has a low five-year overall survival rate (1,2). In
total, ~85% of patients with LC have non-small cell LC (NSCLC), of
which ~50% are lung adenocarcinoma (LUAD) (3,4). Owing
to the lack of effective early diagnosis methods, patients with
LUAD are often diagnosed late and thus miss the optimal time for
disease intervention, leading to high morbidity and mortality rates
(5). Therefore, the discovery of
new premonitory biomarkers and remedial goals for this disease is
urgently needed.
Integrins, a type of transmembrane connector, form a
bidirectional connection between the extracellular matrix and
intracellular actin skeleton. Integrins can promote cells to give
feedback to the external milieu (6), including during proliferation,
differentiation and migration (7).
According to previous studies, there are 24 different heterodimers
of integrins (8,9). Integrin subunit β4 (ITGB4), a
laminin-5 receptor, is a widely-studied integrin, and its effects
on tumor progression have attracted attention (10). ITGB4 was reported to participate in
tumor cachexia in glioma following in vitro and animal
experiments (11). Moreover, it was
demonstrated that ITGB4 expression was upregulated in
hepatocellular carcinoma (HCC) tumor samples, compared with
adjacent non-tumor tissues. Following suppression of ITGB4
expression, the proliferation, colony-forming ability and
invasiveness of HCC cells were reduced (12). In NSCLC, the ITGA6/B4 heterodimer
interacts directly with the receptor tyrosine kinase, MET, to
promote tumor invasion (13,14).
Previously, a systematic bioinformatic analysis of NSCLC was
performed using a series of databases and it was found that ITGB4
was aberrantly expressed in NSCLC, suggesting its potential
significance in this disease (15).
However, the expression pattern of ITGB4 and how it exerts its role
remains unclear.
In the present study, ITGB4 was identified as an
important hub gene in the initiation and development of LUAD using
online data and patient tissue samples. The roles of ITGB4 in A549
and PC9 cells (two LUAD cell lines) were explored and it was found
that downregulation of ITGB4 attenuated LUAD cell proliferation,
promoted cell apoptosis and inhibited colony formation, migration
and invasion. ITGB4 mechanisms were also preliminarily explored
using high throughput sequencing. The findings of the present study
highlighted the oncogenic function of ITGB4 in LUAD and uncovered
the fundamental scheme underlying progression of this disease.
Materials and methods
Patient enrolment
In the present study, 18 LUAD and adjacent normal
tissues were obtained surgically from April 2023 to May 2023, which
included 10 men and 8 women, with an age range of 58 to 77 years.
Inclusion criteria for patients were as follows: i) Individuals
diagnosed with LUAD, excluding other forms of lung cancer; ii)
patients confirmed through pathological examinations; and iii)
individuals willing to actively participate. Exclusion criteria for
patients were as follows: i) Individuals with additional health
conditions, including chronic diseases; and ii) patients unable to
cooperate effectively with researchers. Written consent from all
participants involved in the study was acquired. All experiments
involving human subjects were carried out in The First Affiliated
Hospital of Anhui Medical University. Protocols involving the
obtained tissues were approved (approval no. Quick-PJ 2023-04-36,
Hefei, China) by The Medical Ethical Committee of The First
Affiliated Hospital of Anhui Medical University.
Microarray experiments and data
processing
To screen for critical genes that may be involved in
LUAD progression, three paired human LUAD tissues were collected
for microarray experiments. ITGB4-knocked down A549 cells were also
used for microarray experiments to explore downstream signaling
pathways. All microarrays were performed using Affymetrix U133 Plus
2.0 arrays (Novogene Co., Ltd.). The kits used were RNA-Quick
Purification Kit (cat. no. RN001; EZBioscience) and
NovoScript®Plus All-in-one 1st Strand cDNA Synthesis
SuperMix (gDNA Purge) (cat. no. E047; Novoprotein).
Screening for differentially expressed
genes (DEGs)
Data were normalized (16) and DEGs between NSCLC and adjacent
normal tissues were analyzed using the limma package (http://www.bioconductor.org/). The following criteria
were used to determine significant DEGs: Fold change ≥2 or ≤0.5 and
P<0.05.
Bioinformatics analysis
The ITGB4 expression pattern in LUAD in The Cancer
Genome Atlas (TCGA) dataset, TCGA-LUAD, was obtained through the
GEPIA2 online tool (http://gepia2.cancer-pku.cn/#index, accession number:
LUAD-TCGA). Pan-cancer analysis was also conducted using the GEPIA2
online tool. In the prognostic analysis, the prognostic data were
downloaded from cBioPortal for Cancer Genomics website [datasets:
Lung Adenocarcinoma (TCGA, Firehose Legacy) and Lung Adenocarcinoma
(MSK, 2021)]. The ITGB4 median expression level was used as the
cut-off, with patients with expression levels above the median
assigned to the high expression group and patients with expression
levels below the median assigned to the low expression group. The
prognosis between the different groups was then evaluated using the
Kaplan-Meier method and the log-rank test. P<0.05 was considered
to indicate a statistically significant difference. Online
immunohistochemistry (IHC) data were obtained directly from the
Human Protein Atlas (HPA) database (www.proteinatlas.org/). UALCAN was used to obtain the
presentation data and survival information of the TCGA-LUAD dataset
(17). Furthermore, ITGB4
expression was analyzed using the ONCOMINE database (18,19).
Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/eg/) and Reactome pathway
analyses, involving commentary, visualization and integrated
discovery, were also conducted (https://reactome.org/). The analysis of
protein-protein interactions (PPI) and modules was conducted using
the STRING (Search Tool for the Retrieval of Interacting Genes)
database. The current approach involved uploading the list of DEGs
to the STRING website (www.string-db.org/) to assess protein interactions,
where interactions with an experimentally validated score exceeding
0.4 were considered significant. For module and hub gene
identification, Cytoscape (version 3.9.0; http://cytoscape.org/) software with molecular complex
detection (MCODE) criteria (score >3 and nodes >4) was
employed.
Cell culture
The human LUAD cell lines, A549 and PC9, were
purchased from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. These cells were respectively cultured
in Ham's F-12K medium and Dulbecco's Modified Eagle Medium with 10%
fetal bovine serum (FBS; all from Wuhan Servicebio Technology Co.,
Ltd.) and 1% penicillin and streptomycin, in a humidified
atmosphere at 37°C with 5% CO2.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was obtained using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA
synthesis was then conducted using a PrimeScript™ RT reagent kit
(Takara Bio, Inc.) according to the manufacturer's instructions.
The thermocycling conditions for qPCR were as follows: Initial
denaturation (95°C for 60 sec), then the 40 cycling steps (95°C for
20 sec, annealing for 20 sec at 60°C, and extension at 72°C for 30
sec). TB Green® Fast qPCR Mix (Takara Bio, Inc.) was
then used to quantify the ITGB4 and GAPDH (internal control)
expression levels. The primers used in the qPCR were as follows:
GAPDH forward, 5′-AGCCACATCGCTCAGACAC-3′ and reverse,
5′-GCCCAATACGACCAAATCC-3′; and ITGB4 forward,
5′-GCAGATCTCCGGTGTACACAAG-3′ and reverse, 5′-GCTTTTTCCCGGCATTGG-3′.
mRNA expression was quantified using the 2−ΔΔCq method
(20).
IHC
IHC was conducted in accordance with standard
laboratory protocols. In short, the paraffin-embedded tissue
sections (5 µm) were deparaffinized using xylene and hydrated in an
ethanol gradient. The sections were then incubated in 3% BSA (cat.
no. A9647; Sigma-Aldrich; Merck KGaA) blocking solution, with
gentle shaking at 37°C for 30 min. Then, the sections were
incubated with rabbit monoclonal antibody against ITGB4 (1:300;
cat. no. 14803; Cell Signaling Technology, Inc.) for 1 h, followed
by the HRP-conjugated secondary antibody (1:10,000; cat. no.
ab205718; Abcam) working solution at 37°C for 30 min. After further
dehydration, the slices were sealed with neutral gum. Microscopic
examination was performed, and images were acquired. ITGB4 staining
was then scored by two independent observers (including one
pathologist) to determine the expression levels. A positive
reaction was scored using four graded categories, depending on the
intensity of the staining and the percentage of positively stained
cells. The sum of the intensity and percentage scores determined
the final score.
RNA interference (RNAi) and
transfection
RNAi in A549 and PC9 cell lines was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) as the transfection reagent. The sequences of the
small interfering (si)RNAs (Sangon Biotech Co., Ltd.) used were as
follows: siITGB4#1 sense, 5′-CCACAGAGCUGGUGCCCUATT-3′ and
antisense, 5′-UAGGGCACCAGCUCUGUGGTT-3′; siITGB4#2 sense,
5′-CAGAGAAGCAGGUGGAACATT-3′ and antisense,
5′-UGUUCCACCUGCUUCUCUGTT-3′; and si-NC sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′. The concentration of siRNA used was 20
µM. The duration of siRNA transfection was 48 h at 37°C and
subsequent experiments were performed immediately after
transfection. Western blot analysis and RT-qPCR were conducted to
verify successful transfection of siRNA.
Cell proliferation, colony formation
and apoptosis assays
Cell Counting Kit-8 (CCK-8) was used to detect cell
proliferation. In short, cells were carefully placed at a density
of 2×104 cells per well (0.1 ml) in 96-well plates and
allowed to incubate overnight at 37°C. The number of adherent
ITGB4-knocked down LUAD cells was calculated at 0, 24, 48 and 72 h
using 10 µl of CCK-8 (cat. no. C0037; Beyotime Institute of
Biotechnology) and incubated for 1 h at 37°C. The number of cells
was then determined using a micro titer plate reader at 450 nm
wavelength. To assess the colony formation ability, cells were
diluted to 500 cells/well in a 6-well plate. After 10 days, the
resulting colonies were stained using a 0.1% crystal violet
staining solution (Sangon Biotech Co., Ltd.). The minimum number of
cells forming a colony was 50 cells and ImageJ software (version:
1.42q; National Institutes of Health) was used to quantify
colonies. An Annexin V-FITC Apoptosis Detection kit (cat. no.
CA1020; Beijing Solarbio Science & Technology Co., Ltd.)
combined with flow cytometry (model, FC500; Beckman Coulter, Inc.;
analysis software, FlowJo 10; FlowJo LLC) and Hoechst staining
(cat. no. C0003; Beyotime Institute of Biotechnology; cells were
stained for 5 min in room temperature) were performed to detect
cell apoptosis.
Migration and invasion assays
The invasion and migration abilities of the cells
were measured using Transwell inserts (8 µm) with or without
Matrigel, respectively. The plates were precoated with Matrigel at
4°C. After coating, they were incubated at 37°C for 3 h. A total of
5×104 cells/well were seeded into the upper chambers of
the inserts and 500 µl culture medium containing 20% FBS was added
to the lower chambers. After 48 h of incubation at 37°C, the
non-invasive cells were carefully wiped away using a cotton-tipped
gauze, whereas the invasive cells were stained using 0.1% crystal
violet staining solution (Sangon Biotech Co., Ltd.) for 2 h at room
temperature. Images of five randomly selected fields were captured
by an inverted light microscope (Olympus Corporation) and the
number of migratory or invasive cells was calculated.
Wound healing assays
Cells at a density of 1×105 cells/well
were transfected with the relevant siRNA. Once the cells adhered to
the plate, a wound was made by scratching the cells with a
micropipette tip. All cells were serum-starved (no FBS) during the
wound healing assay. Images of the wound were then recorded using a
light microscope at 0 or 24 h after wounding.
In vivo tumor growth assays
A total of 14 female NOD/SCID mice (6 weeks-old;
weight 28–22 g) were purchased from the Animal Center of Shanghai.
Mice were kept in an SPF animal room with a constant temperature of
25°C, a relative humidity of 40–70%, a 12/12-h light/dark cycle and
free access to food and water. Mice were subcutaneously injected on
the back with cells (106 cells in 100 µl PBS) to produce
xenograft tumors in Hefei Normal University. Tumor growth was
monitored every 3 days before the tumor could be detected. After
that, the tumor growth was monitored every day when the tumor could
be detected. In the followed protocol, if the tumor weight in mice
reached 10% of the body weight, or the size of the tumor in any
dimension exceeded 15 mm, all the mice would be euthanized. If not,
the mice would be sacrificed 54 days after cell injection. After 54
days, all the 14 mice were euthanized using CO2 (35%
vol/min) asphyxiation in chamber (630×480×500 mm). The mice were
exposed to CO2 for at least 1 additional min after
breathing ceased. The methods for confirming animal death included:
Cessation of heartbeat, cessation of breathing, stiffness in the
animal and dilated pupils. The tumor volumes were calculated as
previously reported (21). The
protocol was approved (approval no. HFNU-2023-TK61-1,) by The
Medical Ethical Committee of Hefei Normal University (Hefei,
China), and followed the principles outlined in the Declaration of
Helsinki (2013) for all human or animal experimental
investigations. All animal welfare considerations were taken to
minimize suffering and distress. The tumor weight in mice should
not exceed 10% of the body weight, and the size of the tumor in any
dimension should not exceed 15 mm.
Immunoblotting
Cells were lysed using RIPA protein extraction
buffer (cat. no. P0013B) with PSMF (cat. no. ST505; Beyotime
Institute of Biotechnology) and 1X SDS loading buffer. Protein
concentration was determined using bicinchoninic acid (BCA) method.
A total of 20 µg protein was loaded per lane. The proteins were
transferred to polyvinylidene difluoride (PVDF) membrane. Blocking
was conducted using QuickBlock™ blocking buffer (cat. no. P0252;
Beyotime Institute of Biotechnology) for 15 min at room
temperature. TBST with 1% Tween 20 was used for washing. The
membrane was incubated at 4°C overnight with the following primary
antibodies: anti-ITGB4 antibody (1:1,000; cat. no. 14803) and
anti-GAPDH (1:1,000; cat. no. 2118; both from Cell Signaling
Technology, Inc.). Subsequently, membranes were incubated at room
temperature for 2 h with HRP-conjugated anti-rabbit IgG secondary
antibody (1:5,000; cat. no. 7074; Cell Signaling Technology, Inc.).
Protein bands were visualized using a ChemiDoc XRS
chemiluminescence detection and imaging system (Bio-Rad
Laboratories, Inc.).
Statistical analysis
Data were analyzed using SPSS v20 software (IBM
Corp.). The results were presented as the mean ± standard
deviation. Student's paired t-test was used for RT-qPCR, CCK-8,
migration and invasion analyses. Survival curves were plotted from
the date of operation using the Kaplan-Meier method and were
compared using the log-rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
ITGB4 is a critical hub gene in
LUAD
In microarray experiments using the collected human
LUAD tissues, 167 DEGs were identified (Table SI), among which 164 DEGs were
upregulated and three were downregulated (Fig. 1A and B, Table SII). Furthermore, the 167 DEGs were
analyzed using UALCAN and it was found that 14 genes were
associated with poor survival (P<0.01; Table I). Then, the interactions between
the DEGs were studied and the top 20 identified genes were ranked
by interaction level through protein-protein interaction network
analysis using STRING website and Cytoscape software (Figs. 1C and S1). In addition, 1,028 genes upregulated
in LUAD tissues were identified using data from TCGA dataset
(Fig. 1C). After integrated
bioinformatical analysis, only three genes, ITGB4, B3GNT3 and
CDKN2A, were identified from the aforementioned lists (Fig. 1D). ITGB4 was selected for further
study due to its importance in LUAD pathophysiology with rare
mechanistical study (15).
 | Table I.List of the survival-related genes in
non-small cell lung cancer. |
Table I.
List of the survival-related genes in
non-small cell lung cancer.
| Gene symbol | Log2 FC | P-value | Adjusted
P-value | Overall survival in
The Cancer Genome Atlas |
|---|
| RHOV | 4.93 |
8.99×10−9 |
7.86×10−6 | 0.0001 |
| TNS4 | 5.15 |
1.02×10−7 |
5.71×10−5 | 0.0001 |
| GJB3 | 5.72 |
8.70×10−7 |
3.72×10−4 | 0.0001 |
| FAM83A | 6.27 |
2.06×10−4 |
2.74×10−2 | 0.0001 |
| CCDC34 | 1.54 |
2.81×10−4 |
3.54×10−2 | 0.0010 |
| CBLC | 3.62 |
2.02×10−5 |
5.01×10−3 | 0.0012 |
| HNF4G | 4.48 |
2.40×10−5 |
5.68×10−3 | 0.0014 |
| STYK1 | 2.87 |
3.91×10−4 |
4.55×10−2 | 0.0019 |
| ITGB4 | 2.15 |
1.87×10−4 |
2.56×10−2 | 0.0022 |
| CDKN2A | 2.52 |
1.31×10−5 |
3.45×10−3 | 0.0023 |
| FAM111B | 2.89 |
4.10×10−4 |
4.68×10−2 | 0.0037 |
| NGEF | 5.33 |
5.61×10−8 |
3.76×10−5 | 0.0040 |
| P2RY6 | 3.87 |
2.54×10−7 |
1.23×10−4 | 0.0053 |
| B3GNT3 | 5.15 |
1.34×10−9 |
1.68×10−6 | 0.0066 |
ITGB4 is upregulated in tumor tissues
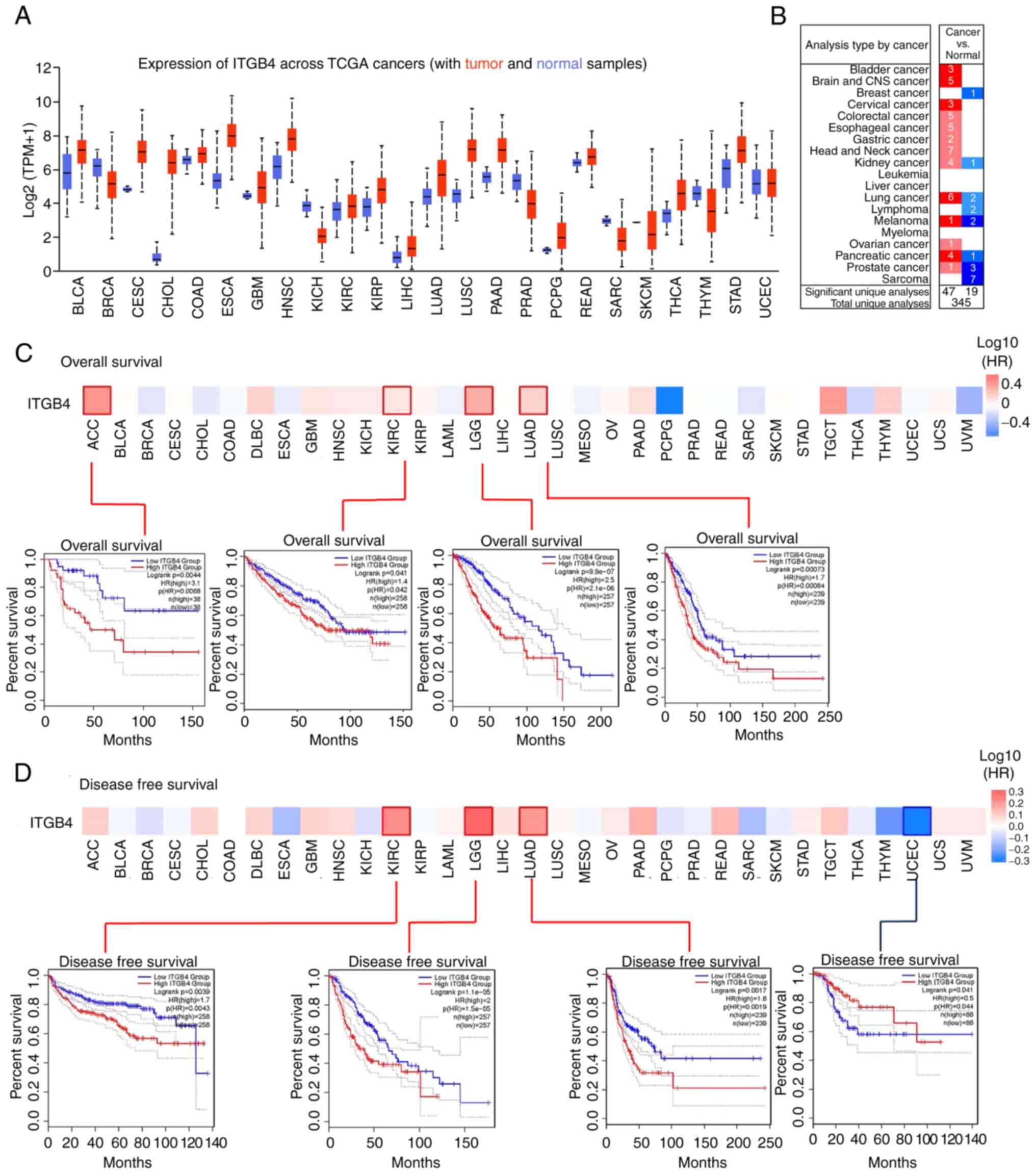
and is associated with poor survival in a pan-cancer analysis
To further understand the role of ITGB4 in cancer
development, a pan-cancer analysis was first conducted. The
difference in ITGB4 expression between tumor and adjacent normal
tissues in different tumor types was explored using TCGA database.
It was found that ITGB4 was markedly upregulated in multiple types
of cancer tissues, including LUAD (Fig.
2A). The same result was also obtained following analysis using
the ONCOMINE database (Fig. 2B). In
the prognostic analysis, the cases were divided into high and low
expression groups according to the ITGB4 expression level. The
results indicated that a high ITGB4 expression level was associated
with poor overall survival for adrenocortical carcinoma, kidney
renal clear cell carcinoma (KIRC), low grade glioma (LGG) and LUAD
(Fig. 2C). It was also identified
that high ITGB4 expression was associated with poor disease-free
survival for KIRC, LGG and LUAD (Fig.
2D). These data indicated the possible oncogenic role of
ITGB4.
ITGB4 is upregulated in LUAD tissues
and is associated with poor survival
Next, the role of IGTB4 in LUAD was focused on. The
copy number and mRNA expression level of ITGB4 in LUAD was analyzed
and it was revealed that gain and amplification significantly
promoted ITGB4 expression (Fig.
3A). Data from TCGA (Fig. 3B)
and HPA (Fig. 3C) online datasets
confirmed the upregulated ITGB4 mRNA and protein expression levels
in LUAD. In prognostic analyses, it was found that elevated ITGB4
expression predicted an adverse clinical outcome in LUAD (Fig. 3D), which also suggested the
oncogenic role of ITGB4 in LUAD. Moreover, these results were
validated using the collected LUAD tissues. The results of the
RT-qPCR analysis suggested that ITGB4 expression was significantly
higher in LUAD tissues (Fig. 3E),
which was also confirmed by IHC (Fig.
3F).
Knockdown of ITGB4 expression
suppresses proliferation and migration and promotes apoptosis of
LUAD cells
To further confirm the oncogenic role and improve
understanding of its biological function, the expression of ITGB4
was knocked down in A549 and PC9 cells using specific siRNAs. The
transfection results demonstrated that the two siRNAs (siITGB4#1
and #2) significantly decreased the ITGB4 expression level
(Figs. 4A and S2A). Downregulated ITGB4 expression also
significantly restrained A549 and PC9 cell proliferation (Fig. 4B) and colony formation (Fig. 4C). Beyond the in vitro
experiments, an in vivo tumorigenesis nude mouse model was
also constructed. It was found that the tumor volume of the
ITGB4-knocked down cells group was significantly smaller than that
of the control group (Fig. 4D).
Moreover, apoptosis assays demonstrated that ITGB4 knockdown also
induced cell apoptosis (Figs. 4E
and S2B). Wound healing and
Transwell assays indicated that ITGB4 downregulation markedly
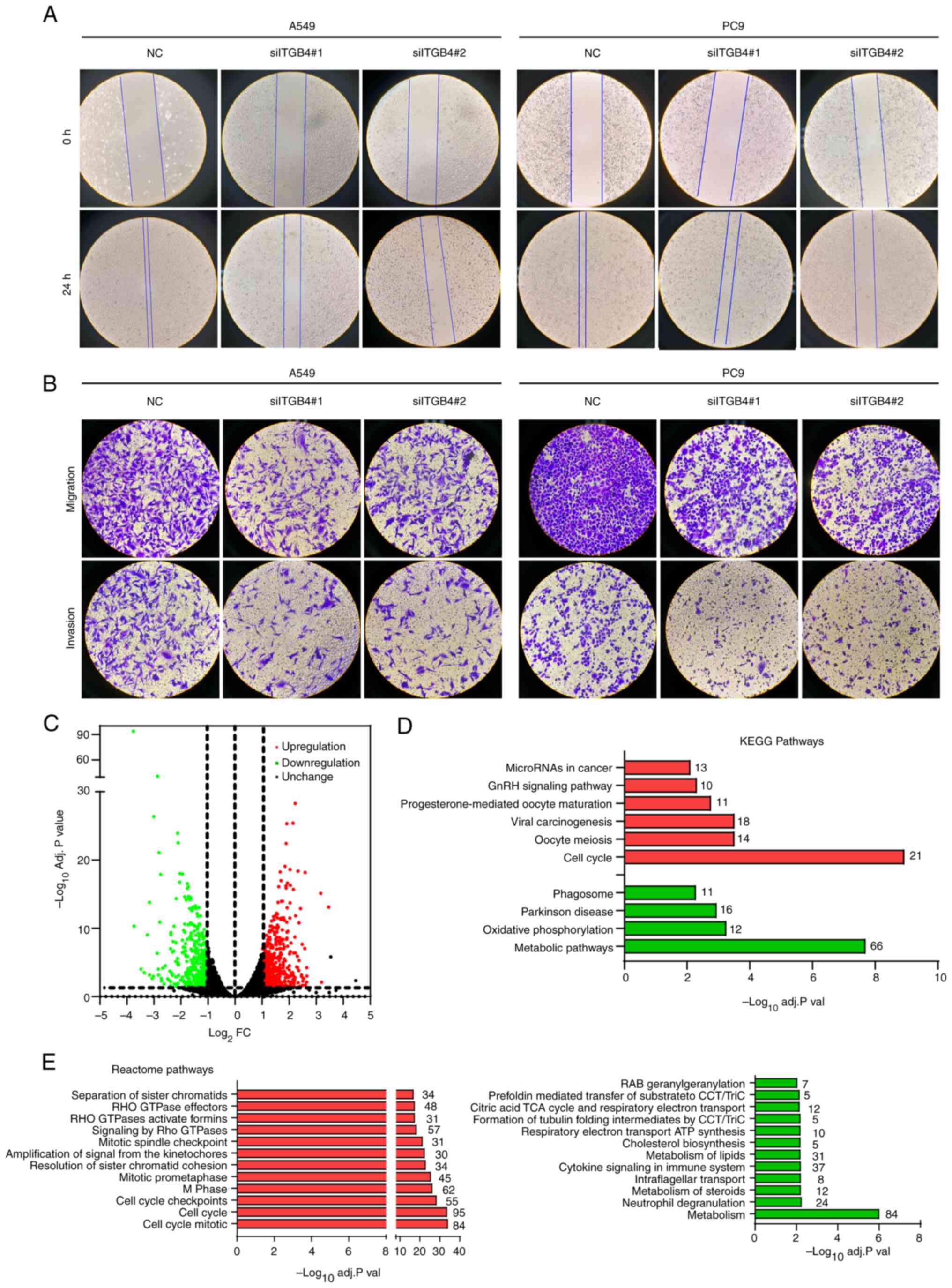
inhibited LUAD cell migration and invasion (Fig. 5A and B).
ITGB4-regulated pathway analysis
Next, the underlying mechanisms were explored. High
throughput sequencing and DEG analysis was performed using
ITGB4-knocked down A549 cells (Table
SIII). A total of 917 DEGs were identified in ITGB4-knocked
down A549 cells, including 522 upregulated and 395 downregulated
genes (Fig. 5C). Pathway enrichment
analysis was subsequently conducted using these DEGs. In the KEGG
analysis of upregulated DEGs, cell cycle, oocyte meiosis and viral
carcinogenesis pathways were identified. Meanwhile, metabolic
pathways, oxidative phosphorylation and Parkinson's disease were
identified for the downregulated DEGs (Fig. 5D). In the Reactome analysis
demonstrated in Fig. 5E, the
enriched pathways for upregulated DEGs consisted of cell cycle
mitotic, cell cycle and cell cycle checkpoint pathways. The
enriched pathways for downregulated DEGs consisted of metabolism,
neutrophil degranulation and metabolism of steroids pathways.
Notably, the cell cycle and metabolism pathways were all identified
in the KEGG and Reactome pathway enrichment analyses, indicating
the potential mechanism underlying the oncogenic influence of ITGB4
on LUAD.
Discussion
LUAD remains one of the most frequently diagnosed
types of cancer worldwide. ITGB4 has been shown to have critical
roles in numerous types of cancer (22,23),
with functions in migration, epithelial-mesenchymal transition,
infringement and diversion (12,19,24).
Moreover, ITGB4 is also a possible prognostic marker in breast
cancer (25). Previously, a
systematic bioinformatics analysis of the correlation between ITGB4
and NSCLC was reported (15).
However, the biological function of ITGB4 in LUAD remains poorly
understood.
In the present study, ITGB4 was identified as a
survival-related gene with aberrant expression in LUAD by combining
RNA sequencing and TCGA-LUAD data. Furthermore, the oncogenic roles
of ITGB4 in LUAD were also confirmed, including roles in promoting
cell proliferation, colony formation, migration and invasion, and
inhibiting cell apoptosis. The roles of ITGB4 in LUAD are similar
to those in other cancer types, such as colorectal (26), pancreatic (27) and prostate (28) cancer. A recent study demonstrated
that ITGB4-targeted cancer immunotherapies could inhibit tumor
progression, and two approaches for immunological targeting of
ITGB4 were explored in breast and head and neck cancer models
(29). Moreover, immunological
targeting of ITGB4 also enhanced the efficacy of anti-programmed
death ligand 1 checkpoint blockade in these models. Since, as
demonstrated in the present study, the ITGB4 expression level is
significantly associated with disease progression and the outcome
of LUAD, ITGB4 has the potential to be used as a predictive gene
and therapeutic target for LUAD disease prognosis in the future.
Due to the high morbidity and mortality rates of LUAD, new disease
prognosis prediction methods and therapeutic targets are of great
significance for improved treatment of this disease.
In the present study, KEGG and Reactome analyses
demonstrated that the ITGB4-regulated genes were greatly enriched
in metabolism and cell cycle-related pathways. It is a
well-accepted theory that cancer is a metabolic disease. The
uncontrolled, unlimited and accelerated proliferation of cancer
cells requires large amounts of energy, which forces cells to
develop ways to derive more energy from metabolism (30). The bridge from metabolism to the
cell cycle is typically autophagy and oxidative phosphorylation,
which were identified pathways in the present study. The mechanism
of metabolic rewiring has been revealed to be related to
ITGB4-involved autophagy (31).
Next, the direct downstream genes or signaling pathways of ITGB4
should be identified to uncover the mechanisms related to ITGB4
promoted LUAD progression. It should also be confirmed whether
ITGB4 influences cell metabolism and the cell cycle through such
genes or pathways.
The present study does have certain limitations. The
present study is only a preliminary exploration using retrospective
data and in vitro and in vivo assays. The results did
not reveal the definitive mechanisms for ITGB4-promoted LUAD
progression. Further clinical and basic research are required for
further exploration.
In summary, the biological function of ITGB4 in LUAD
was reported in the present study, indicating a vital role of ITGB4
in LUAD progression. Combining the results of a previous study
(32) with the results of the
present study, ITGB4 could be a novel therapeutic target for this
highly malignant cancer.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL conceived and supervised the study. YP designed
the experiments. SM performed the experiments. YL, NK and YP
analyzed the data. XL and SM wrote the manuscript. XL and NK
confirm the authenticity of all the raw data. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The study on human tissues was approved (approval
no. Quick-PJ 2023-04-36) by The Ethics Committee of The First
Affiliated Hospital of Anhui Medical University (Hefei, China), and
written informed consent was acquired from all patients before
sample collection. Animal experiments were approved (approval no.
HFNU-2023-TK61-1) by The Medical Ethical Committee of Hefei Normal
University (Hefei, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Martin P and Leighl NB: Review of the use
of pretest probability for molecular testing in non-small cell lung
cancer and overview of new mutations that may affect clinical
practice. Ther Adv Med Oncol. 9:405–414. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dong HX, Wang R, Jin XY, Zeng J and Pan J:
LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via
inhibiting hsa-mir-22-3p. J Cell Physiol. 233:4126–4136. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jan YH, Lai TC, Yang CJ, Huang MS and
Hsiao M: A co-expressed gene status of adenylate kinase 1/4 reveals
prognostic gene signature associated with prognosis and sensitivity
to EGFR targeted therapy in lung adenocarcinoma. Sci Rep.
9:123292019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bianconi D, Unseld M and Prager GW:
Integrins in the Spotlight of Cancer. Int J Mol Sci. 17:20372016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Slack-Davis JK and Parsons JT: Emerging
views of integrin signaling: Implications for prostate cancer. J
Cell Biochem. 91:41–46. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brakebusch C, Bouvard D, Stanchi F, Saki T
and Fassler R: Integrins in invasive growth. J Clin Invest.
109:999–1006. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hynes RO: Integrins-Versatility,
modulation, and signaling in cell adhesion. Cell. 69:11–25. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang L, Dong Z, Zhang Y and Miao J: The
roles of integrin β4 in vascular endothelial cells. J Cell Physiol.
227:474–478. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma B, Zhang L, Zou YJ, He R, Wu Q, Han C
and Zhang B: Reciprocal regulation of integrin β4 and KLF4 promotes
gliomagenesis through maintaining cancer stem cell traits. J Exp
Clin Cancer Res. 38:232019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li XL, Liu L, Li DD, He YP, Guo LH, Sun
LP, Liu LN, Xu HX and Zhang XP: Integrin β 4 promotes cell invasion
and epithelial-mesenchymal transition through the modulation of
Slug expression in hepatocellular carcinoma. Sci Rep. 7:404642017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Trusolino L, Bertotti A and Comoglio PM: A
signaling adapter function for alpha6beta4 integrin in the control
of HGF-dependent invasive growth. Cell. 107:643–654. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hoshino A, Costa-Silva B, Shen TL,
Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di
Giannatale A, Ceder S, et al: Tumour exosome integrins determine
organotropic metastasis. Nature. 527:329–335. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu P, Wang Y, Wu Y, Jia Z, Song Y and
Liang N: Expression and prognostic analyses of ITGA11, ITGB4 and
ITGB8 in human non-small cell lung cancer. Peerj. 7:e82992019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smyth GK, Michaud J and Scott HS: Use of
within-array replicate spots for assessing differential expression
in microarray experiments. Bioinformatics. 21:2067–2075. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rhodes DR, Kalyana-Sundaram S, Mahavisno
V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ,
Kincead-Beal C, Kulkarni P, et al: Oncomine 3.0: Genes, pathways,
and networks in a collection of 18,000 cancer gene expression
profiles. Neoplasia. 9:166–180. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang C, Liu J, Xiong B, Yonemura Y and
Yang X: Expression and prognosis analyses of forkhead box A (FOXA)
family in human lung cancer. Gene. 685:202–210. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qin Z, Xiang C, Zhong F, Liu Y, Dong Q, Li
K, Shi W, Ding C, Qin L and He F: Transketolase (TKT) activity and
nuclear localization promote hepatocellular carcinoma in a
metabolic and a non-metabolic manner. J Exp Clin Cancer Res.
38:1542019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tagliabue E, Ghirelli C, Squicciarini P,
Aiello P, Colnaghi MI and Menard S: Prognostic value of alpha 6
beta 4 integrin expression in breast carcinomas is affected by
laminin production from tumor cells. Clin Cancer Res. 4:407–410.
1998.PubMed/NCBI
|
|
23
|
Grossman HB, Lee C, Bromberg J and Liebert
M: Expression of the alpha6beta4 integrin provides prognostic
information in bladder cancer. Oncol Rep. 7:13–16. 2000.PubMed/NCBI
|
|
24
|
Gan L, Meng J, Xu M, Liu M, Qi Y, Tan C,
Wang Y, Zhang P, Weng W, Sheng W, et al: Extracellular matrix
protein 1 promotes cell metastasis and glucose metabolism by
inducing integrin β4/FAK/SOX2/HIF-1α signaling pathway in gastric
cancer. Oncogene. 37:744–755. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Diaz LK, Cristofanilli M, Zhou X, Welch
KL, Smith TL, Yang Y, Sneige N, Sahin AA and Gilcrease MZ: beta4
integrin subunit gene expression correlates with tumor size and
nuclear grade in early breast cancer. Mod Pathol. 18:1165–1175.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang X, Wang J, Wang M, Xuan M, Han S, Li
C, Li M, Sun XF, Yu W and Zhao Z: ITGB4 as a novel serum diagnosis
biomarker and potential therapeutic target for colorectal cancer.
Cancer Med. 10:6823–6834. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Meng X, Liu P, Wu Y, Liu X, Huang Y, Yu B,
Han J, Jin H and Tan X: Integrin beta 4 (ITGB4) and its
tyrosine-1510 phosphorylation promote pancreatic tumorigenesis and
regulate the MEK1-ERK1/2 signaling pathway. Bosn J Basic Med Sci.
20:106–116. 2020.PubMed/NCBI
|
|
28
|
Wilkinson EJ, Woodworth AM, Parker M,
Phillips JL, Malley RC, Dickinson JL and Holloway AF: Epigenetic
regulation of the ITGB4 gene in prostate cancer. Exp Cell Res.
392:1120552020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ruan S, Lin M, Zhu Y, Lum L, Thakur A, Jin
R, Shao W, Zhang Y, Hu Y, Huang S, et al: Integrin β4-Targeted
cancer immunotherapies inhibit tumor growth and decrease
metastasis. Cancer Res. 80:771–783. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sena LA and Chandel NS: Physiological
roles of mitochondrial reactive oxygen species. Mol Cell.
48:158–167. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kubli DA and Gustafsson AB: Mitochondria
and mitophagy: The yin and yang of cell death control. Circ Res.
111:1208–1221. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhuang Z, Zhou R, Xu X, Tian T, Liu Y, Liu
Y, Lian P, Wang J and Xu K: Clinical significance of integrin αvβ6
expression effects on gastric carcinoma invasiveness and
progression via cancer-associated fibroblasts. Med Oncol.
30:5802013. View Article : Google Scholar : PubMed/NCBI
|