Introduction
Pancreatic cancer (PC) is an heterogeneous disease
with a poor prognosis (1).
Globally, PC incidence is associated with various factors such as
smoking, family history of chronic pancreatitis, increasing age,
male sex and diabetes mellitus. Patients with PC always possess a
dismal prognosis. It is estimated that 90% of tumors are diagnosed
at an advanced stage after spreading beyond the pancreas, and 50%
of tumors have systemic metastasis (2). Hence, understanding the biological and
molecular mechanisms of PC and developing a specific target for
early PC treatment is essential.
Musashi 2 (MSI2) is one of the RNA-binding proteins
originally identified in stem and progenitor cells (3). MSI2 promotes multiple critical
biological processes relevant to the development of numerous cancer
types, such as PC (4–7). MSI2 induced malignant progression and
metastasis of PC and high expression of MSI2 contributed to the
migration and invasion of PC cells (4,8,9). As a
member of the transcription factor protein group, Forkhead Box F2
(FOXF2) is a mesenchymal transcription factor belonging to the
Forkhead Box (FOX) family. FOXF2 functionally promotes cell
differentiation and suppresses the mesenchymal transformation of
adjacent epithelial cells during embryonic development (10). Evidence has shown that the roles of
FOXF2 are complex and controversial across diverse cancers
(11). FOXF2 manifests
tumor-promoting effects on rhabdomyosarcoma (12). However, FOXF2 inhibits the
progression of colorectal (13),
cervical (14) and ovarian cancer
(15). FOXF2 promotes
proliferation, invasion and metastasis in triple-negative breast
cancer (16), but inhibits the
progression of HER2-positive breast cancer (17). However, the role of FOXF2 in PC has
not been elucidated. Based on the bioinformatic prediction
(https://jaspar.genereg.net/), the
promoter MSI2 region can be bound by the FOXF2 at two binding
sites: 5′-tcataataaacaat-3′ and 5′-tgataataaacacg-3′. Studies
should explore how FOXF2 regulates MSI2 to affect the progression
and metastasis of PC. The present study determined the expression
level of FOXF2 in clinical PC tissues and investigated its roles in
proliferation, apoptosis, invasion and migration of PC cells in
vitro. The effect of FOXF2 on tumor formation in vivo
was also evaluated, and the potential regulating mechanism of FOXF2
in PC was illustrated. These results provided detailed insights
into the functions of FOXF2 in PC and highlighted that targeting
the FOXF2-MSI2 axis might be a promising therapeutic strategy for
PC.
Materials and methods
Bioinformatics analyses
The dataset GSE16515 (18) was downloaded from Gene Expression
Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and was analyzed
through the GEO2R online analysis tool. Differentially expressed
genes were screened out with criteria of: log2FoldChange >2.5
and P<0.01. The potential transcription factor was predicted
using the HumanTFDB 3.0 database (http://bioinfo.life.hust.edu.cn/HumanTFDB#!). Venn
diagram and heatmap analysis were conducted using the R package
(version 4.0.2). The JASPAR website (http://jaspar.genereg.net/) was used to predict
potential-binding sites between FOXF2 and the promoter of MSI2. The
survival prediction and correlation analysis were performed using
the Gene Expression Profiling Interactive Analysis (GEPIA) database
(http://gepia.cancerpku.cn/). The UALCAN
database (http://ualcan.path.uab.edu) showed
the expression of FOXF2 in PC tissues.
Reagents and antibodies
The following chemical reagents were used in the
present study: BCA assay kit (cat. no. P0009; Beyotime Institute of
Biotechnology), Cell Counting Kit-8 (CCK-8; cat. no. C0037;
Beyotime Institute of Biotechnology), Hoechst staining kit (cat.
no. C0003; Beyotime Institute of Biotechnology), ECL detection kit
(BIOSS), Lipo3000 kit (cat. no. L3000015; Invitrogen; Thermo Fisher
Scientific, Inc.), 5-ethynyl-20-deoxyuridine (EdU) assay kit (cat.
no. KGA335; Nanjing KeyGen Biotech Co., Ltd.), Annexin V-FITC/PI
dual-staining kit (cat. no. KGA108; Nanjing KeyGen Biotech Co.,
Ltd.), Cell Cycle Detection Kit (cat. no. C1052; Beyotime Institute
of Biotechnology), Dual Luciferase Reporter Gene Assay Kit (cat.
no. KGAF040; Nanjing KeyGen Biotech Co., Ltd.) and chromatin
immunoprecipitation (ChIP) Assay Kit (cat. no. P2078; Beyotime
Institute of Biotechnology).
The following commercially primary antibodies were
used: anti-Ki-67 (1:50; cat. no. AF0198; Affinity Biosciences),
anti-FOXF2 (1:500; cat. no. D260341; Sangon Biotech Co., Ltd.),
anti-NUMB (1:200; cat. no. D122795; Sangon Biotech Co., Ltd.),
anti-MSI2 (1:200; cat. no. D198948; Sangon Biotech Co., Ltd.),
anti-cyclin D1 (1:1,000; cat. no. bs-20596R; BIOSS), anti-CDK2
(1:1,000; cat. no. bs-10726R; BIOSS), anti-p-CDK2 (1:1,000; cat.
no. bs-3483R; BIOSS), anti-p-RB (1:2,000; cat. no. bsm-52197R;
BIOSS), anti-Cleaved caspase-3 (1:500; cat. no. bs-20364R; BIOSS),
anti-Bax (1:500; cat. no. bs-0127R; BIOSS), anti-Bad (1:500; cat.
no. bs-0892R; BIOSS), anti-Bcl-xl (1:1,000; cat. no. bsm-52024R;
BIOSS), anti-Bcl-2 (1:1,000; cat. no. bsm-52304R; BIOSS) and
anti-β-actin (1:500; cat. no. bs-0061R; BIOSS). The secondary
antibodies included: Goat anti-rabbit IgG conjugated with
horseradish peroxidase (IgG-HRP; 1:500; cat. no. 31460; Thermo
Fisher Scientific, Inc.), rabbit anti-mouse IgG-HRP (1:10,000; cat.
no. bs-0377R-HRP; BIOSS) and goat anti-rabbit IgG-HRP (1:10,000;
cat. no. bs-40295G-HRP; BIOSS).
Human tissues
Fresh samples of PC and adjacent normal tissue
(n=29), and clinical paraffin samples from 78 cases of patients
with PC were obtained in the First Hospital of China Medical
University (Shenyang, China) from September 8, 2021 to April 7,
2022. The expression of FOXF2 and MSI2 in cancer and adjacent
normal tissue was detected by reverse transcription-quantitative
PCR (RT-qPCR). Clinical paraffin samples were used for
immunohistochemical (IHC) staining of FOXF2, followed by analyzing
the association between FOXF2 expression and clinicopathological
characteristics of patients with PC. The characteristics of the
cases were collected from the hospital medical records.
Pathological staging was carried out based on the eighth edition of
with American Joint Committee on Cancer. All experiments using
human tissue were approved [approval no. (2021) 113] by the Medical
Science Research Ethics Committee of The First Affiliated Hospital
of China Medical University (Shenyang, China). Written informed
consent for the collection of tissue samples was provided by all
patients.
Cell lines and culture
The human PC cell line AsPC-1 (Procell Life Science
& Technology Co., Ltd.) was maintained in RPMI-1640 medium
(Beijing Solarbio Science & Technology Co., Ltd.) containing
10% fetal bovine serum (FBS; Zhejiang Tianhang Biotechnology Co.,
Ltd.). PANC-1 (iCell; http://m.icellbioscience.com/) was maintained in DMEM
medium (Wuhan Servicebio Technology Co., Ltd.) containing 15% FBS.
These PC cell lines were all cultured at 37°C with 5%
CO2.
Plasmid construction and RNA
interference
Small interfering RNA (siRNA) targeting FOXF2
(siFOXF2), MSI2 (siMSI2) and untargeted interfering RNA (siNC) were
synthesized by JTSBIO Co., Ltd. For establishment of stable cell
line with FOXF2 knockdown, the (shFOXF2) was inserted to pRNA-H1.1
plasmid. The untargeted shRNA (shNC) served as negative control.
The construction of plasmid of shFOXF2 was completed by Anhui
General Biotechnology Co., Ltd. pcDNA3.1 was used for
FOXF2-overexpressing plasmid (FOXF2) construction and empty vector
was used as negative control (vector). FOXF2-overexpressing
plasmids were purchased form Zhejiang Tianyuan Biotechnology Co.,
Ltd. Transfections were performed in 6-well plates when the cells
were ~70% confluent. For each well, a mixture with 125 µl Opti-MEM,
2.5 µg plasmids or 75 pmol siRNA, and 5 µl Lipofectamine 3000
reagent was prepared. This mixture was added to a solution
containing 7.5 µl Lipofectamine 3000 and 125 µl Opti-MEM. After a
15-min incubation, the solution was added drop-wise to the cells
for 48 or 24 h in a 37°C incubator. Transfection was carried out
according to the Lipo3000 kit manufacturer's instructions (cat. no.
L3000015; Invitrogen; Thermo Fisher Scientific, Inc.). The
sequences of all siRNAs used in the present study were as follows:
siFOXF2-1, 5′-GCUUCAUCAAGCUGCCUAATT-3′; siFOXF2-2,
5′-GCGAGUUCAUGUUCGAGGATT-3′; siMSI2, 5′-AGUGGAAGAUGUAAAGCAATT-3′;
and siNC, 5′-UUCUCCGAACGUGUCACGUTT-3′.
Western blotting (WB)
The total protein was extracted with radio
immunoprecipitation assay (RIPA) lysis buffer (cat. no. P0013B;
Beyotime Institute of Biotechnology,) containing 1% PMSF (cat. no.
ST506; Beyotime Institute of Biotechnology), fully lysed for 5 min
on ice, and then centrifuged at 10,000 × g for 3 min at 4°C. The
concentration of total protein was detected by BCA assay kit. Next,
equal volumes of protein lysate (15 µl, 15–30 µg protein) were
separated by 8–14% SDS-PAGE and electrically transferred into
polyvinylidene difluoride membranes (Thermo Fisher Scientific,
Inc.). The membranes were then blocked with 5% BSA (Biosharp Life
Sciences) for 60 min at room temperature and incubated with primary
antibodies overnight at 4°C. Subsequently, the membranes were
incubated with secondary antibodies at 37°C for 40 min.
Immunoreactive protein bands were visualized with an ECL detection
kit (cat. no. C05-07004; BIOSS) and the images were analyzed by gel
image processing system.
RT-qPCR
RT-qPCR assay was carried out with SYBR Green (cat.
no. SY1020; Beijing Solarbio Science & Technology Co., Ltd.) as
previously described with slight modifications (8). In brief, extracted RNA was reverse
transcribed to cDNA by using a kit (cat. no. D7160L; Beyotime
Institute of Biotechnology,) according to the manufacturer's
instructions, and then it was analyzed using a real time
fluorescence quantitative PCR instrument (Bioneer Corporation). The
thermocycling conditions for qPCR were as follows: 94°C for 5 min,
94°C for 15 sec, 60°C for 25 sec and 72°C for 30 sec with 40
cycles, followed by 72°C for 5.5 min, 40°C for 2.5 min, melting
60°C to 94°C, every 1.0°C for 1 sec and 25°C for 1–2 min. The data
were calculated using the 2-ΔΔCq method (19) and presented as relative expression
fold change. The value in controls was arbitrarily set as 1. The
primers were synthesized by Sangon Biotech Co., Ltd. and sequences
were as follows: FOXF2 forward, 5′-CAGGGCTGGAAGAACTCGG-3′ and
reverse, 5′-CGGTGGTACATGGGCTTGA-3′; MSI2 forward,
5′-CCCAGCAAGTGTAGATAAAG-3′ and reverse, 5′-GTGACAAAGCCAAACCC-3′;
and β-actin forward, 5′-CACTGTGCCCATCTACGAGG-3′ and reverse,
5′-TAATGTCACGCACGATTTCC-3′.
CCK-8 assay
The cell proliferation was measured by using CCK-8
assay kit according to the manufacturer's instructions. After 24 h
transfection, cells (4×103/well) were seeded in 96-well plates with
five replicates and incubated at 37°C with 5% CO2 for 0, 24, 48 and
72 h. The co-transfected cells were incubated at same conditions
for 48 h. Each well was added with 10 µl CCK-8 solution and
incubated at 37°C for 48 h. The results were determined by the
optical density values of absorbance at 450 nm.
EdU staining assay
According to protocol of the EdU assay kit, cells
(5×104/well) were cultured with preheated EdU solution in 24-well
plates at 37°C for 2 h. Next, the cells were fixed with 4%
paraformaldehyde for 15 min and then incubated with 0.5% Triton
X-100 at room temperature for 20 min. After being dyed with
2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI, 1
µg/ml) for 5 min, the stained cells were observed under a
fluorescence microscope (Olympus Corporation).
Cell cycle analysis
Cells were washed with cold phosphate-buffered
saline and fixed in 75% ethanol overnight at 4°C. Next, the cells
were dyed using 25 µl propidium iodide (PI) solution and incubated
in darkness for 30 min with 10 µl RNase A at 37°C, followed by
detection using NovoCyte flow cytometer (ACEA Bioscience, Inc.) and
analysis by a NovoExpress software (NovoExpress 1.4.1; Agilent
Technologies, Inc.).
Cell apoptosis analysis
Cell apoptosis was analyzed by using the Annexin
V-FITC/PI dual-staining kits according to the manufacturer's
protocol and it was finally detected using NovoCyte flow
cytometer.
Hoechst staining
The transfected cells (1×105/well) were seeded in
12-well plates, incubated at 37°C with 5% CO2 for 48 h, and stained
with Hoechst staining solution for 5 min. The apoptotic cells were
observed under an inverted fluorescent microscope (Olympus
Corporation).
Wound healing assay
Cell migration was detected by wound healing assay.
Briefly, the confluent cells were serum-starved and treated with 1
µg/ml Mitomycin C (cat. no. M0503; Sigma-Aldrich; Merck KGaA) at
37°C for 1 h. Subsequently, cells were scratched using a pipette
tip and cultured at 37°C with 5% CO2 for 24 h. The distance
migrated by the cells into the wound was measured.
Transwell assay
The invasive ability of cells was determined by
Matrigel-coated Transwell inserts (0.4µm) (LABSELECT, http://www.labselect.cn/index/product/details/language/cn/product_id/149.html).
Matrigel was precoated on the inserts at 37°C for 2 h in the
24-well plate. After 24 h transfection, the cell suspension (5×104)
was seeded into the upper chamber of this system with no medium.
The lower chamber was added RPMI-1640 or DMEM medium containing 10%
FBS. After incubation for 24 h, they were fixed by 4%
paraformaldehyde at room temperature (25°C) and stained with 0.4%
crystal violet. The number of invasive cells was counted under a
light microscope in five randomly selected fields.
Luciferase reporter assay
A total of two potential binding sites were
predicted followed by designing two reporter plasmids to explore
how FOXF2 binds to the MSI2 promoter regions: pGL3-MSI2 promoter
(−1987/+11) and pGL3-MSI2 promoter (−1824/+11). The pRL-TK and
pGL3-basic plasmids were both from Fenghui Biotechnology Co., Ltd.
Luciferase reporter plasmid constructions were synthesized by Anhui
General Biotechnology Co., Ltd. For the luciferase reporter assay,
293 cells (purchased frOm iCell company) were co-transfected with
the reporter plasmids and the plasmid of FOXF2 or vector by using a
Lipo3000 kit. After being cultured for 48 h, luciferase activity
was measured by the dual luciferase reporter gene assay kit
according to the manufacturer's instructions. The ratio of firefly
luciferase intensity/Renilla luciferase intensity indicated
a relative luciferase activity.
ChIP-PCR assay
ChIP-PCR assay was performed using a ChIP Assay Kit.
Briefly, the collected cells were lysed in SDS lysis buffer and
then sonicated to obtain DNA fragments. Next, protein-DNA complexes
were respectively precipitated by IgG or FOXF2 antibody (1:100),
followed by complex elution. The immunoprecipitated DNA was
amplified with 2X Taq PCR MasterMix (cat. no. PC1150; Beijing
Solarbio Science & Technology Co., Ltd.). The thermocycling
conditions for qPCR were as follows: 95°C for 5 min, 95°C for 20
sec, 55°C for 20 sec and 72°C for 30 sec with 40 cycles, followed
by 72°C for 2 min, 25°C for 5 min. The sequences of a pair primer
specific for the MSI2 promoter were as follows: Forward,
5′-CTGTTGTCTGCATTTTG-3′ and reverse, 5′-GTCTCCCTGTTCCCTAA-3′).
Agarose gel electrophoresis (2.0%; containing gold view nucleic
acid dye) was performed on electrophoresis apparatus (Beijing Liuyi
Instrument Factory), and the images was captured by Gel Imaging
System (WD-9413B; Beijing Liuyi Biotechnology).
Xenograft tumor model
Male BALB/C nude mice (aged 4 months; weight, 20–25
g) were used to establish xenograft tumor model. Animals were
housed in a 12/12-h of light/dark cycle at 22°C with 45–55%
humidity and adaptively provided with free access to water and
food. All animal experiments were performed according to the
National Institutes of Health Guide for the Care and Use of
Laboratory Animals. All animal studies were approved (approval no.
KT2021051) by the Laboratory Animal Welfare and Ethics Committee of
China Medical University (Shenyang, China). A total of 24 mice were
used in the present study. Subcutaneous injection with PC cells
(5×106 cells) resuspended in 200 µl PBS, was used to establish the
PC mouse model (20). The nude mice
were randomly divided into four groups (6 mice per group) that
received subcutaneous injection (at dorsal region) of
vector-transfected AsPC-1 cells, FOXF2-transfected AsPC-1 cells,
shNC-transfected PANC-1 cells and shFOXF2-transfected PANC-1 cells,
respectively. From day 7 after injection, tumor volume was measured
every three days. The tumor volume was calculated as follows:
Volume=0.5 × long diameter × short diameter2. Mice bearing
subcutaneous tumors were euthanized upon reaching humane endpoints
of tumor size. A tumor diameter exceeding 17 mm was considered as
humane endpoint. Mice were euthanized by carbon dioxide
asphyxiation (40% vol/min flow rate in the chamber). Death was
confirmed when the mice were immobile, ceased breathing and with
dilated pupils. The mice were observed for a further 5 min to
confirm their death. The tumor body was removed for measurement of
tumor weight and pathological assessment.
Hematoxylin and eosin (H&E)
staining and IHC staining
H&E staining was performed according to standard
procedures and the results of staining were observed under a light
microscope. For IHC staining, the tissues were fixed in 10%
formalin overnight at 25°C, and embedded in paraffin. Next, 5-µm
paraffin sections were deparaffinized by xylene and rehydrated in
95, 85 and 75% ethanol, each for 3 min. Next, the sections were
subjected to antigen-retrieval with citrate buffer at 95°C for 10
min. After endogenous peroxidase removal by 3% H2O2, the sections
were blocked with 1% BSA for 15 min at room temperature and
incubated overnight with anti-Ki-67 (1:50) or anti-FOXF2 (1:50) at
4°C. A secondary antibody goat anti-rabbit IgG-HRP (1:500) was
utilized to incubate these sections at 37°C for 1 h. The sections
were visualized by diaminobenzidine (DAB) solution (Maxim
Biomedical, Inc.) and finally observed under a light microscope.
Quantitative evaluation of the IHC staining was performed based on
the staining density and stained proportion. The score of stained
proportion was based on a scale of 0–3 point (0, ≤10%; 1, 11–25%;
2, 26–50%; and 3, >51%). The staining density was scored as 0
points, weak staining as 1 point, intermediate staining as 2
points, and strong staining as 3 points. The IHC score was
calculated as percentage score × intensity score. FOXF2 staining in
tissues was classified into two categories (low and high
expression): The specimens with an IHC score 0–4 were defined as
the low FOXF2 expression group and specimens with an IHC score
>4 as the high FOXF2 expression group. Ki-67 index was
calculated as the percentage of Ki-67-positive cells. The specimens
were graded by the Ki-67 index according to the WHO 2010
classification (G1: Ki-67 Index, <3%; G2: Ki-67 Index, 3–20%;
and G3 NET/NEC: Ki-67 Index, >20%) (21).
Statistical analysis
All the experiments were performed with at least
three independent replicates and the data were expressed as the
mean ± SD. Statistical analyses were performed with GraphPad Prism
8 software (GraphPad Software Inc.; Dotmatics). Pearson's
correlation analysis was performed for the expression of MSI2 and
FOXF2. Two-tailed unpaired t-test was used to analyze two groups.
One-way ANOVA followed by a Tukey's test or two-way ANOVA followed
by a Sidak post hoc test was performed for comparing more than two
groups. The Chi-square test and Fisher's exact test were used to
analyze the association between the expression level of FOXF2 and
pathological characteristics in pancreatic cancer. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of FOXF2 in PC
MSI2 is a tumor-promoting factor in PC and a
previous study by the authors elucidated several downstream
regulatory mechanisms of MSI2 in PC development (4). The potential transcription factors
that were predicted to be bound with MSI2 were obtained and the
upstream regulatory mechanism of MSI2 was explored. Moreover, the
differentially expressed genes form PC the dataset GSE16515 were
also screened with criteria of: │log2FoldChange│ >2.5 and
P<0.01. A total of 541 differentially expressed genes were
selected and 291 potential transcription factors were predicted.
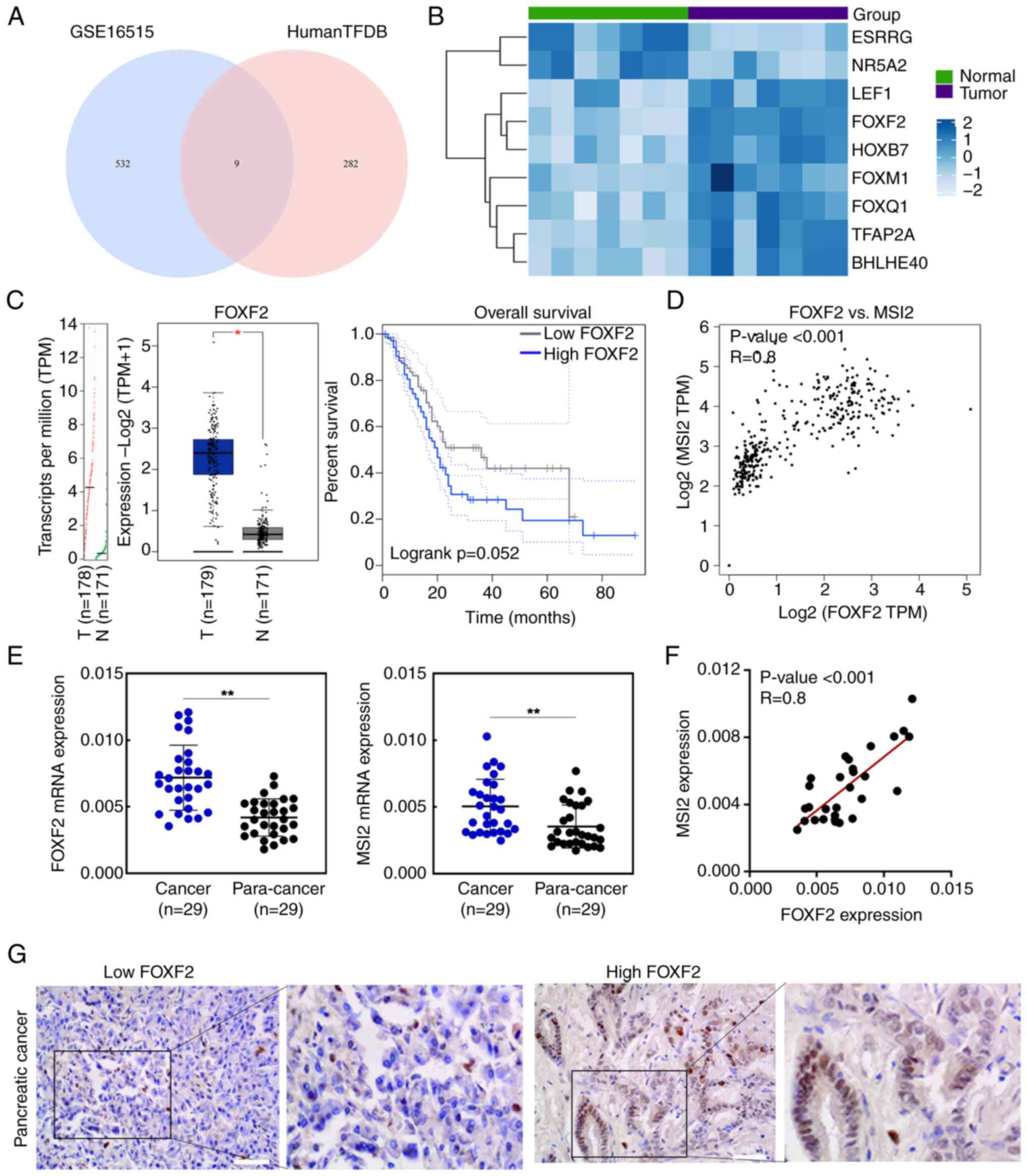
Based on Venn intersection analysis, 9 genes were identified
(Fig. 1A); 2 genes downregulated
and 7 genes upregulated. Among them, FOXF2 was the most markedly
upregulated gene (Fig. 1B).
However, its roles in PC have not been explored. From GEPIA
database, it was also found that the expression level of FOXF2 was
significantly higher in PC tissue than normal tissue (P<0.05)
and high expression of FOXF2 was associated with worse overall
survival in patients with PC (Fig.
1C). Furthermore, the correlation analysis indicated that FOXF2
expression was positively related with MSI2 expression (R=0.8,
P<0.001) (Fig. 1D).
Subsequently, the expression of FOXF2 and MSI2 was detected in 29
pairs of PC tissues and adjacent normal tissues by RT-qPCR. As
shown in Fig. 1E, both the
expression level of FOXF2 and MSI2 were higher in PC tissues than
in adjacent normal tissues (P<0.01). Consistent with the result
from GEPIA database, FOXF2 expression was positively correlative
with MSI2 expression in these paired PC tissues (R=0.8, P<0.001)
(Fig. 1F). Additionally, clinical
samples were analyzed to explore the relationship between FOXF2
expression and clinicopathological characteristics including age,
sex, TNM staging and differentiation. The representative IHC images
of low or high expression level of FOXF2 in clinical PC tissues
were shown in Fig. 1G. Among of 78
specimens, 36 cases were identified as high level of FOXF2 and 42
cases as low level of FOXF2. It was found that FOXF2 expression was
significantly associated with T stage of PC and high expression
level of FOXF2 was associated with an advanced T stage (larger
tumor) (Table I), suggesting that
FOXF2 might contribute to the progression of PC. Besides, patients
with high FOXF2 expression tended to have a higher Ki-67 index. No
significant association was observed between FOXF2 expression and
differentiation in the clinical samples collected. The UALCAN
database indicated that FOXF2 is highly expressed in the PC tissue
with grade 3 compared with normal samples (Fig. S1), but most of the collected
specimens were moderately differentiated, which is responsible for
this absence of association in the present study.
 | Table I.Association between the expression
level of FOXF2 and pathological characteristics in pancreatic
cancer. |
Table I.
Association between the expression
level of FOXF2 and pathological characteristics in pancreatic
cancer.
|
|
| Expression level of
FOXF2 |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristics | Number of
cases | High (36) | Low (42) | P-value |
|---|
| Age, years |
|
|
| 0.082 |
|
<60 | 32 | 11 | 21 |
|
|
≥60 | 46 | 25 | 21 |
|
| Sex |
|
|
| 0.064 |
|
Male | 41 | 23 | 18 |
|
|
Female | 37 | 13 | 24 |
|
| T stage |
|
|
| 0.0029 |
| T1 +
T2 | 68 | 27 | 41 |
|
| T3 +
T4 | 10 | 9 | 1 |
|
| N stage |
|
|
| 0.738 |
| N0 | 60 | 29 | 31 |
|
| N1 | 16 | 6 | 10 |
|
| N2 | 2 | 1 | 1 |
|
| American Joint
Committee on |
|
|
| 0.892 |
| Cancer TNM
staging |
|
|
|
|
|
I/II | 75 | 34 | 41 |
|
|
III | 3 | 2 | 1 |
|
|
Differentiation |
|
|
| 0.066 |
|
High | 12 | 9 | 3 |
|
|
Middle | 53 | 23 | 30 |
|
|
Low | 13 | 4 | 9 |
|
| Ki-67 index |
|
|
| 0.0003 |
| G1 | 13 | 0 | 13 |
|
| G2 | 12 | 4 | 8 |
|
| G3 | 53 | 32 | 21 |
|
FOXF2 accelerates PC cell
proliferation
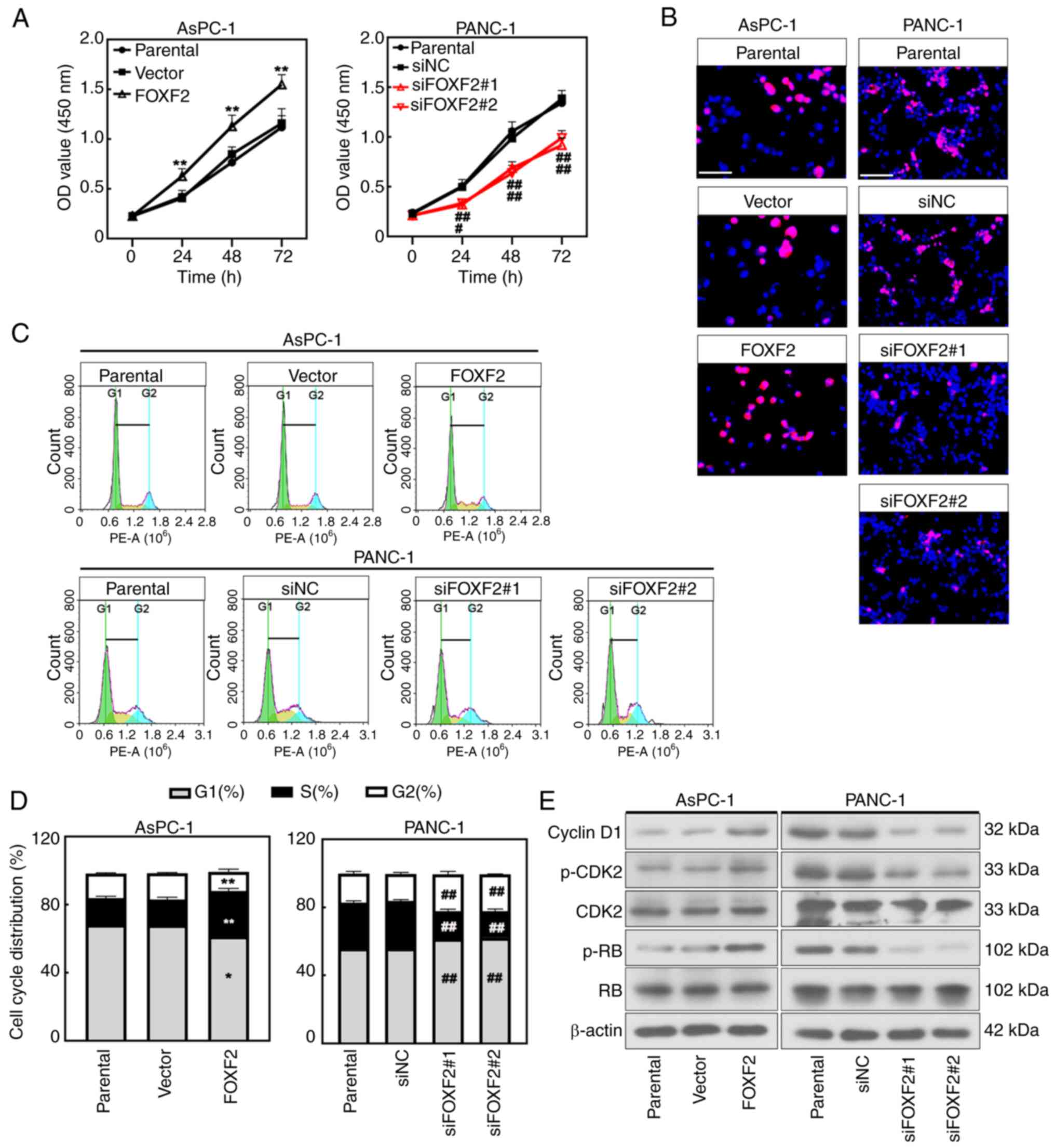
FOXF2 was efficiently silenced or overexpressed
within AsPC-1 cells or PANC-1 cells at mRNA and protein levels
after transfection (P<0.01) (Fig.
S2A and B). CCK-8 assay revealed that the proliferation of the
cells with FOXF2 overexpression was significantly higher than in
vector-transfected cells (P<0.01), and FOXF2 knockdown
significantly decreased proliferation of PANC-1 cells (P<0.05)
(Fig. 2A). Consistently, the EdU
staining results presented that FOXF2 overexpression augmented
proliferation and FOXF2 knockdown decreased it (Fig. 2B). The cell cycle analysis suggested
that the proportion of cells in G1/G2 phase declined and the
proportion in S phase was increased after FOXF2 overexpression
(P<0.01). Compared with the siNC-transfected cells, the
percentage of cells in G1/G2 phase was increased and the percentage
in S phase was decreased in the FOXF2-silenced cells (P<0.01)
(Fig. 2C and D). It was indicated
that FOXF2 contributed to the G1-S phase transition in PC cells.
Cyclin D1, CDK2, p-CDK2 and p-RB are major cell cycle regulatory
proteins and primarily involve G1/S phase transition (22). The expression level of these
proteins was increased after FOXF2 overexpression while it was
decreased in the FOXF2-slienced cells (Fig. 2E). The aforementioned results
indicated that FOXF2 promotes the proliferation of PC cells by
accelerating G1-S phase transition.
FOXF2 knockdown induces apoptosis of
PC cells
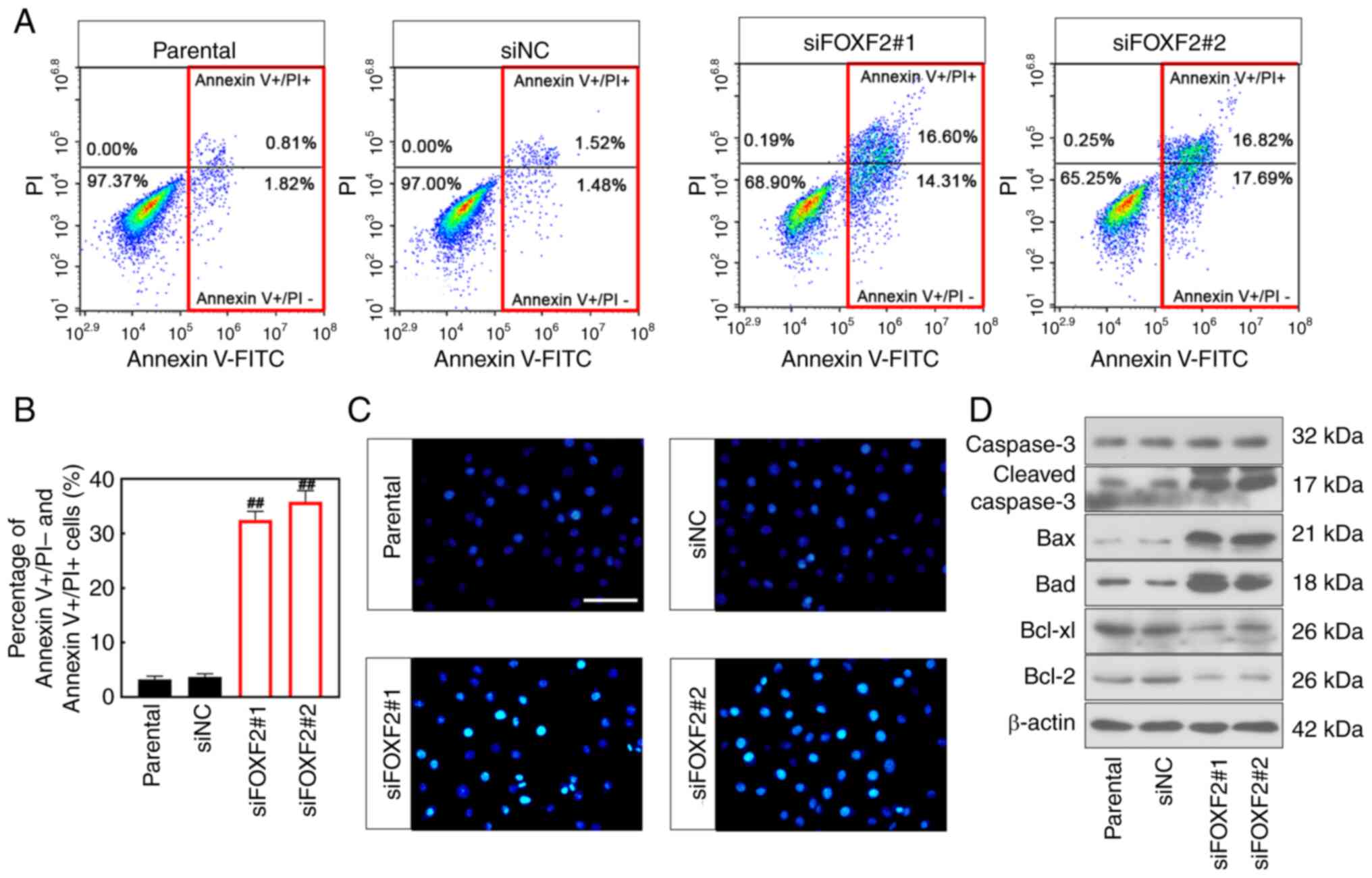
Flow cytometric analysis showed that cell apoptosis
was increased after FOXF2 was silenced (P<0.01) (Fig. 3A and B). Consistently, Hoechst
staining manifested that FOXF2 knockdown increased the number of
apoptotic cells (Fig. 3C). Besides,
the expression of anti-apoptotic proteins (Bcl-2 and Bcl-xl) was
downregulated in the FOXF2-silenced cells. The expression of
pro-apoptotic proteins (Bad, Bax and Cleaved capase-3) was
upregulated in the cells with FOXF2 knockdown (Fig. 3D). Collectively, it was indicated
that knockdown of FOXF2 induces apoptosis of PC cells.
FOXF2 promotes migration and invasion
of PC cells
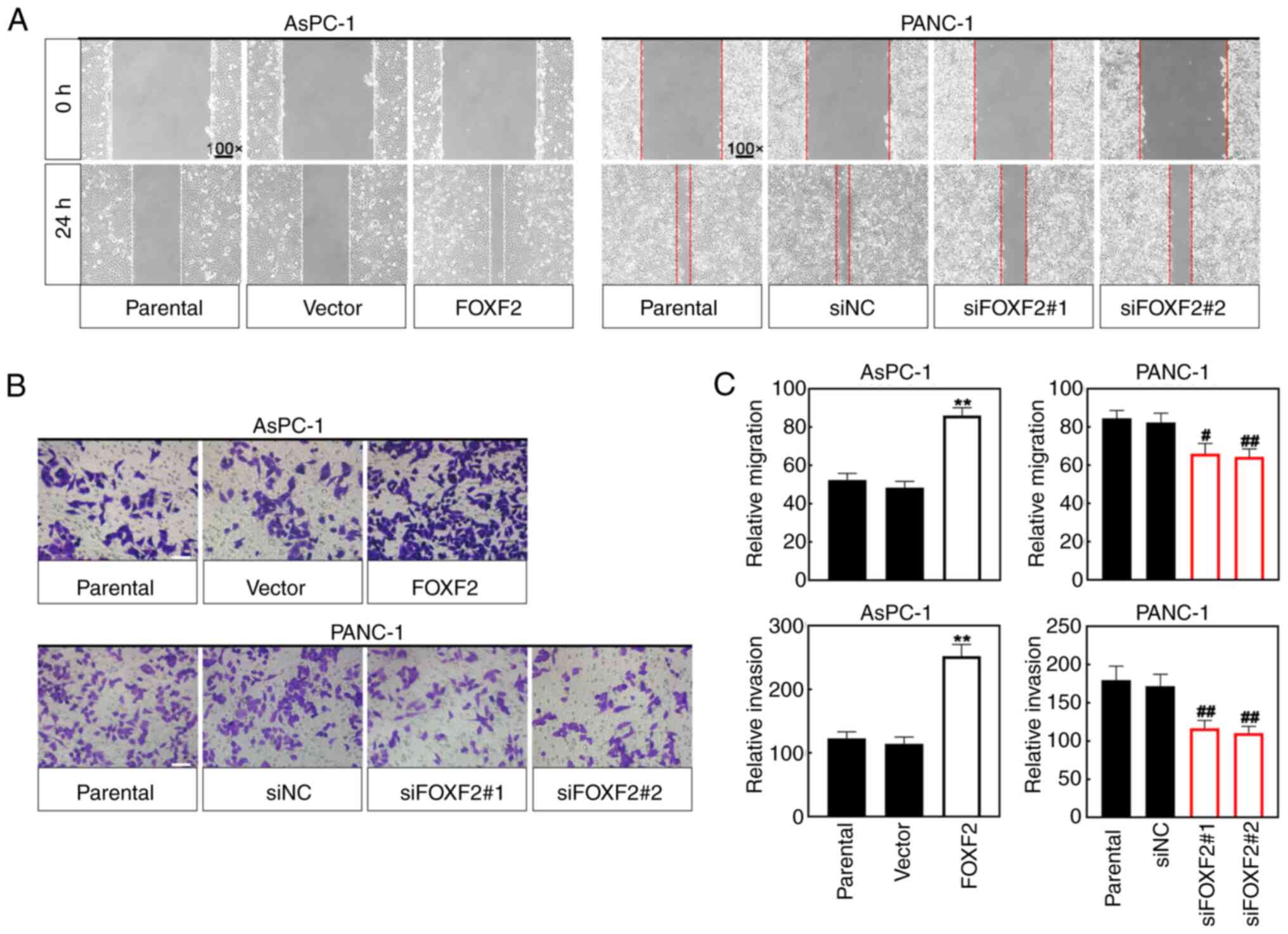
Cell migration was assessed using the wound healing
assay. The wound closure area of FOXF2-overexpresseing cells was
less wide than that of vector-transfected cells, and the wound
closure area of FOXF2-silenced cells was smaller than that of
siNC-transfected cells (Fig. 4A).
Transwell assay was performed for cell invasion. As shown in
Fig. 4B and C, FOXF2 overexpression
increased the number of invasive cells and FOXF2 knockdown
decreased it. These results suggested that FOXF2 may act as a
tumor-progressing factor to enhance the migration and invasion of
PC cells.
FOXF2 promotes malignant behavior of
PC cells possibly by regulating MSI2
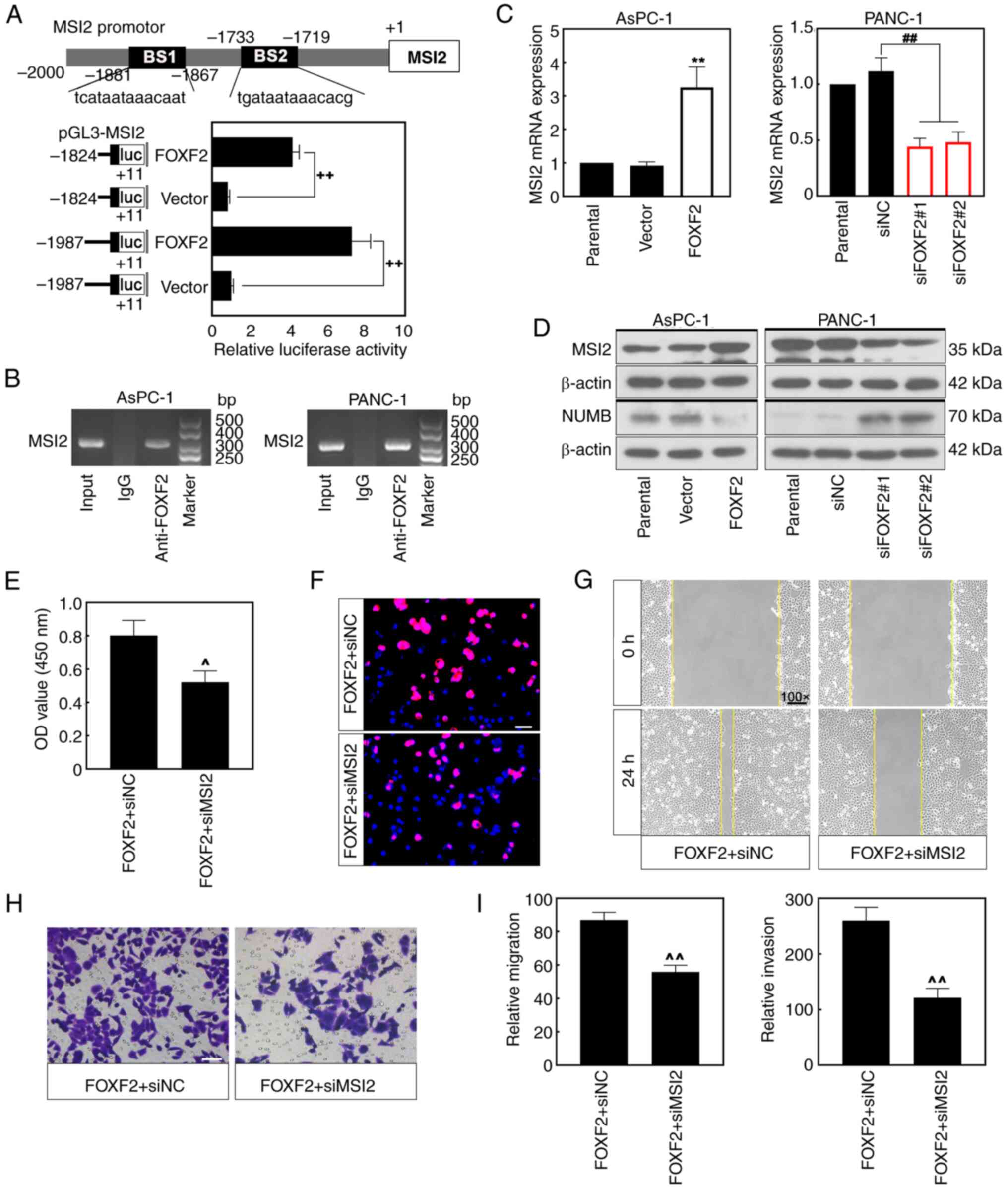
Analysis through the bioinformatics prediction
website (https://jaspar.genereg.net/)
predicted that FOXF2 could be a potential regulator of MSI-2. A
total of two luciferase reporter plasmids with the fragment of MSI2
promotor region were constructed to explore how FOXF2 regulates
MIS2 in PC cells. Luciferase assays revealed that FOXF2 bound to
the MSI2 promoter and FOXF2 overexpression significantly enhanced
the luciferase activity of MSI2 reporter plasmid (P<0.01)
(Fig. 5A). Moreover, this result
was validated within PC cells by ChIP assay (Fig. 5B). The mRNA expression of MSI2 was
increased in the FOXF2-overexpressing cells and decreased in the
FOXF2-silenced cells (P<0.01) (Fig.
5C). MSI2 protein expression showed similar trends as mRNA
level (Fig. 5D). NUMB protein, a
tumor suppressor that is negatively regulated by MSI2, was markedly
decreased in the cells with FOXF2 overexpression and increased in
the cells with FOXF2 knockdown (Fig.
5D). AsPC-1 cells were co-transfected with using
FOXF2-overexpressing plasmid and interference sequence siMSI2 to
investigate the regulatory mechanism between FOXF2 and MSI2. CCK-8
assay and EdU staining revealed that MSI2 silencing reversed the
promoting effect of FOXF2 on proliferation of PC cells (Fig. 5E and F). Additionally, MSI2
knockdown weakened the facilitation of FOXF2 to the migration and
invasion of PC cells (Fig. 5G-I).
These results indicated that FOXF2 can bind to the promoter region
of MSI2 and promote proliferation, migration and invasion of PC
cells possibly by regulating MSI2.
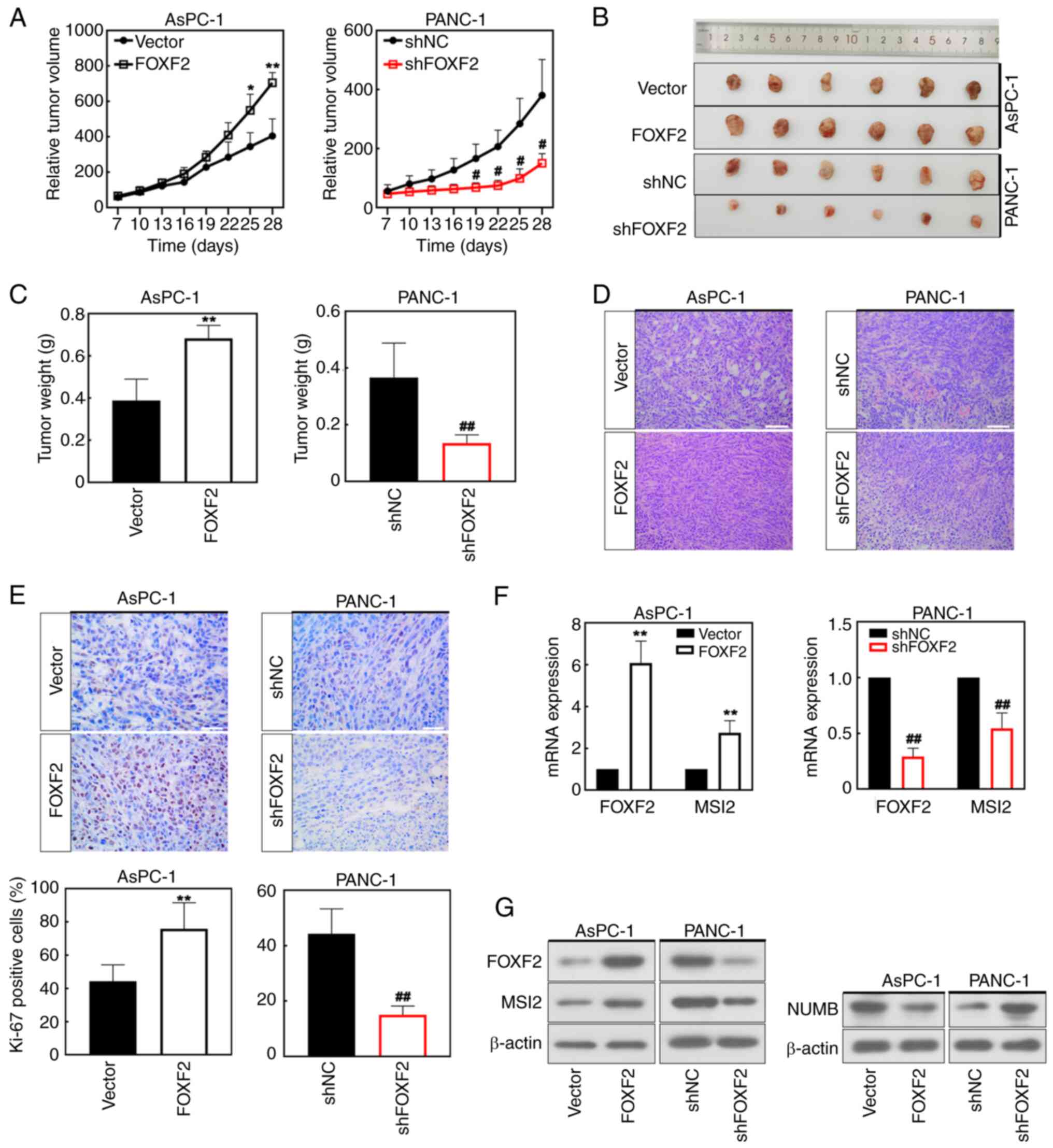
FOXF2 accelerates tumor formation
A xenograft tumor model of PC was constructed to
validate the roles of FOXF2 in the development of tumor in
vivo. Mice were injected with the FOXF2-transfected AsPC-1
cells or shFOXF2-transfected PANC-1 cells, respectively. FOXF2
overexpression accelerated the tumor growth while FOXF2 knockdown
showed opposite effects (Fig. 6A and
B). Results of tumor weight shared the same alteration with
those of tumor volume (Fig. 6C).
H&E staining demonstrated that FOXF2 maintained the integrity
of PC cells and FOXF2 silencing induced the destruction of cell
structure (Fig. 6D). In addition,
IHC staining indicated that FOXF2 overexpression improved Ki-67
expression (Fig. 6E), suggesting
that FOXF2 accelerated the formation of pancreatic tumor in
vivo. The results of WB and RT-qPCR further revealed that MSI2
expression was significantly increased in the FOXF2-overexpressing
mice and decreased in FOXF2-silenced mice (P<0.01) (Fig. 6F). NUMB protein expression showed
opposite trends. FOXF2 overexpression downregulated NUMB while
FOXF2 knockdown upregulated it in PC tissues (Fig. 6G). Hence, MSI2 could be the target
of FOXF2 to promote tumor growth of PC.
Discussion
PC is one of the cancer types with high mortality
and featured with insidious onset and atypical early symptoms
(23). PC is confused with other
digestive diseases, presenting with upper abdominal discomfort,
lower back pain, dyspepsia, or diarrhea (24). Current treatments are challenging to
benefit all patients because of the inter-individual heterogeneity
of PC (4). The average survival is
only ~2 years even for patients with PC with surgical resection
(25). Therefore, an improved
understanding of the molecular mechanisms involved in PC
progression is urgently required.
The roles of FOXF2 are well elucidated across
various tumors. However, neither the cellular roles nor the
regulatory mechanism of FOXF2 in PC has been reported. The current
study observed that FOXF2 was highly expressed in PC tissues,
particularly in patients with PC at advanced T stage. The present
results indicated that high expression of FOXF2 accelerated the
proliferation of PC cells in vitro and in vivo.
Similar results were reported in rhabdomyosarcoma cells (12). Additionally, FOXF2 significantly
prevented the apoptosis of PC cells. The migration and invasion of
PC cells were suppressed by FOXF2 knockdown. Consistently, the
promotive effects of FOXF2 on migration and invasion capability
were reported in breast cancer cells and lung cancer cells
(26,27). On the contrary, FOXF2 inhibited the
malignant behavior of colorectal cancer cells (28). FOXF2 suppressed G1-S cell-cycle
transition of gastric cancer cell and induced cell apoptosis
(22). Therefore, the effect of
FOXF2 on tumors depends on the type of cells. The current findings
suggested that FOXF2 exerts promotive effects on the growth of
PC.
Accumulating evidence elucidated that MSI2
positively regulates the initiation and progression of cancer.
Kudinov, Deneka, Nikonova, Beck, Ahn, Liu, Martinez, Schultz,
Reynolds, Yang, Cai, Yaghmour, Baker, Egleston, Nicolas, Chikwem,
Andrianov, Singh, Borghaei, Serebriiskii, Gibbons, Kurie, Golemis
and Boumber (7) reported that MSI2
overexpression enhanced invasion of non-small cell lung cancer
cells. MSI2 promoted migration and invasion of PC cells and thus
accelerating the progression of PC (4,29). As
a transcription factor, FOXF2 involves in the pathogenesis of
various tumors through regulating the transcription of downstream
genes (30,31). Thus, FOXF2 initiated the
transcription of MSI2 in PC cells, and FOXF2 silencing reduced MSI2
expression at transcriptional and translational levels in
vitro and in vivo. Additionally, MSI2 silencing
significantly reversed the promotive effects of FOXF2 on
proliferation, invasion and migration of PC cells. These findings
suggested that MSI2 was the downstream target of FOXF2 in PC. It
was previously reported that MSI2 promoted the development and
progression of PC by downregulating NUMB protein (8). The present study also found that FOXF2
knockdown upregulated NUMB at the protein level in vitro and
in vivo. NUMB is a tumor suppressor in several types of
tumors (32). The expression of
NUMB was downregulated in the esophageal cancer cells, and
deficiency of NUMB was associated with poor prognosis and more
aggressive tumors in breast and lung cancer (33,34).
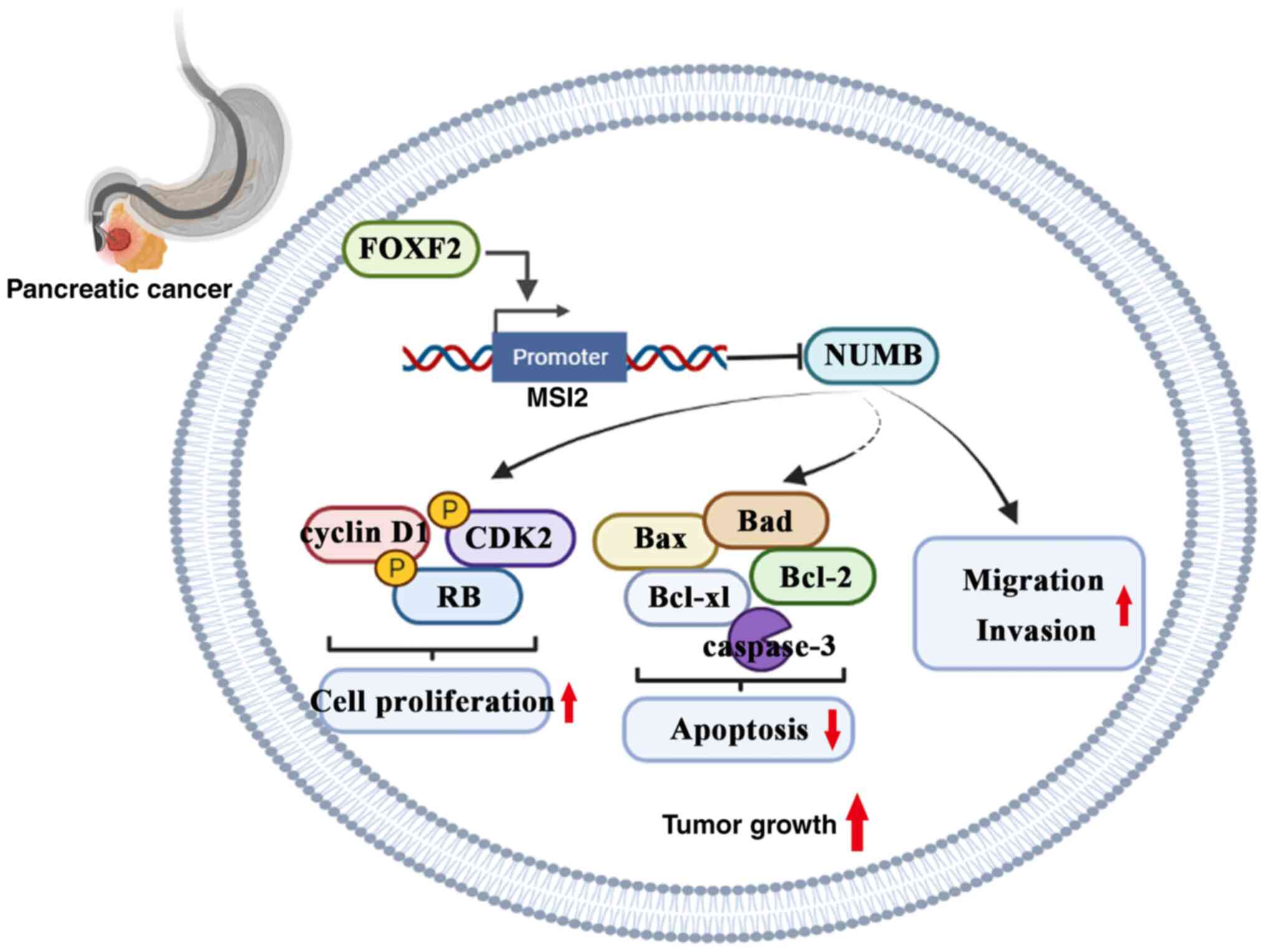
Collectively, FOXF2 promotes malignant behavior of PC cells
possibly through mediating the transcription of MSI2 to
downregulate NUMB protein (Fig. 7).
Additionally, a previous study by the authors demonstrated that
MSI2 promotes invasion and migration of PC cells by upregulating
wtp53 protein (9). The
ZEB1-ERK/MAPK and ISYNA1-p21/ZEB-1 signaling pathways are involved
in the MSI2 regulatory mechanism to facilitate development of PC
(4,29). Therefore, FOXF2 could act as an
upstream regulator involving in these pathways, which could be
explored and verified in further research.
FOXF2 binds to the promoter of MSI2 and promotes its
transcriptional expression. The cell migration and invasion are
decreased by MSI2 knockdown. Therefore, it was postulated that
FOXF2 promoted PC progression possibly via regulating MSI2
transcription. Nevertheless, the upstream regulatory mechanisms of
FOXF2 should be elucidated in further researches. In addition to
being a target gene of microRNA, FOXF2 is upregulated by
specificity protein 1 (SP1), myc-associated zinc-finger protein
(MAZ), TGF-β and MCM3AP-AS1 (35–38).
Safe, Shrestha, Mohankumar, Howard, Hedrick and Abdelrahim
(39) found that SP1 accelerates
cell proliferation and migration of PC cells. MAZ is reported to
promote invasiveness of PC cells (40). MCM3AP-AS1 promotes cell migration in
PC (41). These factors may cause
the upregulation of FOXF2 in PC and FOXF2 perhaps mediates their
tumor-promoting effect on PC. Further in-depth studies are needed
to ascertain the precise molecular regulation of FOXF2 in the
development of PC. Besides, an insufficient sample size of clinical
specimens limited the exploration of FXOF2 expression and tumor
grade. These issues are the current limitations of the present
study, and which will be improved in future research.
In summary, FOXF2 promotes proliferation, migration
and invasion of PC cells. MSI2 was a transcription regulatory
target of FOXF2. FOXF2 accelerates the pancreatic tumor development
and progression possibly by regulating an MSI2-NUMB interaction.
These findings provide a novel therapy target for PC treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Social Development
Program from Shenyang Science and Technology Bureau, China (grant
no. F20-205-4-033).
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article.
Authors' contributions
MD and YTM contributed to the study conception and
design. BHZ, JS and JTT performed material preparation, experiments
and analysis. BHZ wrote the first draft of the manuscript and all
authors commented on previous versions of the manuscript. All
authors read and approved the final manuscript. BHZ and MD confirm
the authenticity of all the raw data.
Ethics approval and consent to
participate
All animal studies were approved (approval no.
KT2021051) by the Laboratory Animal Welfare and Ethics Committee of
China Medical University (Shenyang, China). All experiments using
human tissue were approved [approval no. (2021) 113] by the Medical
Science Research Ethics Committee of The First Affiliated Hospital
of China Medical University (Shenyang, China). Written informed
consent for the collection of tissue samples was provided by all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vincent A, Herman J, Schulick R, Hruban RH
and Goggins M: Pancreatic cancer. Lancet. 378:607–620. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wood LD, Canto MI, Jaffee EM and Simeone
DM: Pancreatic cancer: Pathogenesis, screening, diagnosis, and
treatment. Gastroenterology. 163:386–402. e12022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nakamura M, Okano H, Blendy JA and Montell
C: Musashi, a neural RNA-binding protein required for Drosophila
adult external sensory organ development. Neuron. 13:67–81. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sheng W, Shi X, Lin Y, Tang J, Jia C, Cao
R, Sun J, Wang G, Zhou L and Dong M: Musashi2 promotes EGF-induced
EMT in pancreatic cancer via ZEB1-ERK/MAPK signaling. J Exp Clin
Cancer Res. 39:162020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dong P, Xiong Y, Hanley SJB, Yue J and
Watari H: Musashi-2, a novel oncoprotein promoting cervical cancer
cell growth and invasion, is negatively regulated by p53-induced
miR-143 and miR-107 activation. J Exp Clin Cancer Res. 36:1502017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kharas MG, Lengner CJ, Al-Shahrour F,
Bullinger L, Ball B, Zaidi S, Morgan K, Tam W, Paktinat M, Okabe R,
et al: Musashi-2 regulates normal hematopoiesis and promotes
aggressive myeloid leukemia. Nat Med. 16:903–908. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kudinov AE, Deneka A, Nikonova AS, Beck
TN, Ahn YH, Liu X, Martinez CF, Schultz FA, Reynolds S, Yang DH, et
al: Musashi-2 (MSI2) supports TGF-β signaling and inhibits claudins
to promote non-small cell lung cancer (NSCLC) metastasis. Proc Natl
Acad Sci USA. 113:6955–6960. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sheng W, Dong M, Chen C, Li Y, Liu Q and
Dong Q: Musashi2 promotes the development and progression of
pancreatic cancer by down-regulating Numb protein. Oncotarget.
8:14359–14373. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sheng W, Dong M, Chen C, Wang Z, Li Y,
Wang K, Li Y and Zhou J: Cooperation of Musashi-2, Numb, MDM2, and
P53 in drug resistance and malignant biology of pancreatic cancer.
FASEB J. 31:2429–2438. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang S, Li GX, Tan CC, He R, Kang LJ, Lu
JT, Li XQ, Wang QS, Liu PF, Zhai QL and Feng YM: FOXF2 reprograms
breast cancer cells into bone metastasis seeds. Nat Commun.
10:27072019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He W, Kang Y, Zhu W, Zhou B, Jiang X, Ren
C and Guo W: FOXF2 acts as a crucial molecule in tumours and
embryonic development. Cell Death Dis. 11:4242020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Milewski D, Pradhan A, Wang X, Cai Y, Le
T, Turpin B, Kalinichenko VV and Kalin TV: FoxF1 and FoxF2
transcription factors synergistically promote rhabdomyosarcoma
carcinogenesis by repressing transcription of p21Cip1 CDK
inhibitor. Oncogene. 36:850–862. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hauptman N, Jevšinek Skok D, Spasovska E,
Boštjančič E and Glavač D: Genes CEP55, FOXD3, FOXF2, GNAO1, GRIA4,
and KCNA5 as potential diagnostic biomarkers in colorectal cancer.
BMC Med Genomics. 12:542019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang J, Zhang C, Sang L, Huang L, Du J
and Zhao X: FOXF2 inhibits proliferation, migration, and invasion
of Hela cells by regulating Wnt signaling pathway. Biosci Rep.
38:BSR201807472018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang A, Jin C, Li H, Qin Q and Li L:
LncRNA ADAMTS9-AS2 regulates ovarian cancer progression by
targeting miR-182-5p/FOXF2 signaling pathway. Int J Biol Macromol.
120:1705–1713. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lo PK: FOXF2 differentially regulates
expression of metabolic genes in non-cancerous and cancerous breast
epithelial cells. Trends Diabetes Metab. 1:10.15761/TDM.1000103.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lo PK, Lee JS, Liang X and Sukumar S: The
dual role of FOXF2 in regulation of DNA replication and the
epithelial-mesenchymal transition in breast cancer progression.
Cell Signal. 28:1502–1519. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pei H, Li L, Fridley BL, Jenkins GD,
Kalari KR, Lingle W, Petersen G, Lou Z and Wang L: FKBP51 affects
cancer cell response to chemotherapy by negatively regulating Akt.
Cancer Cell. 16:259–266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xia G, Wang H, Song Z, Meng Q and Huang X
and Huang X: Gambogic acid sensitizes gemcitabine efficacy in
pancreatic cancer by reducing the expression of ribonucleotide
reductase subunit-M2 (RRM2). J Exp Clin Cancer Res. 36:1072017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou Q and Gallo JM: Differential effect
of sunitinib on the distribution of temozolomide in an orthotopic
glioma model. Neuro Oncol. 11:301–310. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Higashimori A, Dong Y, Zhang Y, Kang W,
Nakatsu G, Ng SSM, Arakawa T, Sung JJY, Chan FKL and Yu J: Forkhead
Box F2 suppresses gastric cancer through a novel
FOXF2-IRF2BPL-β-catenin signaling axis. Cancer Res. 78:1643–1656.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Torphy RJ, Fujiwara Y and Schulick RD:
Pancreatic cancer treatment: Better, but a long way to go. Surg
Today. 50:1117–1125. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu H, Wei M, Xu J, Hua J, Liang C, Meng
Q, Zhang Y, Liu J, Zhang B, Yu X and Shi S: PARP inhibitors in
pancreatic cancer: Molecular mechanisms and clinical applications.
Mol Cancer. 19:492020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Heinrich S and Lang H: Neoadjuvant therapy
of pancreatic cancer: Definitions and benefits. Int J Mol Sci.
18:16222017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang T, Wan JG, Liu JB and Deng M:
MiR-200c inhibits metastasis of breast tumor via the downregulation
of Foxf2. Genet Mol Res. 16:gmr160389712017. View Article : Google Scholar
|
|
27
|
Kundu ST, Byers LA, Peng DH, Roybal JD,
Diao L, Wang J, Tong P, Creighton CJ and Gibbons DL: The miR-200
family and the miR-183~96~182 cluster target Foxf2 to inhibit
invasion and metastasis in lung cancers. Oncogene. 35:173–186.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen J, Ding J, Wang Z, Zhu J, Wang X and
Du J: Identification of downstream metastasis-associated target
genes regulated by LSD1 in colon cancer cells. Oncotarget.
8:19609–19630. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou L, Sheng W, Jia C, Shi X, Cao R, Wang
G, Lin Y, Zhu F, Dong Q and Dong M: Musashi2 promotes the
progression of pancreatic cancer through a novel ISYNA1-p21/ZEB-1
pathway. J Cell Mol Med. 24:10560–10572. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li T, Huang S, Yan W, Zhang Y and Guo Q:
FOXF2 regulates PRUNE2 transcription in the pathogenesis of
colorectal cancer. Technol Cancer Res Treat.
21:153303382211187172022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu JT, Tan CC, Wu XR, He R, Zhang X, Wang
QS, Li XQ, Zhang R and Feng YM: FOXF2 deficiency accelerates the
visceral metastasis of basal-like breast cancer by unrestrictedly
increasing TGF-β and miR-182-5p. Cell Death Differ. 27:2973–2987.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Choi HY, Seok J, Kang GH, Lim KM and Cho
SG: The role of NUMB/NUMB isoforms in cancer stem cells. BMB Rep.
54:335–343. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Colaluca IN, Tosoni D, Nuciforo P,
Senic-Matuglia F, Galimberti V, Viale G, Pece S and Di Fiore PP:
NUMB controls p53 tumour suppressor activity. Nature. 451:76–80.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pece S, Serresi M, Santolini E, Capra M,
Hulleman E, Galimberti V, Zurrida S, Maisonneuve P, Viale G and Di
Fiore PP: Loss of negative regulation by Numb over Notch is
relevant to human breast carcinogenesis. J Cell Biol. 167:215–221.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tian HP, Lun SM, Huang HJ, He R, Kong PZ,
Wang QS, Li XQ and Feng YM: DNA methylation affects the
SP1-regulated transcription of FOXF2 in breast cancer cells. J Biol
Chem. 290:19173–19183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu ZH, Lun SM, He R, Tian HP, Huang HJ,
Wang QS, Li XQ and Feng YM: Dual function of MAZ mediated by FOXF2
in basal-like breast cancer: Promotion of proliferation and
suppression of progression. Cancer Lett. 402:142–152. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Meyer-Schaller N, Heck C, Tiede S, Yilmaz
M and Christofori G: Foxf2 plays a dual role during transforming
growth factor beta-induced epithelial to mesenchymal transition by
promoting apoptosis yet enabling cell junction dissolution and
migration. Breast Cancer Res. 20:1182018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dai W, Zeng W and Lee D: lncRNA MCM3AP-AS1
inhibits the progression of colorectal cancer via the
miR-19a-3p/FOXF2 axis. J Gene Med. 23:e33062021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Safe S, Shrestha R, Mohankumar K, Howard
M, Hedrick E and Abdelrahim M: Transcription factors specificity
protein and nuclear receptor 4A1 in pancreatic cancer. World J
Gastroenterol. 27:6387–6398. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Maity G, Haque I, Ghosh A, Dhar G, Gupta
V, Sarkar S, Azeem I, McGregor D, Choudhary A, Campbell DR, et al:
The MAZ transcription factor is a downstream target of the
oncoprotein Cyr61/CCN1 and promotes pancreatic cancer cell invasion
via CRAF-ERK signaling. J Biol Chem. 293:4334–4349. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu X, Zheng Q, Zhang Q, Zhang S, He Y and
Guo W: MCM3AP-AS1: An indispensable cancer-related LncRNA. Front
Cell Dev Biol. 9:7527182021. View Article : Google Scholar : PubMed/NCBI
|