Introduction
PCa is the leading non-cutaneous malignant tumors in
males in China (1). PCa is the most
common type of cancer affecting males (20%) and accounts for 6.8%
of male cancer-related deaths worldwide. According to statistics,
in 2020, the number of new cases of PCa reached 1.41 million and
caused 375,000 deaths. The number of new cases and deaths from PCa
is expected to nearly double by the year 2040 (2). In 2019, the number of cases of PCa in
China reached 150,000 and there were 54,000 related deaths
(3). In recent years, technological
progress and the development of new methods have largely led to an
increased understanding of the underlying molecular mechanisms that
promote tumor growth and progression (4). The advent of current drug and therapy
combinations has enriched the treatment options for PCa, providing
more treatment options for patients with PCa (5). However, there are certain challenges
for clinical practitioners. The early stages of PCa often present
with subtle or asymptomatic manifestations, which may be easily
overlooked. The emergence of PARP inhibitors has brought the
diagnosis and treatment of PCa into an era of precision, and has
individualized treatment possible based on the results of genetic
testing, which is the future trend of treatment development
(6). The combination of an AKT
(PI3K signaling gene) inhibitor and abiraterone has been shown to
improve radiological progression-free survival in patients with
metastatic castration-resistant PCa (mCRPC) and tumor suppressor
gene PTEN deletion (7).
Cyclin-dependent kinases 4 and 6 (CDK4/6) are activated during the
G1-S checkpoint of the cell, which promotes cancer cell
proliferation. A phase III study [CYCLONE-03 (NCT05288166)], which
is a phase III, randomized, double-blind, placebo-controlled study,
using either abemaciclib (a selective CDK4/6 inhibitor) or a
placebo, plus abiraterone and prednisone is investigating the
treatment of high-risk metastatic hormone-sensitive PCa (8).
Currently, the main treatment strategies for
patients with PCa are surgery, endocrine therapy, radiotherapy,
chemotherapy, cryotherapy and immunotherapy. Some patients are
often diagnosed with advanced-stage or metastatic PCa due to a lack
of clear early clinical signs and awareness of PCa screening. Even
if PCa is treated promptly at an early stage, it continues to
progress and subsequently evolves into CRPC (9).
N6-methyladenosine (m6A)
methylation, a modification of RNA molecules, was first discovered
in 1974 (10). It is mainly
distributed around stop codons, 3′UTRs, and long internal exons,
and occurs mainly in RRACH sequences (where R=A or G, H=A, C, or U)
(11). M6A is the most
abundant modification of higher eukaryotic messenger RNAs(mRNAs),
including methyltransferases, demethylases, and
m6A-binding proteins, which play essential roles in the
various biological functions of RNA (12). In addition to mRNAs, m6A
methylation modifications are present on different non-coding RNAs.
Modification by methylation of m6A is a dynamic
reversible modification similar to DNA and histones. It is involved
in the whole life cycle of RNA translation, processing, and
degradation (13). The association
between m6A methylation modification and tumorigenesis
and development has gradually received attention in recent years;
however, m6A methylation modification and PCa have not
been extensively studied.
Recent research has demonstrated that m6A
methylation modification, which plays a crucial role in the tumor
development process, and related genes influenced by it, may be key
targets for cancer diagnosis and treatment (14). It has been suggested that
m6A and its associated regulators can become novel
prognostic indicators and novel therapeutic targets in clinical
practice (15). The present review
focuses on the role of m6A regulators in the progression
of PCa and discusses new directions for future research on
m6A therapy.
PCa
PCa stems from the epithelial tissue of the prostate
and is a disease that mainly affects middle-aged and older males
(16). The main feature of PCa is
the abnormal division of prostate cells, leading toabnormal
prostate cell hyperplasia (17).
PCa is the most common non-cutaneous cancer with an estimated
1,600,000 cases and 366,000 related deaths annually (18). According to a study published in
2022, PCa has become the third most common and fifth most lethal
type of cancer among the diagnosed cases of cancer in the USA
(19). In China, the incidence of
PCa in the population has been increasing each year due to
improvements in living conditions in recent years (19).
According to the fifth edition of the World Health
Organization classification of urological and male reproductive
system tumors, PCa can be divided into the following: Prostate
ductal adenocarcinoma, neuroendocrine PCa and adenoid cystic
carcinoma (20). Ductal
adenocarcinoma does not occur alone and is frequently intermingled
with acinar adenocarcinoma. Both are typically driven by
aberrations in speckle type BTB/POZ protein, forkhead box (FOX)A1
and other molecules, and have similar androgen receptor (AR)
expression levels (20). The
studies by Lotan et al (21), Gillard et al (22) and Schweizer et al (23) have demonstrated that ERG fusion
rearrangements and molecular abnormalities are more common in
ductal adenocarcinoma than in ductal carcinoma, as well as
mutations in the WNT signaling pathway genes, catenin beta-1 and
adenomatous polyposis coli. PIN-like carcinomas, characterized by
large discrete glands lined with flattened or tufted epithelium,
and a high frequency of activation mutations in the RAF/RAS
pathway, were reclassified as acinar subtypes. It has been found
that transdifferentiation in PIN-like carcinomas is associated with
the deletion of TP53, RB1 and PTEN, and epigenetic alterations in a
specific genomic environment. Neuroendocrine adenocarcinoma has the
histological and immunohistochemical features of simple small-cell
or, less commonly, large-cell neuroendocrine carcinoma, which has a
mixture of tumors with high-grade components. p53
immunohistochemical staining is often positive in neuroendocrine
carcinoma, while prostate-specific antigen (PSA) and prostate acid
phosphatase are usually lost. Adenoid cystic (basal cell) carcinoma
histologically presents as an adenoid cystic pattern with hyaline
globules (inspissated secretion); a basal pattern comprising small
solid nests of basal cells; or a mixture of both. Fluorescence
in situ hybridization reveals the fusion of the MYB-NFIB
gene in more adenoid cystic carcinoma (20). The study by Epstein et al
(24) demonstrated that intraductal
carcinoma of the prostate was caused by carcinogenesis of the
prostate gland and ductal epithelium and/or intraductal spread of
aggressive PCa. Microcystic adenocarcinoma is a benign variant of
acinar PCa, which is easily confused with benign atrophied
glandular cystic changes. Pleomorphic giant cell adenocarcinoma is
a rare type of PCa, which is a giant cell with pleomorphic and
relatively homogeneous nuclei. In addition, a rare new variant of
neuroendocrine tumor, termed NE PCa, is not dissimilar to that of
large cell neuroendocrine carcinoma of other organs in a way of
morphology (25). A new
classification method for PCa is emerging that relies mainly on
molecular markers of different PCa subtypes for fine
classification, which helps to personalize the description of PCa
rather than relying solely on morphological information (26,27).
Currently, there is no uniform standard for
prognostic markers for PCa. Novel biomarkers improve risk
stratification for PCa diagnosis and treatment (28). A previous study found that
RNA-binding protein was one of the meaningful biomarkers of PCa.
The high expression of small nuclear ribonucleoprotein polypeptide
A' in RNA-binding proteins in PCa tissue was positively associated
with the Gleason score and pathological TNM stage, which is
critical for determining the prognosis of patients with PCa
(29). The expression patterns of
endosomal genes are also interesting in a new indicator for
predicting PCa prognosis. The expression of adaptor protein,
phosphotyrosine interacting with PH domain and leucine zipper 1
(APPL1), Ras-related protein Rab-5A (RAB5A), early endosome antigen
1 (EEA1), programmed cell death 6-interacting protein (PDCD6IP),
nicotinamide adenine dinucleotide oxidase 4 (NOX4) and sortilin 1
(SORT1) in malignant prostate tissue differs from that in benign or
normal prostate tissue (30). Serum
total testosterone and 25-hydroxyvitamin D have been reported to
predict the prognosis of patients with PCa (31,32).
However, the study by Holt et al (31) did not find sufficient evidence that
prostate cancer prognosis is affected by serum vitamin D levels
measured following diagnosis. Izumi et al (32) demonstrated that both low and high
serum TT levels indicated a poor prognosis of patients with
PCa.
At present, the clinical indicators for the
diagnosis of PCa are a PSA value ≥4 ng/ml and a positive digital
rectal test (33). A previous study
found that in patients undergoing a radical prostatectomy, a higher
body mass index (BMI) was associated with a higher prostate weight
and PSA, as well as higher pT staging and pathological Gleason
score (34). A higher percentage of
fatty tissue around the prostate has been shown to be significantly
associated with a higher Gleason score (35). BMI constitutes another risk factor
in addition to PSA. Moreover, in individuals with abdominal
obesity, the larger the waist circumference, the greater the linear
association between the risk of developing PCa and BMI (36). However, some scholars argue that the
role of BMI is unclear and that there is a lack of valid evidence
to support BMI as a risk factor for males with PCa (37). However, PCa involves a variety of
risk factors and complex mechanisms that require further
research.
M6A
Recently, the epigenetics of m6A
modification has attracted increasing attention from scholars.
M6A modification directly affects the expression levels
of genes that regulate a variety of physiological and pathological
processes in the body, and ultimately affect the occurrence and
development of tumors (38). It has
been found that m6A RNA methylation modification causes
cancer cells to metabolize and reorganize by altering molecules and
pathways associated with tumor metabolism, meeting the growth needs
of cancer cells, and maintaining the balance of the surrounding
tissue environment. The m6A modification not only
participates in all phases of the RNA cycle but also modulates
non-coding RNA (39). Similar to
DNA or protein methylation, m6A methylation modification
is dynamically and reversible regulated by different types of
regulators, among which the molecules that play a biological role,
mainly include methyltransferases (writers), demethylases (erasers)
and m6A-binding proteins (readers) (40,41).
Among these, writers are mainly methyltransferase complexes
composed of methyltransferase-like (METTL)3, METTL14 and their
cofactor, Wilm's tumor 1-associated protein (WTAP), accompanied by
other necessary proteins, which play the role of methylation
modification. The demethylation function of erasers works mainly
through fat mass and obesity-associated protein (FTO) and
alpha-ketoglutarate-dependent dioxygenase homolog (ALKBH)5. In
addition, some unknown members of the ALKB subfamily may contain
m6A demethylase function (42). M6A functions mainly by
recruiting m6A-binding proteins, of which the readers
are effector proteins, the most well-known of which are the YTH
N6-methyladenosine RNA binding protein (YTHDF) family
and the insulin-like growth factor 2 mRNA binding protein (IGF2BP)
family without the Yth domain (43,44).
It has been shown that the various regulatory factors involved in
the modification of m6A are closely related to the
occurrence and development of cancer, and play an essential role in
this process (38) (Fig. 1).
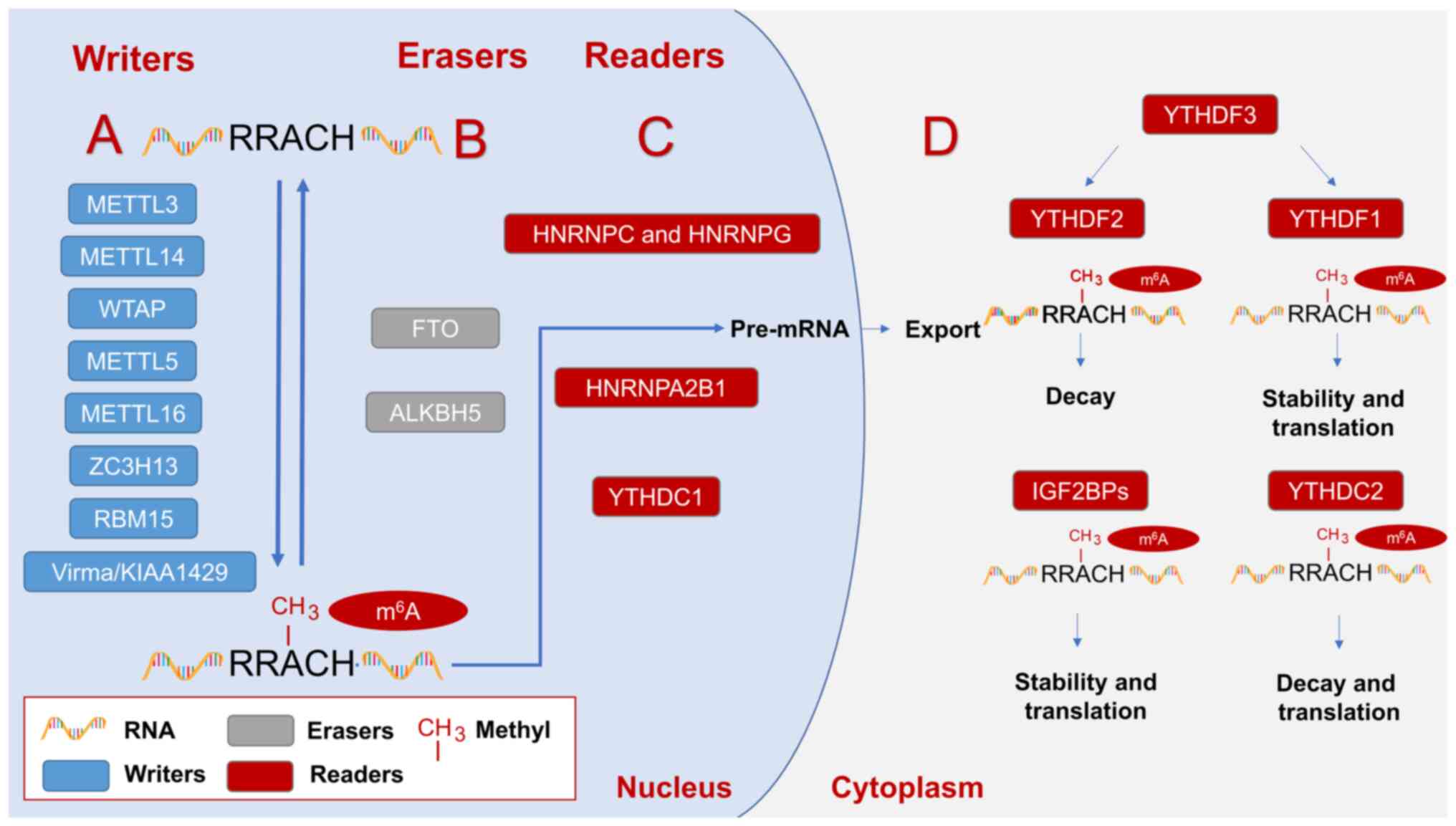 | Figure 1.Regulators involved in m6A
modification and play an influential role in regulating the
occurrence and development of prostate cancer. (A)
Methyltransferases mainly include METTL3, METTL14, WTAP, METTL5,
METTL15, ZC3H13, RBM15, Virma/KIAA1429, etc., which play the role
of RNA m6A methylation modification. (B) Demethylase
mainly includes FTO and ALKBH5, which plays the role of removing
m6A methylation modification. (C) The m6A
binding proteins in the nucleus mainly include YTHDC1, HNRNPA2B1,
HNRNPC and HNRNPG, which can recognize the methylation modification
of m6A. (D) The cytoplasmic m6A binding
proteins mainly include YTHDF1-3, IGF2BPs and YTHDC2, which can
recognize the methylation modification of m6A. Writers,
erasers, and readers work together to form the reversible
modification of m6A. Readers can be divided into readers
(YTHDC1, HNRNPA2B1, HNRNPC, and HNRNPG) in the nucleus and readers
(YTHDF1-3, IGF2BPs, YTHDC2) in the cytoplasm according to the
different intracellular localization. Readers within the nucleus
are mainly involved in the splicing of precursor mRNA and the
delivery of RNA from the nucleus into the cytoplasm. The role of
readers in the cytoplasm is not the same. IGF2BPs play a role in
improving the stability of m6A methylation modification
mRNA and promoting m6A methylation modification mRNA
translation. YTHDC2 plays a role in promoting the decay and
translation of m6A methylation-modified mRNA. YTHDF3 can
play a role in promoting the decay and translation processes of
m6A methylation-modified mRNA, respectively, by
affecting YTHDF1 and YTHDF2. METTL, methyltransferase-like; WTAP,
Wilm's tumor 1-associated protein; ZC3H13, Zinc finger CCCH
domain-containing protein 13; RBM15, RNA binding motif protein 15;
Virma, Vir like m6A methyltransferase associated;
m6A, N6-methyladenosine; FTO, fat mass and
obesity-associated protein; ALKBH, alpha-ketoglutarate-dependent
dioxygenase homolog; YTHD, YTH N6-methyladenosine RNA
binding protein; IGF2BP, insulin-like growth factor 2 mRNA binding
protein; HNRNP, heterogeneous nuclear ribonucleoprotein
protein. |
M6A writers
M6A refers to the methylation
modification of the sixth nitrogen atom of adenine (45). As an epigenetic marker that acts
primarily on RNA, m6A methylation modification relies on
the m6A methyl-conversion enzyme. It has been
demonstrated that m6A methyltransferases consist of a
catalytic subunit m6A-METTL complex and a regulatory
subunit m6A-METTL associated complex, also known as a
‘writer’ (46). The m6A
methyltransferase complex consists of two protein substances,
METTL3 and METTL14, and contains WTAP, KIAA1429 (Virilizer), Hakai,
RNA binding motif protein 15 (RBM15), METTL16 and additional
co-regulatory subunits (45,47).
The METTL3-METTL14 complex exhibits more potent in vitro
methyltransferase activity than any single protein. Thus, METTL3
and METTL14 are the core components of the m6A writer
complex. Although METTL14 lacks the catalytic activity of
methyltransferase and is a pseudo methyltransferase in the complex,
it plays an integral role in maintaining the activity of the writer
complex. METTL14 plays an essential role in maintaining the
structural integrity of the binary complex of METTL3-METTL14
complex to improve the catalytic activity of the m6A
writer complex. Recombinant METTL3 monomers exhibit weak
methyltransferase catalytic activity. METTL3 exhibits a significant
increase in methyltransferase catalytic activity when METTL3 and
METTL14 with methyltransferase domains form heterodimeric complexes
(48). WTAP has a unique
localization role in recruiting METTL3 and METTL14 into the nuclear
spot. In addition, WTAP participates in the m6A
methylation modification process as part of the m6A
methyltransferase complex, along with METTL3, METTL14, and other
methyltransferases (49). RBM15, as
a member of a family of adapter proteins that contain RNA binding
motifs, primarily recruits m6A methylated RNA into
U-rich regions (50). Vir like
m6A methyltransferase associated (Virma), also known as
KIAA1429, recruits the WTAP-METTL3-METTL14 complex by binding to
WTAP. Alternatively, Virma may interact with plant cleavage factors
linked to m6A methylation and polyadenylation
mechanisms, participating in mRNA processing (51). METTL5 is an 18srRNA m6A
methyltransferase that acquires metabolic stability by forming
parallel β zippers between the backbone atoms and heterodimers with
the tRNA methyltransferase homolog 112. METTL16 can directly
methylate mRNA containing the UAC m6A GAGAA motif
(52). Zinc refers to the CCCH
domain-containing protein 13 (ZC3H13) and E3 ubiquitin-protein
ligase Hakai, which interacts with the methyltransferase complex to
affect the RNA methylation process (48) (Table
I).
 | Table I.Functions of m6A
‘writers’. |
Table I.
Functions of m6A
‘writers’.
| Regulator | Effect on
m6A modification | Role | (Refs.) |
|---|
| METTL3 | Methyltransferase
activity | Activator | (45,47,48) |
| METTL14 | Maintain the
structural integrity of binary complexes | Activator | (48) |
| WTAP | METTL3 and METTL14
are recruited into nuclear spots and involved in m6A
methylation | Activator | (49) |
| Virma/KIAA1429 | Recruitment of
WTAP-METTL3-METTL14 complex and participate in mRNA processing | Activator | (51) |
| RBM15 | m6A
methyl bodies are recruited into U-rich regions | Activator | (50) |
| ZC3H13 | Interacts with
methyltransferase complex components and affects methylation
pathways | Activator | (48) |
| METTL5 | 18S rRNA
m6A methyltransferase | Activator | (52) |
| METTL16 | Methylation of
specific sequences of mRNA | Activator | (52) |
M6A erasers
Thus far, only two types of m6A
demethylase have been identified: FTO and ALKBH5. FTO, also known
as ALKBH9, belongs to the non-heme KGFe(II)/α-KG-dependent family
of dioxygenase ALKB (ABH1-9) and is the m6A demethylase
of the first eukaryotic mRNA enzyme. The role of FTO in
adipogenesis and tumorigenesis is related to its m6A
demethylase activity. It can also interact with melanocortin
receptor 4 (MC4R) through m6A modification to control
the proliferation, migration, and the invasion of PCa cells
(53).
FTO has been reported to play a key role as a
demethylase in a various types of cancer. FTO is responsible for
the dynamic modification of m6A and mediates
m6A and the N6,2-O-dimethyladenosine (m6Am)
demethylation of poly(A) RNA. The demethylation of m6A
is first mediated when FTO is in the nucleus, and demethylation of
m6A and m6Am is first mediated when FTO is in
the cytoplasm. FTO demethylase promotes abnormal m6A
modification in PCa. This suggests that FTO has a tumor-suppressing
effect in PCa (54). In addition,
FTO depletion first significantly increases the m6A
levels of chloride intracellular channel protein 4 (CLIC4) mRNA,
and subsequently inhibits PCa proliferation and transfer by
reducing mRNA stability and promoting CLIC4 mRNA degradation
(55).
ALKBH5, by removing m6A methylation,
leads to the erasure of m6A methylation modifications,
returning m6A to its previously unregulated state. Of
note, seven m6A-associated crosstalk genes, including
ALKBH5, are differently expressed in PCa and periodontitis. These
genes have significantly increased expression levels in several
signaling pathways, including nuclear plasticity transport,
ubiquitin-mediated protein breakdown, p53 signal transduction,
cellular senescence, and transcriptional regulation disorders
(56). The expression of the ALKBH5
gene with abnormal copy number changes is strongly associated with
the prognosis of PCa (57). This
suggests that the pattern of ALKBH5 copy number variation is
significantly associated with relapse-free survival in PCa
(56). Furthermore, it has been
found that FTO and ALKBH5 are negatively associated with the
Gleason grade and are less well expressed in PCa (58). The abnormal expression of FTO or
ALKBH5 primarily affects m6A levels, which then, through
complex biological mechanisms, affect certain biological processes
of tumorigenesis and development (Table II).
 | Table II.Functions of m6A
‘erasers’. |
Table II.
Functions of m6A
‘erasers’.
| Regulator | Effect on
m6A modification | Role | (Refs.) |
|---|
| FTO | Remove
m6A modifications and promote RNA decay | Inhibitor | (53–55) |
| ALKBH5 | Remove
m6A modifications | Inhibitor | (56–58) |
M6A readers
M6A methylation modifications perform
their corresponding biological functions with the involvement of
the writer and eraser and with the recognition of m6A
modifications by m6A recognition proteins. IYT521-B
homologous (YTH) family proteins were the first m6A
reader proteins to be discovered. It was found that m6A
readers are primarily involved in the occurrence and development of
cancer by regulating the metabolism of targeted RNA, including RNA
splicing, output, translation and degradation, which result in
changes in the biological function of RNA. At present, a total of
five proteins containing the YTH domain in the human genome have
been found and divided into three types of m6A reader
proteins: YTH m6A-binding protein 1–3 (YTHDF1-3), YTH
domain 1 (YTHDC1) and YTH domain 2 (YTHDC2) (59).
The degradation of mRNA and tje translation of YTH
family members, with different reader proteins, play their
respective roles through different pathways. YTHDF1 may recruit
argonaute 2 protein and miRNA via the YTH domain and interact to
form P-body (mRNA degradation centers in yeast cells and animal
cells, also known as cytoplasmic bodies, Dcp bodies, or GW bodies)
to degrade mRNA. Moreover, YTHDF1 facilitates the translation of
m6A-modified mRNA (10,60).
In addition, YTHDF1 recognizes m6A-modified target genes
through multiple mechanisms. Thus, YTHDF1 improves the stability of
RNA and thus promotes expression (61). YTHDF2 relies primarily on
m6A modifications to regulate signaling pathways in
cancer cells. YTHDF2 promotes the degradation of targeted mRNA
transcripts by recruiting the CCR4-NOT deadenylase complex. YTHDF2
can also promote tumor cell proliferation by binding to tumor
suppressors, triggering a downstream cascade that can do the
opposite by interacting with oncogenes (62). YTHDF2 also affects various aspects
of RNA metabolism, including mRNA degradation and ribosome pre-RNA
processing (10). The YTHDF family
protein DF1-3 is the dominant cytosolic m6A-binding
protein and is considered to mediate the action of m6A
in cells. All three DF proteins contribute to the destabilization
of mRNA and together, mediate the degradation of mRNAs containing
m6A (63). YTHDF3
functions synergistically with YTHDF1 to promote protein synthesis
and mediates the decay of methylated mRNA by affecting YTHDF2.
YTHDF1-3 cooperates and plays a crucial role in facilitating the
metabolism of m6A-modified mRNA in the cytoplasm
(64). YTHDC1 is also the only
m6A reader in the YTH protein family that is localized
in the nucleus. YTHDC1 is a protein that interacts with splicing
factors that regulate RNA splicing. Its m6A-dependent
functions include selective polyadenylation and the nuclear
production of m6A-modified mRNAs, which control the
maturation of intranuclear mRNA. Recent research has demonstrated
that there is a close association between chromatin-associated
RNAs, non-coding RNAs, and regulatory RNAs, which can control the
expression of genes within cells (65). It has been shown that YTHDC1 plays a
crucial role in cellular functions, such as cancer cell
proliferation. YTHDC1 may also have the potential to promote the
efficacy of tumor immunotherapy (66). YTHDC2 plays its biological role in
using its distinct RNA binding domain to bind to targeted
m6A RNA and bridge between ribosomes, which reduces the
abundance of related mRNAs and increases the translation efficiency
of related mRNAs (67,68).
The reader protein includes not only the YTH
structural protein family, but also the heterogeneous nuclear
ribonucleoprotein protein (HNRNP) family and IGF2BP1, IGF2BP2, and
IGF2BP3. The HNRNP family and IGF2BP1-3 together recognize
m6A-modified fragments in RNA (69). HNRNPC is regulated as an
m6A switch, which affects the abundance and selective
splicing of target RNAs by altering their binding activity. HNRNPC
can also facilitate the conversion of fresh heteronuclear RNAs into
mature mRNAs, and can stabilize the structure of mRNAs and control
their translation process (70).
The RGG motif of HNRNPG directly binds to the phosphorylated
carboxyl terminal domain of RNA polymerase II (RNAPII). The
interaction between the phosphorylated carboxyl terminal domain and
the new RNA leads to the co-transcription of HNRNPG and RNAPII.
Finally, selective splicing of new RNA (71). HNRNPA2B1 specifically recognizes and
directly binds with elevated affinity to RNAs that share the
m6A-modified RNAs containing the m6A
co-sequence RGm6ACH. M6A can boost the
binding capability of HNRNPA2B1 to certain sites, which enhances
its nuclear event capability. In addition, HNRNPA2B1 recruits the
microprocessor complexes, Drosha and DGCR8, to facilitate primary
miRNA processing by binding m6A to primary miRNA
transcripts (72). IGF2BPs first
identify m6A-modified mRNA and improve the stability of
the mRNA target which facilitates storage, translation, and gene
expression output. IGF2BPs may exert oncogenic effects on cancer
cells by enhancing the stability of methylated mRNAs of oncogenic
targets (73,74) (Table
III).
 | Table III.Functions of m6A
‘readers’. |
Table III.
Functions of m6A
‘readers’.
| Regulator | Effect on
m6A modification | (Refs.) |
|---|
| YTH domain
family |
|
|
|
YTHDF1 | Promotes mRNA
degradation and translation | (10,60,61,63) |
|
YTHDF2 | Degrades mRNA and
affects RNA metabolism | (10,62,63) |
|
YTHDF3 | Mediated methylated
mRNA decay | (63,64) |
|
YTHDC1 | Regulates splicing
factors for RNA splicing and controls intranuclear mRNA
maturation | (65,66) |
|
YTHDC2 | Facilitate mRNA
translation | (67,68) |
| HNRNP family |
|
|
| HNRNPC
and HNRNPG | Promote the
maturation of nuclear RNAs and stabilize the structure of mRNAs and
control their translation process | (69,70,71) |
|
HNRNPA2B1 | Recruitment of
microprocessor complexes to facilitate primary microRNA
processing | (72) |
|
IGF2BP1-3 | Promotes stability,
storage, and translation of mRNA targets and causes cancer | (73,74) |
M6A induces specific drug
resistance
Recent research has demonstrated that
chemotherapeutic resistance in cancer is associated with
m6A RNA methylation, which leads to the abnormal
expression of various targets and pathways. For example, the
resistance of lung adenocarcinoma to the clinically used drugs
nicotine and oseltamivir is due to an increase in MET-TL7B content
in the cancer tissue, which enables m6A expression and
reactive oxygen species (ROS) clearance dependence (75). The development of resistance to
gefitinib in lung adenocarcinoma has also been found to be
associated with the ribonucleic acid cleavage of
m6A-modified circASK1 produced by YTHDF2 (76). Resistance to the chemotherapeutic
drug, cisplatin, in esophageal squamous epithelial carcinoma has
also been found to be associated with m6A. The stability
of the CASC8 transcription process is enhanced by the
m6A demethylation induced by ALKBH5, which induces drug
resistance in esophageal squamous epithelial carcinoma (77). In addition, resistance to cisplatin
in intrahepatic cholangiocarcinoma promotes the degradation of
CDKN1B mRNA via YTHDF2 in an m6A-dependent manner
(78). Tamoxifen is a conventional
chemotherapy drug for breast cancer (79). Breast cancer develops tamoxifen
resistance due to an increased ROS production and p38 activation.
One of the reasons for this mechanism is that AK4mRNA translation
is enhanced by METTL3-mediated m6A overexpression
(80). HNRNPA2B1 activates the
ser/thr kinase growth factor signaling pathway in an
m6A-dependent manner to abnormally regulate downstream
targets, leading to tamoxifen resistance in cancer tissue.
Resistance to temozolomide in glioblastoma multiforme arises from
the transcription of histone-associated genes modified by
METTL3-mediated m6A (81). Strong resistance to tyrosine kinase
inhibitors in clear cell renal cell carcinoma is regulated by the
YTHDC1-mediated m6A-dependent YTHDC1/ANXA1 axis
(82). In addition, the decreased
sensitivity of PCa to enzalutamide is due to the methylation of
nuclear receptor subfamily 5 group A member 2 (NR5A2), which is
caused by the low expression of METTL3 (83). The mechanism of m6A
development and enhancement of cancer resistance is receiving
increasing attention, which provides insight for future drug
development and potential therapeutic targets which can be used to
reduce resistance.
Role of m6A regulators in
PCa
PCa is associated with five key m6A
methylation regulators according to clinical data analysis. These
regulators are tRNA methyltransferase activated subunit 11-2,
nuclear RNA output factor 1, YTHDF1, HNRNPG and HNRNPA2B1, which
integrate novel prognostic features that independently predict PCa
prognosis. In the tumor microenvironment, three different modes of
m6A regulation have been found in PCa through the
identification of m6A regulatory molecules. Each
m6a regulatory mode has a different proportion of C3
immune subtypes. It has been suggested that m6A
regulation in PCa is closely related to the tumor immune
microenvironment. In addition, patients with PCa present with an
increased expression of m6A ‘writers’ or a decreased
expression of ‘erasers’ (84).
Thus, the m6A methylation regulator manipulates the
occurrence and progression of PCa.
METTL3 in PCa
It has been shown that an increased expression of
METTL3 may have a tumor-promoting effect (85). Increased expression levels of METTL3
can promote the proliferation, migration and invasion of PCa by
promoting ARHGDIA expression, and leading to an upregulation of the
total m6A methylation modification level in PCa tissue
(86). In addition, the
overexpression of METTL3 increases the m6A content, and
promotes the growth and invasion of PCa cells through Sonic
hedgehog (SHH)-GLI family zinc finger 1 (GLI1) signaling (87). The m6A methylation of
lymphoid enhancer-binding factor 1 mRNA is mediated by METTL3.
Lymphoid enhancer-binding factor 1 enhances the activity of the
Wnt/β-catenin pathway, promotes the proliferation of prostate
cancer cells and inhibits cell differentiation (88). METTL3 promotes the maturation of
pre-miRNAs by upregulating the m6A content and
interacting with the microprocessor protein, DGCR8, to mediate
m6A modification, which recognizes pre-miR-182 (89). The m6A modification of
the METTL3-mediated long non-coding RNA (lncRNA) MALAT1 can also
promote PCa cell growth and transfer by activating PI3K/AKT
signaling (90). METTL3 increases
the m6A level of MYC mRNA transcription and enhances MYC
expression, which leads to the occurrence and development of PCa
(91). METTL3 can also induce
m6A modification on kinesin superfamily protein 3C
(KIF3C) by increasing the stability of IGF2-binding protein 1 to
KIF3C-mRNA. KIF3C is overexpressed in PCa, which promotes its
growth, migration and invasion during the m6A-dependent
miR-320d/METTL3 (92). The tiny
lipid molecule, lipoxin A4, in PCa cells promotes the polarization
of M2-like macrophages by inhibiting the METTL3-medidated
activation of STAT6, which produces effects that enhance tumor
metastasis and growth activity. M6A levels were reduced
in tumor-associated macrophages in PCa patients. METTL3 drives M1
macrophage polarization through the methylation of STAT1 mRNA,
which exerts antitumor effects (93). METTL3 levels in patients with PCa
are up-regulated, promoting cell proliferation, migration and
invasion in PCa via a variety of mechanisms (Fig. 2).
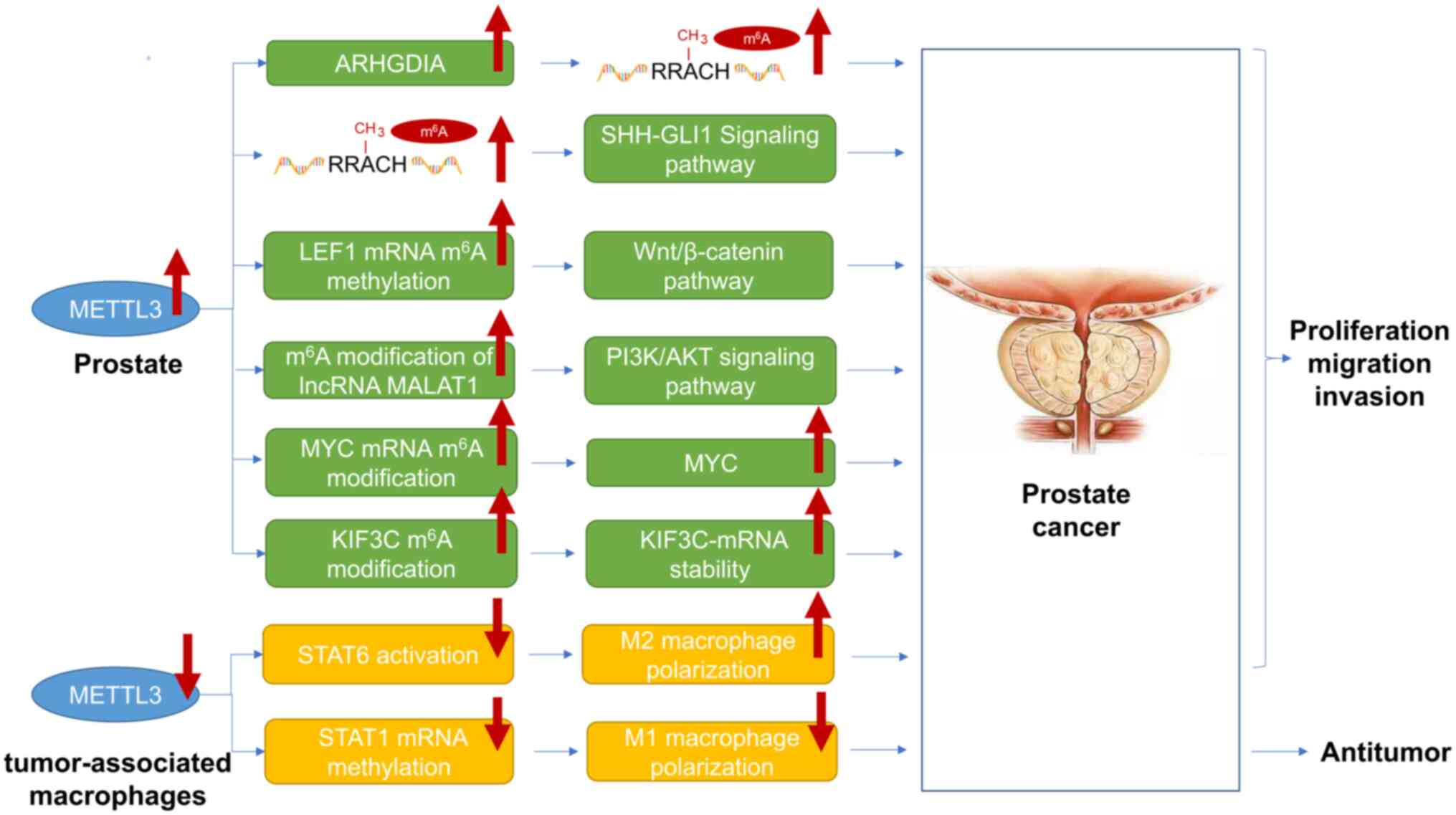 | Figure 2.MEETL3 plays a crucial role in the
progression of prostate cancer. METTL3 expression levels increase
in prostate cancer cells, but decrease in tumor-associated
macrophages. METTL3 plays a role in the malignant progression of
prostate cancer mainly through the following six pathways: i) An
increase in the content of ARHGDIA promotes an increase in the
total m6A level; ii) an increase in the total
m6A level activates the SHH-GLI1 signaling pathway; and
iii) an increase in the m6A modification of LEF1 mRNA
activates Wnt/β-catenin signaling pathway; iv) lncRNA MALAT1
m6A modification increases the activation of the
PI3K/AKT signaling pathway; v) the increased modification of MYC
mRNA m6A leads to an increase in the MYC content; vi)
the KIF3C m6A modification increases the stability of
KIF3C mRNA. In addition, METTL3 is expressed at low levels in
tumor-related macrophages, promoting the proliferation, migration,
and metastasis of prostate cancer by inhibiting the activation of
STAT6 and leading to the activation of M2 macrophages. The
low-level expression of METTL3 in tumor-associated macrophages
exerts its anti-prostate cancer effect by inhibiting the
methylation of STAT1 mRNA and promoting the activation of M1
macrophages. METTL, methyltransferase-like; ALKBH,
alpha-ketoglutarate-dependent dioxygenase homolog; SHH, Sonic
hedgehog; GLI1, -GLI family zinc finger 1; KIF3C, kinesin
superfamily protein 3C. |
WTAP in PCa
WTAP is a writer for m6A methylation
modification in PCa tissue (58).
In PCa, WTAP has been shown to promote cell proliferation and
metastasis by binding to the corresponding androgen receptor.
STAT1, FOXO1, Interferon regulatory factor 1, glucocorticoid
receptor and PPARγ transcription factor binding sites were
identified in the promoter region of the WTAP gene. WTAP expression
may be affected by these tumor-associated transcripts to promote
tumorigenesis (94). WTAP may play
a role in the processing of androgen-responsive circular RNA
(circRNA) biogenesis. circRNAs can be used as non-invasive markers
for PCa diagnosis and prognosis (95).
FTO in PCa
A lack of FTO attenuates growth rates and elevated
levels of FTO expression can lead to weight gain due to an
increased energy intake; FTO was first identified as a gene
associated with weight and obesity. It has been shown that FTO can
function as an eraser of m6A methylation modifications,
which manipulate m6A methylation reversals and
participate in dynamically reversible m6A modifications
(54). It was found that FTO is
downregulated in PCa. FTO and ALKBH5 are inversely correlated with
the Gleason classification of PCa (58). It has been suggested that FTO is an
oncogene in PCa (96). FTO
m6A demethylase inhibits the invasion and migration of
PCa cells by regulating total m6A levels. When FTO is
present in the nucleus, it first promotes the demethylation of
m6A. It first promotes the demethylation of
m6A and m6Am when FTO is present in the
cytosol. It has been shown that FTO in PCa cells is primarily found
in the nucleus. mRNA decay experiments have also shown that the
knockout of FTO does not affect the stability of the target mRNA in
PCa cell lines. Increased levels of FTO expression may be
associated with obesity-associated FTO single nucleotide
polymorphisms, and promote tumorigenesis and progression (54). In addition, the m6A
modification is positively associated with the degree of tumor
malignancy, which suggests a tumor suppressor effect of FTO in PCa
(54).
It has been shown that FTO can also affect the
occurrence and development of PCa by modulating the MC4R content.
It promotes mRNA stability and modulates nuclear processes, miRNA
processing and retinol-binding protein interactions. FTO is capable
of oxidizing single-stranded DNA and single-stranded RNA in
vitro to demethylate m-3T and m-3U. Moreover, the FTO and MC4R
expression levels exhibit a significant negative correlation. The
high expression of FTO partially modifies the boosting effect of
high MC4R expression on the PCa malignant phenotype (53).
In addition, CLIC4 is one of the functional targets
of FTO-mediated m6A modification by multiple assays. FTO
depletion suppresses PCa proliferation and transfer by increasing
m6A levels of CLIC4 mRNA which decreases mRNA stability.
Functionally, FTO inhibits PCa cell proliferation and metastasis
in vitro and in vivo, which are associated with a
poor prognosis of patients with PCa, while the ectopic expression
of FTO has the opposite effect (55).
The polymorphisms rs9930506 and rs9939609 in the
FTO gene have been found to be associated with obesity and PCa
(97). Both rs9930506 and rs9939609
are associated with high BMI in the European population, while
obesity is associated with a high risk of developing PCa, which
suggests an association between FTO genotypes and the risk of
developing PCa. The mutation of rs9939609 is negatively associated
with the risk of developing PCa and is positively associated with
being overweight. Cases of severe PCa are more likely to occur in
individuals who are overweight and have a mutation in the rs9939609
gene. The prevalence of heterozygous forms of rs9939609 suggests
that its ‘A’ allele may be related to the phenotype of PCa
(97). The rs9939609 A allele has
been found to be associated with cancer risk in PCa cases, which is
not with the occurrence of an increased risk of PCa itself. The
data suggest that the rs9939609 A allele reduces PCa risk and the
likelihood of detecting low-grade PCa, which may increase the
likelihood of elevated-grade PCa (98). In addition, it has been suggested
that FTO rs8050136 polymorphisms are not associated with PCa
(97,99).
ALKBH5 in PCa
ALKBH5 is a member of the m6A ‘eraser’,
which functions as a demethylase. In model-based studies, ALKBH5
has been shown to affect PCa diagnosis and prognosis. Lower levels
of ALKBH5 reduce protein expression in the tumor. It has been
suggested that a reduction in the ALKBH5 level, which is caused by
the deletion of the ALKBH5 amplification gene, is a factor in the
development and progression of PCa (57). The expression of ALKBH5 has been
shown to be significantly increased in patients with PCa compared
to the normal population. Patients with CRPC with bone metastases
have been found to have higher levels of ALKBH5 than patients with
CRPC with lymph node metastases. Furthermore, ALKBH5 is negatively
associated with the Gleason score, which suggests that ALKBH5 may
be an indicator of PCa prognosis (56,58).
The differential methylation of the ALKBH5 CpG site may enhance the
progression of PCa transfer, although the specific molecular
mechanisms remain unknown (100).
YTHDF2 in PCa
YTHDF2 and METTL3 have been found to be expressed
at increased levels in PCa, which suggests a lower overall
survival. The in vitro and in vivo inhibition of
YTHDF2 and METTL3 has been shown to inhibit PCa development. This
suggests an association between YTHDF2/METTL3 and PCa (101). YTHDF2 is overexpressed in PCa and
CRPC with lymph node metastasis and is positively associated with
the Gleason grade in PCa (58). The
C and N terminal domains of YTHDF2 play the roles of specifically
binding to the m6A modification site, recruiting the
CCR4-NOT complex and mediating the localization of the YTHDF2mRNA
complex to the cellular RNA decay site, respectively. METTL3
functions as a core writer-catalyzed m6A modification,
which can promote YTHDF2 to play a regulatory role. YTHDF2 can
directly bind to the m6A modification sites of NK3
homeobox 1 (NKX3-1) and phospholysine phosphohistidine inorganic
pyrophosphate phosphatase (LHPP), and degrade their mRNA, which
indirectly induces AKT phosphorylation modification and promotes
PCa occurrence and development in an m6A-dependent
manner. The increased expression of NKX3-1 suppresses the
development of PCa, while LHPP suppresses AKT phosphorylation
modification to regulate tumor progression (56,101).
In addition, YTHDF2 is a target gene for miR-495 and is negatively
associated with it. miR-495 can bind to the mRNA of YTHDF2. The
knockout of miR-495 has been shown to significantly reduce cell
proliferation and transfer in the DU-145 and PC3 PCa cell lines.
YTHDF2 overexpression attenuates the inhibitory effect of miR-495
on PCa development and induces apoptotic effects (102). In addition, as previously
demonstrated, YTHDF2 can recruit the RNA-binding protein HNRNPD and
bind to ubiquitin specific protease 4 (USP4) mRNA, which induces
mRNA degradation. The reduction in the USP4 level fails to remove
the ubiquitin group in the ELAVL1 protein, which results in a
reduction in the ELAVL1 protein content. It allows ARHGDIA to be
over-expressed and promotes PCa development (86). In addition, the expression levels of
YTHDF2 are increased in PCa tissue and in the DU-145 and PC3 cell
lines, while miR-493-3p expression is decreased, suggesting an
inverse correlation (103). The
target protein of miR-493-3p is YTHDF2, which is also an upstream
factor in the inhibition of PCa cell line development by YTHDF2.
The overexpression of miR-493-3p leads to an increase in
m6A levels, while the inhibition of miR-493-3p leads to
a decrease in m6A levels. The miR-493-3p-mediated
downregulation of YTHDF2 significantly suppresses PCa progression
by increasing the m6A levels. YTHDF2 and miR-493-3p
indirectly alter the occurrence and development of PCa in an
m6A-dependent manner (103).
IGF2BP1 in PCa
It has been shown that the IGF2BP1 protein family
primarily acts as a recognition enzyme for m6A
methylation modification and as a cancer promoter. METTL3 induces
an increase in the level of m6A methylation modification
of KIF3C, improving the stability of IGF2BP1 against KIF3C mRNA and
reducing KIF3C mRNA degradation. KIF3C is expressed at peak levels
in PCa tissues and cell lines, which is positively associated with
PCa growth, migration and invasion. Following the knockdown if
METLL3, the expression of m6A and KIF3C is reduced. The
expression and stability of KIF3C are reduced following the
knockout of IGF2BP1. Thus, IGFBP1 positively modulates the
stability and expression of KIF3C by relying on m6A
(92).
Other m6A regulators in
PCa
It has been shown that METTL14 facilitates the
occurrence and development of PCa in vitro and in
vivo. METTL14 recruits the YTHDF2 protein in an
m6A-dependent manner to inhibit thrombospondin 1 (THBS1)
mRNA expression (104). METTL14
methylates the THBS1 mRNA in an m6A-dependent manner.
Methylated THBS1 mRNA can be recognized by the YTHDF2 protein which
binds to the m6A site in the cytoplasm leading to the
degradation of the THBS1 mRNA. In addition, METTL14 can inhibit the
angiogenesis inhibitory effect of THBS1 in an
m6A-dependent manner, which provides sufficient energy
and oxygen for the growth of PCa tissues and cell lines (104).
In addition, increased METTL14 and ZC3H13
expression levels are positively associated with the number of Th1
cells and Th17 cells, as well as with the mesenchymal fraction and
transforming growth factor-β responses. An increased mesenchymal
fraction of Th1 cell and Th17 cell numbers suggests a good
prognosis in PCa (105).
The expression of KIA1429 and HNRPA2B1 leads to
m6A methylation modification, which indicates a poor
prognosis in PCa. This will lead to increased intracellular
heterogeneity and Th2 cell penetration within PCa tissues and cell
lines, with lower Th17 cell penetration and macrophage M1/M2
polarization (105).
The overexpression of Virma leads to increased
levels of m6A RNA methylation in androgen-independent
PCa cells and CRPCs. It has been shown that the viability and
proliferation of PC-3 cells can be suppressed by Virma inhibition.
The downregulation of Virma attenuates PCa migration and
aggressiveness by inhibiting m6A expression, which
decreases the stability and abundance of lncRNA, and reduces the
malignant phenotype of PCa (106).
The expression levels of YTHDC2 and YTHDF1 are
elevated in PCa and CRPC with lymphoma metastases, suggesting a
poor prognosis (58). In addition,
YTHDF1 overexpression induces the occurrence and development of
PCa. It has been shown that the downstream factor of YTHDF1 is
polo-related kinase1 (PLK1). YTHDF1 recognizes PLK1 mRNA and
interacts with the m6A modification of PLK1 mRNA 3′
non-coding region in an m6A modification-dependent
manner, which improves the translation efficiency of PLK1 mRNA and
increases the level of PLK1. It can promote the activation of the
PI3K/AKT signaling pathway. Finally, YTHDF1 promotes the occurrence
and development of PCa by increasing the PLK1 protein content and
activating the PI3K/AKT signaling pathway (107).
HNRNPA2B1 promotes the genesis and development of
PCa tissue in an m6A-dependent manner, including several
nuclear processes (108). In
addition, HNRNPA2B1 has been shown to be associated with
recurrence-free survival in PCa, which suggests its association
with a poor prognosis (96).
HNRNPA2B1 can also enhance its expression levels through
lncRNA-PCAT6 (PCAT6), which promotes the occurrence and development
of PCa. The knockout of the hnRNPA2B1 gene significantly reduces
the ability of PCa cells to grow and migrate due to PCAT6 (109).
The HNRNPC protein can enhance the stability of the
nucleosome assembly protein (NAP1) L2 mRNA by binding to lncNAP1L6,
which is recruited into the m6A-methylated modified
NAP1L2 mRNA. HNRNPC indirectly promotes the increase of NAP1L2 in
NAP1L2 translational expression products, and induces PCa transfer
and invasion by the activation of the MMP signaling pathway
(110).
The protein IGF2BP2 recognizes
m6A-methylated PCAT6 and enhances RNA stability, which
shortens the half-life of PCAT6. PCAT6 is overexpressed in PCa
tissues with bone metastases and is associated with the poor
prognosis of patients with PCa (111).
IGF2BP3 binds to hsa_circ_0003258 and enhances the
stability of the HDAC4 mRNA, which activates the ERK signaling
pathway and accelerates the transfer and invasion of PCa tissue
(112).
In the process of PCa, multiple m6A
regulators are required to participate in regulation. Each
regulator is interconnected or mutually restricted and jointly
regulates the body's m6A levels. The complex and diverse
regulatory mechanisms make it difficult to study the association
between m6A and PCa.
The m6A and non-coding RNA
modifications
The role of coding RNAs in the development and
metastasis of PCa has been continuously revealed. However, the
majority of the RNAs in the human genome are non-coding RNAs. The
association between non-coding RNAs and PCa has not received ample
attention (113). Non-coding RNAs
are not only abundant, but also versatile (114). For example, non-coding RNAs from
liquid biopsies of patients with PCa have shown some benefits in
terms of diagnosis, prognosis and detection (115). Although determining the role of
non-coding RNA in PCa requires a certain scale and in-depth study,
the results of such studies may be beneficial for the adjuvant
treatment of patients with PCa and the development of novel
therapeutic agents (116). The
potential clinical operability of non-coding RNAs is being explored
(114).
m6A and miRNAs in cancer
metabolism
Existing studies have shown that tumor metabolic
activity is related to miRNAs; however, the metabolic effects of
m6A on the regulation of tumor cells by miRNAs have not
yet been fully elucidated (117).
In mammalian cells, DGCR8 recognizes and acts on pre-miRNAs
undergoing METTL3-induced methylation. Of these, miR-182 can mature
under m6A-dependent METTL3 methylation modification and
promote proliferation, migration, and invasion of prostate cells
(83). In addition, METTL3 in PCa
cells can induce the methylation modification of KIF3C in an
m6A-dependent manner and enhance the stability of KIF3C
mRNA through IGF2BP1 which results in abnormally elevated KIF3Cd
levels and promotes PCa progression. miR-320d can inhibit the
occurrence and progression of PCa by specifically modulating METTL3
expression levels, thereby reducing KIF3C content (92). miR-141-3p inhibits the
m6A methylation modification of PRMT6 by mediating the
low-level expression of ALKBH5. PRMT6 is not inhibited by ALKBH,
which is highly expressed and promotes the occurrence and
development of PCa (118).
Lysine-specific demethylase 5A can inhibit its transcription and
expression by binding to the promoter sequence of miR-495. YTHDF2
can recognize MOB3B mRNA. Due to the low expression level of
miR-495, this leads to the degradation of MOB3B mRNA, thereby
reducing MOB3B expression. The low expression of MOB3B ultimately
promotes malignancy progression in PCa (102). HNRNPA2B1 interacts with primary
miRNA-93 through the oncogenic axis of protein 6 of the
HNRNPA2B1/miR-93-5p/FERM domain to stimulate PCa progression in an
m6A-dependent manner (119).
m6A and lncRNAs in cancer
metabolism
lncRNAs consist of >200 nucleotides, which are
non-coding RNAs that can participate in important processes in
epigenetics, the cell cycle, cell differentiation, and even cancer
cell metabolism (111). MALAT1 is
a long non-coding RNA that contributes to the development and
progression of cancer in cancer cells by promoting the glycolysis
process and inhibiting gluconetics. Studies have linked the lncRNA
MALAT1 and the m6A methyltransferase METTL3 to
malignancy progression in PCa. The m6A modification of
lncRNA MALAT1 can be mediated by METTL3, thereby activating the
PI3K/AKT signaling pathway, which promotes malignancy progression
in PCa (90). Elevated levels of
the lncRNA nuclear enriched abundant transcript (NEAT)1-1 recognize
and activate the activity of CYCLOINL1 and form a different complex
with extreme levels of CDK19 in PCa, which ultimately acts on the
promoter Runt-related transcription factor 2 (RUNX2). NEAT1-1
promotes bone metastasis in PCa through RUNX2 and other related
signaling pathways which can survive, proliferate, and invade the
bone environment (120). The
stability and expression levels of the LncRNA CCAT1/2 transcript
are reduced by the Virma content, as well as the intracellular
m6A levels. Low expression of oncogenic LncRNA CCAT1/2
reduces prostate aggressiveness (106). Furthermore, the m6A
methylation of lncRNA NAP1L2 is mediated by the METTL14/METTL3
complex and stabilized by the HNRNPC protein recruited by
lncNAP1L6. The improvement of metastatic capacity in PCa relies on
increased levels of lncRNA NAP1L2 and YY1 mediated MMP2 and MMP9
transcription (110).
M6A methyltransferase ZC3H13 is modified by lncRNA A1BG
derived from exosomes. The modified LncRNA A1BG is stably
expressed, which inhibits the progression of PCa (121).
m6A and circRNAs in cancer
metabolism
circRNAs are stable closed-loop RNA structures that
are less susceptible to degradation by RNA exonuclease than regular
linear RNAs. In addition, circRNAs compete with endogenous RNAs for
miRNA sites. Although circRNAs and m6A methylation have
been relatively poorly studied in cancer tissue metabolism,
circRNAs have been shown to affect the metabolic activity of cancer
cells and thus, tumor development (122,123). CircPDE5A interferes with the
formation of m6A methylation by recognizing and binding
to specific sites of WTAP, which results in decreased
m6A methylation levels of eukaryotic translation
initiation factor 3 subunit C (EIF3C) mRNA. YTHDF1 reduces the
translation efficiency of the EIF3C mRNA m6A with low
methylation levels, which leads to the inactivation of the MAPK
pathway and the inhibition of PCa development (122). CircFAM126A exhibits a high
expression in PCa and an enhanced transcriptional stability through
m6A modification, promoting PCa progression in
vitro. CircFAM126A mediates calnexin by targeting miR-505-3p.
The low expression of calnexin can inhibit cholesterol synthesis in
PCa cells and the malignant progression of PCa (124). CircDDIT4 is expressed at a low
level as a tumor suppressor in PCa. The modification of circDDIT4
by m6A promotes the biogenesis of circDDIT4. The
methyltransferase complex is composed of WTAP/METTL3/METTL14, which
increases circDDIT4 levels, while FTO exerts the opposite effect
(125). The circRBM33-FMR1 complex
stabilizes PDHA1 mRNA in an m6A-dependent manner and
activates the mitochondrial metabolism of PCa, thereby promoting
PCa progression (126).
PCa and m6A
M6A modification plays a multifaceted
role in the pathogenesis, diagnosis and treatment of PCa. Further
research into the exact mechanisms of m6A dysregulation
in PCa and the development of targeted therapeutic interventions is
required in order to improve patient outcomes in the future.
Pathogenesis
METTL3 plays a direct role in AR expression. The
knockdown of METTL3 leads to the increased expression of the AR
target gene, NKX3.1, and to the decreased expression of PSA. The
knockdown of METTL3 leads to an increase in the key regulatory
factor lysine-specific demethylase-1. Lysine-specific demethylase-1
is involved in the development of PCa and affects the expression
and function of AR (127). The
high expression of HNRNPA2B1 can promote the proliferation and
metastasis of PCa. In a new oncogenic axis HNRNPA2B1/miR-93-5p/FERM
domain-containing protein 6, HNRNPA2B1 promotes the maturation of
miR-93-5p in an m6A-dependent manner. Thus, the
expression of tumor suppressor FERM domain-containing protein 6 is
reduced (119).
As an epithelial-mesenchymal transition (EMT)
regulator in PCa, FTO can inhibit the m6A modification
level of EMT tumor cells. When the FTO gene is knocked out, EMT
occurs in tumor cells, and promotes cell migration and
proliferation (128). In addition,
it has been shown that NAP1L2 and lncNAP1L6 are involved in the
migration, invasion and EMT processes of PCa cells (110).
RNA binding motif 3 over methylated m6A
on catenin β1 (CTNNB1) mRNA in a manner dependent on METTL3.
Alterations in β-catenin signaling can affect stem-like properties
and the self-renewal ability of tumor cells. The stability of
CTNNB1 mRNA was reduced by methylation of m6A. This
results in the inactivation of the Wnt signaling pathway, and
eventually, the stemness remodeling of PCa cells by osteoblasts was
inhibited (129).
Diagnosis
The transfer of PCa is associated with
m6A-modified mRNA. Methylated RNA immunoprecipitation
sequencing (MeRIP-Seq) is a technique that combines RNA-protein
immunoprecipitation and high-throughput sequencing. MeRIP-Seq can
map m6A methylated mRNA (130). The score of m6A
modified mRNA was calculated by the results of MeRIP-Seq. A higher
m6A-modified mRNA score is associated with a shorter
biochemical relapse time in patients with PCa, and m6A
hypomethylation may contribute to PCa initiation. By contrast, the
transfer group exhibits more m6A modification peaks than
the primary group. MeRIP-Seq helps study the prognosis and
diagnosis of PCa (131). MeRIP-seq
can also predict the results of PCa by detecting the content of
m6A methylated lncRNA in PCa tissues and calculating the
lncRNA score modified by m6A (132).
Compared with normal tissues, malignant tissues of
patients with PCa have a lower FTO content and higher
m6A levels. Higher levels of FTO expression have been
detected in patients with PCa with a poor prognosis. This indicates
that the expression level of FTO is associated with the prognosis
of PCa, suggesting that FTO is one of the diagnostic markers of PCa
(133). THBS1 is a tumor
suppressor that can inhibit the proliferation of PCa. In the
nucleus, METTL14 inhibits THBS1 expression in an
m6A-dependent manner, leading to PCa proliferation.
Therefore, METTL14 may be a prognostic marker and an effective
therapeutic target for PCa (104).
METTL3 is highly expressed in tumor cells and is predictive of a
poor prognosis; thus, it is a promising diagnostic and prognostic
marker (134).
Treatment
The methylation of m6A is associated
with immune response, tumor growth and metastasis (135). M6A regulators cluster 3
modulates METTL14 and ZC3H13 expression levels and increases Th1
cells, Th17 cells, mesenchymal fraction and transforming growth
factor-β. Of these, Th1 cells can initiate an antitumor immune
response. The interstitial fraction is inversely related to the
degree of malignancy of the tumor. TGF-β can inhibit the Th1
response and reduce the effect of ICIs. In addition, Th17 cells are
a good prognostic indicator of PCa, which is related to the
efficacy of PD-1 blockade in PCa treatment (105). Additionally, it has been shown
that the m6A methylation regulators HNRNPA2B1 and METTL3
affect the immune microenvironment of PCa (136).
The latest research suggests that the
radiosensitivity of tumors can be modulated by methylation
modifications of m6A, which greatly increase the role of
radiotherapy in cancer. The pathogenesis and progression of bone
metastatic PCa can be inhibited by deletion of the
MLXIPe/KHSRP/PSMD9 regulatory complex in vitro and in
vivo, thereby improving the efficacy of radiotherapy. This
mechanism is achieved by the RNA-binding protein KHSRP, which
simultaneously recognizes m6A on the enhancer RNA and
m6Am on the 5′-UTR, while resisting degradation by the
exonuclease XRN2 (137).
METTL3, FTO, YTHDC1-2, YTHDF1-3 and IGF2BP1-3
proteins generally promote tumorigenesis. METTL3, METTL14, FTO and
ALKBH5 can promote or inhibit the progression of cancer cells.
Similarly, METTL3, FTO and ALKBH5 can alter the susceptibility or
resistance of cancer cells to anticancer treatments (138). By identifying appropriate
treatments that affect the functions leading to the development of
PCa, it may be possible to treat PCa. Treatment with enzalutamide
combined with METTL3 knockdown has been shown to result in
AR-independent upregulation of gastrointestinal-specific gene
features driven by nuclear receptor NR5A2, which result in
enzalutamide resistance. This suggests that NR5A2 and other
downstream pathway genes may be one of the targets for the
treatment of CRPC (83). The
functional inhibition of METTL3 may reduce tumor chemotherapeutic
resistance induced by METTL3 and restore tumor sensitivity to
chemotherapy drugs (134). PCa
photothermal immunotherapy is also a treatment direction for PCa.
Meclofenamic acid, a highly selective FTO inhibitor, can be
combined with a gold nanorod-based nanoplatform to promote
photothermal immunotherapy for PCa (139). Curcumin can inhibit the expression
of m6A-dependent TNF receptor-associated factor 4
induced by ALKBH5 and YTHDF1 (140). In addition, the potentially
beneficial effect of curcumin in reducing PSA in patients with
intermittent androgen deprivation PCa has also been demonstrated in
a clinical trial (141).
Solute carrier family 12 member 5 is a
neuron-specific potassium chloride cotransporter 2. Solute carrier
family 12 member 5 promotes the tumorigenesis and development of
PCa through YTHDC1 and the transcription factor, homeobox B13
(HOXB13). Solute carrier family 12 member 5 inhibitors may be used
in the treatment of PCa (142).
METTL3 knockdown combined with enzalutamide treatment has been
shown to result in the development of resistance to enzalutamide in
PCa cells. This suggests the mechanism by which PCa cells develop
resistance to enzalutamide and may be an effective therapeutic
target (83).
The change in drug delivery has largely enabled
precision therapy. The treatment based on nanotechnology can
improve the systemic toxicity and low efficacy of paclitaxel,
adriamycin, docetaxel and other classical chemotherapy drugs,
providing a new exploration direction for the precise targeted
therapy of PCa (143). Gold
nanoparticles coated with bovine serum albumin can be potentially
cytotoxic to PCa (144).
Multifunctional self-assembly magnetic nanocarriers can effectively
improve the delivery efficiency of prostate tumors in the process
of photothermal therapy, which enhances the efficacy of
photothermal therapy on PCa and plays an antitumor role (145). The microwave-induced expression of
heat shock protein (HSP)70 in prostate tissue and the transfer of
HSP70 to the cell membrane have been studied. The HSP70 antibody is
then coated with nanoparticles and doxorubicin is precisely ablated
and released under near-infrared irradiation, enabling precise drug
therapy (146).
Conclusion and future perspectives
Currently, PCa remains one of the most common types
of cancer worldwide among males, where it accounts for more than a
quarter of cancer diagnoses. Current diagnostic and prognostic
markers for PCa are also diverse. As regards the treatment of PCa,
the this is mainly selected in relation to objective factors, such
as the Gleason score and clinical stage; the corresponding clinical
treatment methods, such as radical surgical treatment, external
radiation therapy, brachytherapy therapy, the experimental local
treatment of PCa, endocrine therapy and chemotherapy are selected
to treat patients (147). While
there are numerous treatments for PCa, they still have their
strengths and weaknesses, and some treatments can even cause damage
to the body. For patients with advanced-stage PCa, treatment can
only prolong survival and relieve symptoms, and cannot completely
cure the disease.
As one of the hot topics of discussion in
epigenetics, the mechanism of m6A and its biological
impact on cancer development is gradually being elucidated. For
example, FTO can promote the proliferation of oral cancer cells by
renewing PD-1 expression (38). The
methylation modification of m6A plays a crucial role in
the occurrence and development of cancer and has led to new insight
and approaches for the diagnosis, treatment and prognosis of PCa.
During m6A modification, methyltransferases,
demethylases and m6A-binding proteins act as three
different types of m6A regulators to modify various
types of specific RNA molecules in the same dynamic and reversible
methylation of various types of RNA molecules as DNA and histones.
Androgen function related gene TRIM68 plays a key role in prostate
cancer progression. It was found that YTHDF1-mediated m6A
modification promoted PCa progression by regulating TRIM68 in PCa
(148). This suggests that
m6A modification may also be involved in the regulation
of tumor-related genes. In addition, the association between
changes in the expression levels of three types of partial
regulators in PCa, and the development and progression of PCa have
been demonstrated (113,127). The three regulators interact to
specifically regulate RNA splicing, translation, stability and
other aspects in an m6A-dependent manner, and promote
specific biological behaviors such as proliferation, migration, and
invasion of cancerous tissues. The level of m6A
methylation modification and the expression content of its
regulatory factors may have different biological effects on
different tumors. As a result, m6A and its regulators
may become targets for PCa diagnosis and treatment, both specific
and non-specific, as well as current prognostic markers.
The changes in PCa caused by m6A
modification have a complex biological mechanism. For example, an
increase in the expression level of the m6A
methyltransferase METTL3 promotes m6A modification and
the expression of the hedgehog pathway GLI1, thereby promoting the
proliferation, migration and invasion of PCa cells (87). In addition, the m6A
demethylase FTO inhibits the development and progression of PCa by
increasing m6A methylation modification levels and
reducing CLIC4 mRNA degradation (55). In addition, the neuron-specific
potassium chloride transporter solute carrier family 12 member 5 in
the nucleus forms a complex with the m6A-binding protein
YTHDC1, which in turn regulates HOXB13 to promote PCa progression,
particularly castration-resistant PCa (142). This suggests that the development
of PCa tissue is regulated by different pathways of the
m6A modification regulator. Thus, all three classes of
m6A-modified regulatory factors are involved in multiple
cellular activities in PCa. The interplay of these three may
together constitute a complex mechanistic network of PCa, whose
specific biological mechanisms need to be further explored.
The present review provides an overview of the
association between m6A methylation modification and
PCa, in an aim to provide new insight and methods for the
prevention, diagnosis, prognosis and treatment of PCa. Existing
research has shown that m6A regulatory agents have
become effective in the clinical prevention and treatment of
cancer. The shift in m6A levels effectively promotes or
suppresses the occurrence and development of tumor tissue.
Restoring the balance of m6A modifications by targeting
specific imbalance modulators could be a novel anticancer strategy
(149). MA2, for example, is the
ethyl ester form of meclofenamic acid and acts as a highly
selective FTO inhibitor, inhibiting the development and progression
of glioblastoma (44). In addition,
STM2457 inhibits METTL3 expression and reduces the level of
m6A modification, which has become a new direction in
the treatment of acute myeloid leukemia (150). However, current research on
micro-molecular drugs targeting the epigenetics of m6A
regulators is still insufficient and needs to be explored in the
long-term. Moreover, m6A modification has both
advantages and disadvantages for tumor development. In the face of
the fact that the mechanism of action of m6A cannot be
adequately elucidated, the lack of a reliable theoretical basis for
the corresponding drug development has become one of the major
limiting factors in the development of micro-molecular drugs for
the regulation of m6A (149). The elucidation of the essential
targets of the m6A regulator in PCa and the treatment of
PCa by correcting abnormal m6A modifications by
targeting the epigenetic action of the m6A regulator may
prove to be a direction for future research and may improve the
diagnosis and treatment of patients with PCa.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Key Medical Discipline of
Hangzhou City (grant no. 2021-21), the Key Medical Discipline of
Zhejiang Province (grant no. 2018-2-3), the Key Laboratory of
Clinical Cancer Pharmacology and Toxicology Research of Zhejiang
Province (grant no. 2020E10021), the Zhejiang Province Medical and
Health Science and Technology Program (grant no. 2023KY933), and
the Zhejiang Traditional Chinese Medicine Science and Technology
Project (grant no. 2023ZL565).
Availability of data and materials
Not applicable.
Authors' contributions
QX and JP conceived the review and critically
revised the manuscript. QX, JP and FT drafted the manuscript. NR,
YY, LR and JP drew the figures and collected the related
references. FG conceived and designed the study and provided
academic leadership and guidance. QX and JP supervised and revised
the manuscript. All authors have read and approved the final
manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wasim S, Lee SY and Kim J: Complexities of
prostate cancer. Int J Mol Sci. 23:142572022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Khan MM, Sharma V and Serajuddin M:
Emerging role of miRNA in prostate cancer: A future era of
diagnostic and therapeutics. Gene. 888:1477612023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen N, Wang Z, Chen M, Ma Q, He Y, Wang
Y, Li X, Qiu M, Shi L, Zhu S, et al: Real-world effectiveness and
safety of goserelin 10.8-mg depot in Chinese patients with
localized or locally advanced prostate cancer. Cancer Biol Med.
20:1047–1059. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mamello S, Keamogetswe R, Paballo M,
Lemohang G, Ayodeji A and Samson M: Prostate cancer review:
Genetics, diagnosis, treatment options, and alternative approaches.
Molecules. 27:57302022. View Article : Google Scholar
|
|
5
|
Corti M, Lorenzetti S, Ubaldi A, Zilli R
and Marcoccia D: Endocrine disruptors and prostate cancer. Int J
Mol Sci. 23:12162022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Giri VN, Morgan TM, Morris DS, Berchuck
JE, Hyatt C and Taplin ME: Genetic testing in prostate cancer
management: Considerations informing primary care. CA Cancer J
Clin. 72:360–371. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Piombino C, Oltrecolli M, Tonni E, Pirola
M, Matranga R, Baldessari C, Pipitone S, Dominici M, Sabbatini R
and Vitale MG: De novo metastatic prostate cancer: Are we moving
toward a personalized treatment? Cancers (Basel). 15:49452023.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
McKay RR, Agarwal N, Matsubara N, Piulats
Rodriguez JM, Smith MR, Todenhöfer T, Zhang T, Balar AV, Schaverien
C, Sherwood S, et al: 1423TiP CYCLONE 3: A phase III, randomized,
double-blind, placebo-controlled study of abemaciclib in
combination with abiraterone plus prednisone in men with high-risk
metastatic hormone-sensitive prostate cancer (mHSPC). Ann Oncol.
33:S1195–S1196. 2022. View Article : Google Scholar
|
|
9
|
Rathi N, McFarland TR, Nussenzveig R,
Agarwal N and Swami U: Evolving role of immunotherapy in metastatic
castration refractory prostate cancer. Drugs. 81:191–206. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Desrosiers R, Friderici K and Rottman F:
Identification of methylated nucleo-sides in messenger RNA from
Novikoff hepatoma cells. Proc Natl Acad Sci USA. 71:3971–3975.
1974. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zheng S, Han H and Lin S:
N6-methyladenosine (m6A) RNA modification in
tumor immunity. Cancer Biol Med. 19:385–397. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu ZX, Li LM, Sun HL and Liu SM: Link
between m6A modification and cancers. Front Bioeng Biotechnol.
6:892018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou Y, Yang J, Tian Z, Zeng J and Shen W:
Research progress concerning m6A methylation and cancer.
Oncol Lett. 22:7752021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen Y, Miao L, Lin H, Zhuo Z and He J:
The role of m6A modification in pediatric cancer. Biochim Biophys
Acta Rev Cancer. 1877:1886912022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Quan C, Belaydi O, Hu J, Li H, Yu A, Liu
P, Yi Z, Qiu D, Ren W, Ma H, et al: N6-Methyladenosine
in cancer immunotherapy: An undervalued therapeutic target. Front
Immunol. 12:6970262021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
De Silva F and Alcorn J: A tale of two
cancers: A current concise overview of breast and prostate cancer.
Cancers (Basel). 14:29542022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schatten H: Brief overview of prostate
cancer statistics, grading, diagnosis and treatment strategies. Adv
Exp Med Biol. 1095:1–14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xia C, Dong X, Li H, Cao M, Sun D, He S,
Yang F, Yan X, Zhang S, Li N and Chen W: Cancer statistics in China
and United States, 2022: Profiles, trends, and determinants. Chin
Med J (Engl). 135:584–590. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kench JG, Amin MB, Berney DM, Compérat EM,
Cree IA, Gill AJ, Hartmann A, Menon S, Moch H, Netto GJ, et al: WHO
Classification of Tumours Fifth edition: Evolving issues in the
classification, diagnosis, and prognostication of prostate cancer.
Histopathology. 81:447–458. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lotan TL, Toubaji A, Albadine R, Latour M,
Herawi M, Meeker AK, DeMarzo AM, Platz EA, Epstein JI, Netto GJ, et
al: TMPRSS2-ERG gene fusions are infrequent in prostatic ductal
adenocarcinomas. Mod Pathol. 22:359–365. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gillard M, Lack J, Pontier A, Gandla D,
Hatcher D, Sowalsky AG, Rodriguez-Nieves J, Vander Griend D, Paner
G and VanderWeele D: Integrative genomic analysis of coincident
cancer foci implicates CTNNB1 and PTEN alterations in ductal
prostate cancer. Eur Urol Focus. 5:433–442. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schweizer MT, Antonarakis ES, Bismar TA,
Guedes LB, Cheng HH, Tretiakova MS, Vakar-Lopez F, Klemfuss N,
Konnick EQ, Mostaghel EA, et al: Genomic characterization of
prostatic ductal adenocarcinoma identifies a high prevalence of DNA
repair gene mutations. JCO Precis Oncol. 3:PO.18.00327.
2019.PubMed/NCBI
|
|
24
|
Epstein JI, Egevad L, Amin MB, Delahunt B,
Srigley JR and Humphrey PA; Grading Committee, : The 2014
International Society of Urological Pathology (ISUP) consensus
Confer-ence on Gleason grading of prostatic carcinoma: Definition
of grading patterns and proposal for a new grading system. Am J
Surg Pathol. 40:244–252. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cimadamore A, Scarpelli M, Raspollini MR,
Doria A, Galosi AB, Massari F, Di Nunno V, Cheng L, Lopez-Beltran A
and Montironi R: Prostate cancer pathology: What has changed in the
last 5 years. Urologia. 87:3–10. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vlajnic T and Bubendorf L: Molecular
pathology of prostate cancer: A practical approach. Pathology.
53:36–43. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rodrigues DN, Butler LM, Estelles DL and
de Bono JS: Molecular pathology and prostate cancer therapeutics:
From biology to bedside. J Pathol. 232:178–184. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nevo A, Navaratnam A and Andrews P:
Prostate cancer and the role of biomarkers. Abdom Radiol (NY).
45:2120–2132. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Penghui Y, Le L, Xintao G, Sun T, Miao J,
Yuan X, Liu J, Wang Z and Liu B: Identification of RNA-binding
protein SNRPA1 for prognosis in prostate cancer. Aging (Albany NY).
13:2895–2911. 2021.
|
|
30
|
Johnson IR, Parkinson-Lawrence EJ, Keegan
H, Spillane CD, Barry-O'Crowley J, Watson WR, Selemidis S, Butler
LM, O'Leary JJ and Brooks DA: Endosomal gene expression: A new
indicator for prostate cancer patient prognosis? Oncotarget.
6:37919–37929. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Holt SK, Kolb S, Fu R, Horst R, Feng Z and
Stanford JL: Circulating levels of 25-hydroxyvitamin D and prostate
cancer prognosis. Cancer Epidemiol. 37:666–670. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Izumi K, Shigehara K, Nohara T, Narimoto
K, Kadono Y and Mizokami A: Both high and low serum total
testosterone levels indicate poor prognosis in patients with
prostate cancer. Anticancer Res. 37:5559–5564. 2017.PubMed/NCBI
|
|
33
|
De Nunzio C, Presicce F, Lombardo R,
Cancrini F, Petta S, Trucchi A, Gacci M, Cindolo L and Tubaro A:
Physical activity as a risk factor for prostate cancer diagnosis: A
prospective biopsy cohort analysis. BJU Int. 117:E29–E35. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Goris Gbenou MC, Peltier A, Schulman CC
and Velthoven R: Increased body mass index as a risk factor in
localized prostate cancer treated by radical prostatectomy. Urol
Oncol. 34:254.e1–e6. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tan WP, Lin C, Chen M and Deane LA:
Periprostatic fat: A risk factor for prostate cancer? Urology.
98:107–112. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Choi JB, Myong JP, Lee Y, Kim I, Kim JH,
Hong SH and Ha US: Does increased body mass index lead to elevated
prostate cancer risk? It depends on waist circumference. BMC
Cancer. 20:5892020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tangen CM, Schenk J, Till C, Goodman PJ,
Barrington W, Lucia MS and Thompson IM: Variations in prostate
biopsy recommendation and acceptance confound evaluation of risk
factors for prostate cancer: Examining race and BMI. Cancer
Epidemiol. 63:1016192019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu H, Wang Y, Xue T, Yang Z, Kan S, Hao
M, Gao Y, Wang D and Liu W: Roles of m6A modification in
oral cancer (Review). Int J Oncol. 62:52023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fang Z, Mei W, Qu C, Lu J, Shang L, Cao F
and Li F: Role of m6A writers, erasers and readers in cancer. Exp
Hematol Oncol. 11:452022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhu H, Jia X, Wang Y, Song Z, Wang N, Yang
Y and Shi X: M6A classification combined with tumor
microenvironment immune characteristics analysis of bladder cancer.
Front Oncol. 11:7142672021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zeng J, Zhang H, Tan Y, Wang Z, Li Y and
Yang X: Genetic alterations and functional networks of m6A RNA
methylation regulators in pancreatic cancer based on data mining. J
Transl Med. 19:3232021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sun T, Wu R and Ming L: The role of m6A
RNA methylation in cancer. Biomed Pharmacother. 112:1086132019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
An Y and Duan H: The role of m6A RNA
methylation in cancer metabolism. Mol Cancer. 21:142022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang G, Sun Z and Zhang N: Reshaping the
role of m6A modification in cancer transcriptome: A review. Cancer
Cell Int. 20:3532020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gu Z, Du Y, Zhao X and Wang C: Diagnostic,
therapeutic, and prognostic value of the m6A writer
complex in hepatocellular carcinoma. Front Cell Dev Biol.
10:8220112022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Su S, Li S, Deng T, Gao M, Yin Y, Wu B,
Peng C, Liu J, Ma J and Zhang K: Cryo-EM structures of human
m6A writer complexes. Cell Res. 32:982–994. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gu J, Zhan Y, Zhuo L, Zhang Q, Li G, Li Q,
Qi S, Zhu J, Lv Q, Shen Y, et al: Biological functions of
m6A methyltransferases. Cell Biosci. 11:152021.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huang J and Yin P: Structural Insights
into N6-methyladenosine (m6A) modification in
the transcriptome. Genomics Proteomics Bioinformatics. 16:85–98.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang Q, Mo J, Liao Z, Chen X and Zhang B:
The RNA m6A writer WTAP in diseases: Structure, roles, and
mechanisms. Cell Death Dis. 13:8522022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Balacco DL and Matthias S: The
m6A Writer: Rise of a machine for growing tasks.
Biochemistry. 58:2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mathoux J, Henshall DC and Brennan GP:
Regulatory mechanisms of the RNA modification m6A and
significance in brain function in health and disease. Front Cell
Neurosci. 15:6719322021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang W, Qian Y and Jia G: The detection
and functions of RNA modification m6A based on m6A writers and
erasers. J Biol Chem. 297:100973. 2021. View Article : Google Scholar
|
|
53
|
Li S and Cao L: Demethyltransferase FTO
alpha-ketoglutarate dependent dioxygenase (FTO) regulates the
proliferation, migration, invasion and tumor growth of prostate
cancer by modulating the expression of melanocortin 4 receptor
(MC4R). Bioengineered. 13:5598–5612. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhu K, Li Y and Xu Y: The FTO
m6A demethylase inhibits the invasion and migration of
prostate cancer cells by regulating total m6A levels.
Life Sci. 271:1191802021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zou L, Chen W, Zhou X, Yang T, Luo J, Long
Z, Wu J, Lv D, Mao X and Cen S: N6-methyladenosine demethylase FTO
suppressed prostate cancer progression by maintaining CLIC4 mRNA
stability. Cell Death Discov. 8:1842022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ding D, Liu G, Gao J and Cao M: Unveiling
the m6A methylation regulator links between prostate cancer and
periodontitis by transcriptomic analysis. Dis Markers.
2022:40300462022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ji G, Huang C, He S, Gong Y, Song G, Li X
and Zhou L: Comprehensive analysis of m6A regulators prognostic
value in prostate cancer. Aging (Albany NY). 12:14863–14884. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wu Q, Xie X, Huang Y, Meng S, Li Y, Wang H
and Hu Y: N6-methyladenosine RNA methylation regulators contribute
to the progression of prostate cancer. J Cancer. 12:682–692. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Dai XY, Shi L, Li Z, Yang HY, Wei JF and
Ding Q: Main N6-methyladenosine readers: YTH family proteins in
cancers. Front Oncol. 11:6353292021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li J, Chen K, Dong X, Xu Y, Sun Q, Wang H,
Chen Z, Liu C, Liu R, Yang Z, et al: YTHDF1 promotes mRNA
degradation via YTHDF1-AGO2 interaction and phase separation. Cell
Prolif. 55:e131572022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zuyao C, Xiaolin Z, Min X and Jing Z: The
roles and mechanisms of the m6A reader protein YTHDF1 in tumor
biology and human diseases. Mol Ther Nucleic Acids. 26:1270–1279.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen X, Zhou X and Wang X: m6A
binding protein YTHDF2 in cancer. Exp Hematol Oncol. 11:212022.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zaccara S and Jaffrey SR: A unified model
for the function of YTHDF proteins in regulating
m6A-modified mRNA. Cell. 181:1582–1595.e18. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu
PJ, Liu C and He C: YTHDF3 facilitates translation and decay of
N6-methyladenosine-modified RNA. Cell Res. 27:315–328.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Widagdo J, Anggono V and Wong JJL: The
multifaceted effects of YTHDC1-mediated nuclear m6A
recognition. Trends Genet. 38:325–332. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yan H, Zhang L, Cui X, Zheng S and Li R:
Roles and mechanisms of the m6A reader YTHDC1 in biological
processes and diseases. Cell Death Discov. 8:2372022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ma C, Liao S and Zhu Z: Crystal structure
of human YTHDC2 YTH domain. Biochem Biophys Res Commun.
518:678–684. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kretschmer J, Rao H, Hackert P, Sloan KE,
Höbartner C and Bohnsack MT: The m6A reader protein
YTHDC2 interacts with the small ribosomal subunit and the 5′-3′
exoribonuclease XRN1. RNA. 24:1339–1350. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Qin S, Liu G, Jin H, Chen X, He J, Xiao J,
Qin Y, Mao Y and Zhao L: The comprehensive expression and
functional analysis of m6A modification ‘readers’ in hepatocellular
carcinoma. Aging (Albany NY). 14:6269–6298. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Mo L, Meng L, Huang Z, Yi L, Yang N and Li
G: An analysis of the role of HnRNP C dysregulation in cancers.
Biomark Res. 10:192022. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek
Ż, Pan JN, He C, Parisien M and Pan T: Regulation of
Co-transcriptional Pre-mRNA Splicing by m6A through the
Low-Complexity Protein hnRNPG. Mol Cell. 76:70–81.e9. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wu B, Su S, Patil DP, Liu H, Gan J,
Jaffrey SR and Ma J: Molecular basis for the specific and
multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun. 9:4202018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu
H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Recognition of RNA
N6-methyladenosine by IGF2BP proteins enhances mRNA stability and
translation. Nat Cell Biol. 20:285–295. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ramesh-Kumar D and Guil S: The IGF2BP
family of RNA binding proteins links epitranscriptomics to cancer.
Semin Cancer Biol. 86:18–31. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Song H, Liu D, Wang L, Liu K, Chen C, Wang
L, Ren Y, Ju B, Zhong F, Jiang X, et al: Methyltransferase like 7B
is a potential therapeutic target for reversing EGFR-TKIs
resistance in lung adenocarcinoma. Mol Cancer. 21:432022.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang T, Liu Z, She Y, Deng J, Zhong Y,
Zhao M, Li S, Xie D, Sun X, Hu X and Chen C: A novel protein
encoded by circASK1 ameliorates gefitinib resistance in lung
adenocarcinoma by competitively activating ASK1-dependent
apoptosis. Cancer Letters. 520:321–331. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wu Q, Zhang H, Yang D, Min Q, Wang Y,
Zhang W and Zhan Q: The m6A-induced lncRNA CASC8 promotes
proliferation and chemoresistance via upregulation of hnRNPL in
esophageal squamous cell carcinoma. Int J Biol Sci. 18:4824–4836.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Huang CS, Zhu YQ, Xu QC, Chen S, Huang Y,
Zhao G, Ni X, Liu B, Zhao W and Yin XY: YTHDF2 promotes
intrahepatic cholangiocarcinoma progression and desensitises
cisplatin treatment by increasing CDKN1B mRNA degradation. Clin
Transl Med. 12:e8482022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Cucciniello L, Gerratana L, Del Mastro L
and Puglisi F: Tailoring adjuvant endocrine therapy in early breast
cancer: When, how, and how long? Cancer Treat Rev. 110:1024452022.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Liu X, Gonzalez G, Dai X, Miao W, Yuan J,
Huang M, Bade D, Li L, Sun Y and Wang Y: Adenylate Kinase 4
modulates the resistance of breast cancer cells to tamoxifen
through an m6A-Based epitranscriptomic mechanism. Mol
Ther. 28:2593–2604. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Li F, Chen S, Yu J, Gao Z, Sun Z, Yi Y,
Long T, Zhang C, Li Y, Pan Y, et al: Interplay of m6 A
and histone modifications contributes to temozolomide resistance in
glioblastoma. Clin Transl Med. 11:e5532021. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Li W, Ye K, Li X, Liu X, Peng M, Chen F,
Xiong W, Wang Y and Zhu L: YTHDC1 is downregulated by the YY1/HDAC2
complex and controls the sensitivity of ccRCC to sunitinib by
targeting the ANXA1-MAPK pathway. J Exp Clin Cancer Res.
41:2502022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Cotter KA, Gallon J, Uebersax N, Rubin P,
Meyer KD, Piscuoglio S, Jaffrey SR and Rubin MA: Mapping of m6A and
its regulatory targets in prostate cancer reveals a METTL3-Low
induction of therapy resistance. Mol Cancer Res. 19:1398–1411.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhang SY and Zeng Y: Research progress of
m6A methylation in prostate cancer. Asian J Androl. 25:166–170.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Haigh DB, Woodcock CL, Lothion-Roy J,
Harris AE, Metzler VM, Persson JL, Robinson BD, Khani F, Alsaleem
M, Ntekim A, et al: The METTL3 RNA Methyltransferase regulates
transcriptional networks in prostate cancer. Cancers (Basel).
14:51482022. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Chen Y, Pan C, Wang X, Xu D, Ma Y, Hu J,
Chen P, Xiang Z, Rao Q and Han X: Silencing of METTL3 effectively
hinders invasion and metastasis of prostate cancer cells.
Theranostics. 11:7640–7657. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Cai J, Yang F, Zhan H, Situ J, Li W, Mao Y
and Luo Y: RNA m6A Methyltransferase METTL3 promotes the growth of
prostate cancer by regulating hedgehog pathway. Onco Targets Ther.
12:9143–9152. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ma XX, Cao ZG and Zhao SL: m6A
methyltransferase METTL3 promotes the progression of prostate
cancer via m6A-modified LEF1. Eur Rev Med Pharmacol Sci.
24:3565–3571. 2020.PubMed/NCBI
|
|
89
|
Wang D, Wang X, Huang B, Zhao Y, Tu W, Jin
X, Shao Y, Zhu Y and Lu G: METTL3 promotes prostate cancer
progression by regulating miR-182 maturation in m6A-dependent
manner. Andrologia. 54:1581–1591. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Mao Y, Li W, Weng Y, Hua B, Gu X, Lu C, Xu
B, Xu H and Wang Z: METTL3-Mediated m6A Modification of lncRNA
MALAT1 facilitates prostate cancer growth by activation of PI3K/AKT
signaling. Cell Transplant. 31:96368972211229972022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Yuan Y, Du Y, Wang L and Liu X: The M6A
methyltransferase METTL3 promotes the development and progression
of prostate carcinoma via mediating MYC methylation. J Cancer.
11:3588–3595. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Honggui M, Facai Z, Quliang Z and Jianquan
H: METTL3-mediated m6A modification of KIF3C-mRNA promotes prostate
cancer progression and is negatively regulated by miR-320d. Aging
(Albany NY). 13:22332–22344. 2021.PubMed/NCBI
|
|
93
|
Jia G, Wang X, Wu W, Zhang Y, Chen S, Zhao
J, Zhao W, Li W, Sun X and Han B: LXA4 enhances prostate cancer
progression by facilitating M2 macrophage polarization via
inhibition of METTL3. Int Immunopharmacol. 107:1085862022.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Wu LS, Qian JY, Wang M and Yang H:
Identifying the role of Wilms tumor 1 associated protein in cancer
prediction using integrative genomic analyses. Mol Med Rep.
14:2823–2831. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Kong Z, Lu Y, Wan X, Luo J, Li D, Huang Y,
Wang C, Li Y and Xu Y: Comprehensive characterization of
Androgen-responsive circRNAs in prostate cancer. Life (Basel).
11:10962021.PubMed/NCBI
|
|
96
|
Su H, Wang Y and Li H: RNA m6A methylation
regulators Multi-Omics analysis in prostate cancer. Front Genet.
12:7680412021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Salgado-Montilla JL, Rodriguez-Caban JL,
Sanchez-Garcia J, Sanchez-Ortiz R and Irizarry-Ramirez M: Impact of
FTO SNPs rs9930506 and rs9939609 in prostate cancer severity in a
cohort of Puerto Rican men. Arch Cancer Res. 5:1482017. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Lewis SJ, Murad A, Chen L, Davey Smith G,
Donovan J, Palmer T, Hamdy F, Neal D, Lane JA, Davis M, et al:
Associations between an obesity related genetic variant (FTO
rs9939609) and prostate cancer risk. PLoS One. 5:e134852010.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Zhao J, Huang X, Yang M, Li M and Zheng J:
Association between the FTOrs8050136 polymorphism and cancer risk:
A meta-analysis. Fam Cancer. 15:145–153. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Zhao S, Geybels MS, Leonardson A, Rubicz
R, Kolb S, Yan Q, Klotzle B, Bibikova M, Hurtado-Coll A, Troyer D,
et al: Epigenome-wide tumor DNA methylation profiling identifies
novel prognostic biomarkers of metastatic-lethal progression in men
diagnosed with clinically localized prostate cancer. Clin Cancer
Res. 23:311–319. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Li J, Xie H, Ying Y, Chen H, Yan H, He L,
Xu M, Xu X, Liang Z, Liu B, et al: YTHDF2 mediates the mRNA
degradation of the tumor suppressors to induce AKT phosphorylation
in N6-methyladenosine-dependent way in prostate cancer. Mol Cancer.
19:1522020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Du C, Lv C, Feng Y and Yu S: Activation of
the KDM5A/miRNA-495/YTHDF2/m6A-MOB3B axis facilitates prostate
cancer progression. J Exp Clin Cancer Res. 39:2232020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Li J, Meng S, Xu M, Wang S, He L, Xu X,
Wang X and Xie L: Downregulation of N6-methyladenosine binding
YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by
elevating N6-methyladenosine levels. Oncotarget. 9:3752–3764. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Wang Y, Chen J, Gao WQ and Yang R: METTL14
promotes prostate tumorigenesis by inhibiting THBS1 via an
m6A-YTHDF2-dependent mechanism. Cell Death Discov. 8:1432022.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Liu Z, Zhong J, Zeng J, Duan X, Lu J, Sun
X, Liu Q, Liang Y, Lin Z, Zhong W, et al: Characterization of the
m6A-Associated tumor immune microenvironment in prostate cancer to
aid immunotherapy. Front Immunol. 12:7351702021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Barros-Silva D, Lobo J, Guimarães-Teixeira
C, Carneiro I, Oliveira J, Martens-Uzunova ES, Henrique R and
Jerónimo C: VIRMA-Dependent N6-Methyladenosine modifications
regulate the expression of long Non-coding RNAs CCAT1 and CCAT2 in
prostate cancer. Cancers (Basel). 12:7712020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Li P, Shi Y, Gao D, Xu H, Zou Y, Wang Z
and Li W: ELK1-mediated YTHDF1 drives prostate cancer progression
by facilitating the translation of Polo-like kinase 1 in an m6A
dependent manner. Int J Biol Sci. 18:6145–6162. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Jiang M, Lu Y, Duan D, Wang H, Man G, Kang
C, Abulimiti K and Li Y: Systematic investigation of mRNA
N6-Methyladenosine machinery in primary prostate cancer. Disease
Markers. 2020:88334382020. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Liu B, Jiang HY, Yuan T, Luo J, Zhou WD,
Jiang QQ and Wu D: Enzalutamide-Induced Upregulation of PCAT6
promotes prostate cancer neuroendocrine differentiation by
regulating miR-326/HNRNPA2B1 axis. Front Onco. 11:6500542021.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zheng Y, Qi F, Li L, Yu B, Cheng Y, Ge M,
Qin C and Li X: LncNAP1L6 activates MMP pathway by stabilizing the
m6A-modified NAP1L2 to promote malignant progression in prostate
cancer. Cancer Gene Ther. 30:209–218. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Lang C, Yin C, Lin K, Li Y, Yang Q, Wu Z,
Du H, Ren D, Dai Y and Peng X: m6A modification of lncRNA PCAT6
promotes bone metastasis in prostate cancer through
IGF2BP2-mediated IGF1R mRNA stabilization. Clin Transl Med.
11:e4262021. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Yu YZ, Lv DJ, Wang C, Song XL, Xie T, Wang
T, Li ZM, Guo JD, Fu DJ, Li KJ, et al: Hsa_circ_0003258 promotes
prostate cancer metastasis by complexing with IGF2BP3 and sponging
miR-653-5p. Mol Cancer. 21:122022. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Han Z, Yi X, Li J, Zhang T, Liao D, You J
and Ai J: RNA m6A modification in prostate cancer: A new weapon for
its diagnosis and therapy. Biochim Biophys Acta Rev Cancer.
1878:1889612023. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Altschuler J, Stockert JA and Kyprianou N:
Non-Coding RNAs set a new phenotypic frontier in prostate cancer
metastasis and resistance. Int J Mol Sci. 22:21002021. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Alahdal M, Perera RA, Moschovas MC, Patel
V and Perera RJ: Current advances of liquid biopsies in prostate
cancer: Molecular biomarkers. Mol Ther Oncolytics. 30:27–38. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Ruiz C, Alborelli I, Manzo M, Calgua B,
Keller EB, Vuaroqueaux V, Quagliata L, Rentsch CA, Spagnoli GC,
Diener PA, et al: Critical evaluation of transcripts and long
noncoding RNA expression levels in prostate cancer following
radical prostatectomy. Pathobiology. 90:400–408. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Heyn GS, Corrêa LH and Magalhães KG: The
impact of adipose Tissue-derived miRNAs in metabolic syndrome,
obesity, and cancer. Front Endocrinol (Lausanne). 11:5638162020.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Li X, Liu B, Wang S, Li J and Ge X:
MiR-141-3p promotes malignant progression in prostate cancer
through AlkB homolog 5-mediated m6A modification of
protein arginine methyltransferase 6. Chin J Physiol. 66:43–51.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Sun M, Shen Y, Jia G, Deng Z, Shi F, Jing
Y and Xia S: Activation of the HNRNPA2B1/miR-93-5p/FRMD6 axis
facilitates prostate cancer progression in an m6A-dependent manner.
J Cancer. 14:1242–1256. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Wen S, Wei Y, Zen C, Xiong W, Niu Y and
Zhao Y: Long non-coding RNA NEAT1 promotes bone metastasis of
prostate cancer through N6-methyladenosine. Mol Cancer. 19:1712020.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Yang Z, Luo Y, Zhang F and Ma L:
Exosome-derived lncRNA A1BG-AS1 attenuates the progression of
prostate cancer depending on ZC3H13-mediated m6A modification. Cell
Division. 19:1712024. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Ding L, Wang R, Zheng Q, Shen D, Wang H,
Lu Z, Luo W, Xie H, Ren L, Jiang M, et al: circPDE5A regulates
prostate cancer metastasis via controlling WTAP-dependent
N6-methyladenisine methylation of EIF3C mRNA. J Exp Clin Cancer
Res. 41:1872022. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin
D, Liu J and Sun Z: The role of N6-methyladenosine (m6A)
modification in the regulation of circRNAs. Mol Cancer. 19:1052020.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Luo L, Li P, Xie Q, Wu Y, Qin F, Liao D,
Zeng K and Wang K: n6-methyladenosine-modified circular RNA family
with sequence similarity 126, member A affects cholesterol
synthesis and malignant progression of prostate cancer cells by
targeting microRNA-505-3p to mediate calnexin. J Cancer.
15:966–980. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Kong Z, Lu Y, Yang Y, Chang K, Lin Y,
Huang Y, Wang C, Zhang L, Xu W, Zhao S and Li Y: m6A-Mediated
biogenesis of circDDIT4 inhibits prostate cancer progression by
sequestrating ELAVL1/HuR. Mol Cancer Res. 21:1342–1355. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Zhong C, Long Z, Yang T, Wang S, Zhong W,
Hu F, Teoh JY, Lu J and Mao X: M6A-modified circRBM33 promotes
prostate cancer progression via PDHA1-mediated mitochondrial
respiration regulation and presents a potential target for ARSI
therapy. Int J Biol Sci. 19:1543–1563. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Zhou X, Zhu H, Luo C, Yan Z, Zheng G, Zou
X, Zou J and Zhang G: The role of RNA modification in urological
cancers: Mechanisms and clinical potential. Discov Oncol.
14:2352023. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Zhao Y, Hu X, Yu H, Sun H, Zhang L and
Shao C: The FTO mediated N6-methyladenosine modification of DDIT4
regulation with tumorigenesis and metastasis in prostate cancer.
Research (Wash D C). 7:03132024.PubMed/NCBI
|
|
129
|
Zhang S, Lv C, Niu Y, Li C, Li X, Shang Y,
Zhang Y, Zhang Y, Zhang Y and Zeng Y: RBM3 suppresses stemness
remodeling of prostate cancer in bone microenvironment by
modulating N6-methyladenosine on CTNNB1 mRNA. Cell Death Dis.
14:912023. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Bhattarai DP and Aguilo F: m6A RNA
immunoprecipitation followed by High-Throughput sequencing to map
N6-Methyladenosine. Methods Mol Biol. 2404:355–362. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Lu J, Chen J, Lin Z, Liu Q, Zhong C, Cai
Z, Jia Z, Zhong W, Liang Y and Cai C: A prognostic signature
consisting of N6-methyladenosine modified mRNAs demonstrates
clinical potential in prediction of biochemical recurrence and
guidance on precision therapy in prostate cancer. Transl Oncol.
33:1016702023. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Liang Y, Yin W, Cai Z, Luo H, Liu Q, Zhong
C, Chen J, Lin Z, Huang Y, Liang Z, et al: N6-methyladenosine
modified lncRNAs signature for stratification of biochemical
recurrence in prostate cancer. Hum Genet. Sep 27–2023.doi:
10.1007/s00439-023-02603-8 (Epub ahead of print). View Article : Google Scholar
|
|
133
|
Azhati B, Reheman A, Dilixiati D and
Rexiati M: FTO-stabilized miR-139-5p targets ZNF217 to suppress
prostate cancer cell malignancies by inactivating the PI3K/Akt/mTOR
signal pathway. Arch Biochem Biophys. 741:1096042023. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Zhou X, Chai K, Zhu H, Luo C, Zou X, Zou J
and Zhang G: The role of the methyltransferase METTL3 in prostate
cancer: A potential therapeutic target. BMC Cancer. 24:82024.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Ye X, Wang R, Yu X, Wang Z, Hu H and Zhang
H: m6A/m1A/m5C/m7G-related methylation modification patterns and
immune characterization in prostate cancer. Front Pharmacol.
13:10307662022. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Zhao Y, Sun H, Zheng J and Shao C:
Analysis of RNA m6A methylation regulators and tumour immune cell
infiltration characterization in prostate cancer. Artif Cells
Nanomed Biotechnol. 49:407–435. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Zhao Y, Wen S, Li H, Pan CW, Wei Y, Huang
T, Li Z, Yang Y, Fan S and Zhang Y: Enhancer RNA promotes
resistance to radiotherapy in bone-metastatic prostate cancer by
m6A modification. Theranostics. 13:596–610. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Lan Q, Liu PY, Bell JL, Wang JY,
Hüttelmaier S, Zhang XD, Zhang L and Liu T: The emerging roles of
RNA m6A methylation and demethylation as critical regulators of
tumorigenesis, drug sensitivity, and resistance. Cancer Res.
81:3431–3440. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Zhu W, Zhao R, Guan X and Wang X: The
emerging roles and mechanism of N6-methyladenosine (m6A)
modifications in urologic tumours progression. Front Pharmacol.
14:11924952023. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Chen Y, Wu R, Chen W, Liu Y, Liao X, Zeng
B, Guo G, Lou F, Xiang Y, Wang Y and Wang X: Curcumin prevents
obesity by targeting TRAF4-induced ubiquitylation in m6A-dependent
manner. EMBO Rep. 22:e521462021. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Choi YH, Han DH, Kim SW, Kim MJ, Sung HH,
Jeon HG, Jeong BC, Seo SI, Jeon SS, Lee HM and Choi HY: A
randomized, double-blind, placebo-controlled trial to evaluate the
role of curcumin in prostate cancer patients with intermittent
androgen deprivation. Prostate. 79:614–621. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Yuan S, He SH, Li LY, Xi S, Weng H, Zhang
JH, Wang DQ, Guo MM, Zhang H, Wang SY, et al: A potassium-chloride
co-transporter promotes tumor progression and castration resistance
of prostate cancer through m6A reader YTHDC1. Cell Death
Dis. 14:72023. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Tian Y, Liu Z, Wang J, Li L, Wang F, Zhu Z
and Wang X: Nanomedicine for combination urologic cancer
immunotherapy. Pharmaceutics. 15:5462023. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Barbezan AB, Rosero WAA, Vieira DP, Rigo
MEZ, da Silva GD, Rodrigues AA, de Almeida LF, da Silva FFA, Rivera
AG, da Silva NG, et al: Radioactive gold nanoparticles coated with
BSA: A promising approach for prostate cancer treatment.
Nanotheranostics. 8:112–126. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Li S, Ma Y, Ma C, Shi L, Li F and Chang L:
NIR–triggerable self-assembly multifunctional nanocarriers to
enhance the tumor penetration and photothermal therapy efficiency
for castration-resistant prostate cancer. Discover Nano. 18:462023.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Liu P, Wu Y, Xu X, Fan X, Sun C, Chen X,
Xia J, Bai S, Qu L, Lu H, et al: Microwave triggered
multifunctional nanoplatform for targeted photothermal-chemotherapy
in castration-resistant prostate cancer. Nano Res. 16:9688–9700.
2023. View Article : Google Scholar
|
|
147
|
Swami U, McFarland TR, Nussenzveig R and
Agarwal N: Advanced prostate cancer: Treatment advances and future
directions. Trends Cancer. 6:702–715. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Nie Q, Wu X, Huang Y, Guo T, Kuang J and
Du C: RNA N6-methyladenosine-modified-binding protein YTHDF1
promotes prostate cancer progression by regulating androgen
function-related gene TRIM68. Eur J Med Res. 28:5522023. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Gao R, Ye M, Liu B, Wei M, Ma D and Dong
K: m6A Modification: A Double-Edged sword in tumor development.
Front Oncol. 11:6793672021. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Xu P and Ge R: Roles and drug development
of METTL3 (methyltransferase-like 3) in anti-tumor therapy. Eur J
Med Chem. 230:1141182022. View Article : Google Scholar : PubMed/NCBI
|
















