Introduction
Cancer is a complex disease that can be described as an uncontrolled and abnormal proliferation of cells. Most of the time these abnormal cells can spread and invade surrounding tissues, even migrating and reaching some other body parts by using bloodstream or lymphatic system (1).
Head and neck squamous cell carcinoma (HNSCC) is a type of complex malignant tumour originating from the mucosal epithelium in the mouth, pharynx, nose, throat and laryngeal region (2). HNSCC has unique anatomical structure and unpredicted tumour microenvironment (3). HNSCC originates from the flat squamous cells, a thin layer of tissue on the surface of head and neck structures (4). If this cancer is limited to the squamous cell layer is called carcinoma in situ and if HNSCC cancer cells proliferate beyond this cell layer and invade deeper tissues, it is classified as invasive (5). Adoption and use of the tumour, lymph node and metastasis (TNM) cancer staging system (Fig. 1) allows clinicians and researchers to categorise complex tumours of the head and neck region in a defined way that can help in treatment plans and disease management (6).
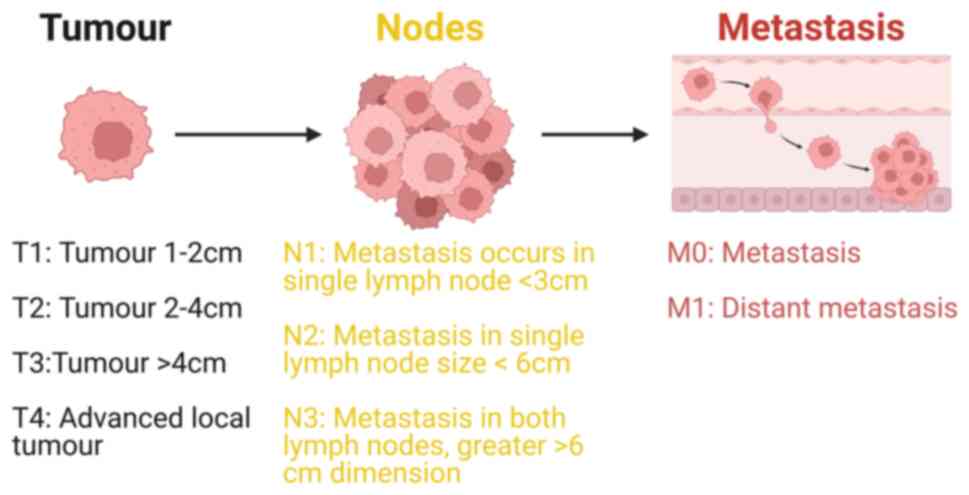 |
Figure 1.
TNM classification system, a widely used staging for HNSCC; the system denotes the size and extent of the tumour. T signifies the size and width of the primary tumour, with higher numbers indicating more extensive or deeper growth within the tissue. T1-T4 indicate the size of the primary tumour, with larger numbers signifying greater tumour size area. N indicates the spread of cancer to nearby lymph nodes. N0 denotes the absence of cancer in nearby lymph nodes. N1-N3 indicate the presence of cancer and its size in lymph nodes with cancer. M designation indicates the presence of cancer metastasis, signifying the spread of cancer from the primary tumour to other areas of the body. M0 indicates no spread of cancer, while M1 indicates that the cancer has spread. Also, by using TNM stage system HNSCC can be categorised as stage I, II, III, or IV. The frequency of different types of head and neck cancer has variability depending on its association with HPV. Stage I or II indicates that the cancer is small and has not metastasised from the original site (head and neck cancer). Stage III or IV signifies that the cancer is larger and has spread to other parts of the body.
|
Risk factors that can be linked with the development of HNSCC are excessive smoking of tobacco products (7) and their derivatives (8), regular and heavy consumption of alcohol (9), especially combined with tobacco use (10–12), human papillomavirus (HPV) infection commonly transmitted through oral sexual activity (13), and prolonged sun exposure. Other risk factors include gastroesophageal reflux disease and laryngopharyngeal reflux disease, as the backflow of stomach acid into the upper airway, may contribute to HNSCC development. Finally, history of previous HNSCC can increase risk of developing another cancer disease in the same or a different region (2).
HNSCC treatment is currently based on radiation therapy (14), removal of cancer tumour (surgery), chemotherapy, targeted therapies and immunotherapy (15). Immunotherapy options for HNSCC include immune checkpoint inhibitors (ICIs), programmed death-1 (PD-1), programmed death-ligand 1 (PD-L1), cytotoxic T lymphocyte antigen 4 (CTLA-4), as well as natural killer cell receptors, such as those with inhibitory motives based on intracytoplasmic tyrosine. Additionally, there are oncolytic viruses capable of selectively infecting tumour cells with specific receptors, leading to their lysis without harming normal host cells (16). The typical approach to combination therapy involves using chemotherapy alongside radiotherapy. Additionally, there is a concurrent utilisation of chemotherapeutics and new compounds tailored to the molecular targets of specific cancer types in combination with radiation. Combination molecular targeted therapy often involves the simultaneous inhibition of EGF receptors with other targets and is a primary focus in combating cancer cell resistance to single epidermal growth factor receptor inhibition (17). All combinatorial therapies can improve response rates and survival outcomes in patients with HNSCC. There is the potential to repurpose anti-diabetic and anti-hypertensive drugs in the treatment of HNSCC as a means of reducing side effects and long-term usage of anticancer drugs or combinatorial therapies for patients. Pioglitazone, metformin and sodium-glucose co-transporter-2 inhibitors are among the anti-diabetic drugs that have been found to have positive effects in treating HNSCC. Additionally, atenolol, Hydralazine, Propranolol, candesartan and captopril are examples of anti-hypertensive agents that have been tested in clinical trials for HNSCC. The repurposed drugs not only enhance safety, tolerability and effectiveness when used in low doses, but are also suitable for long-term use, providing significant advantages over other drug classes in HNSCC treatment (18,19).
Despite the application of advanced treatments for treating HNSCCs (20), the overall 5-year survival rate has not shown significant improvement, and the number of individuals succumbing to HNSCC remains high (15). The problem lies in the fact that HNSCCs are typically detected at advanced stages, contributing considerably to high mortality rates associated with this disease (21). Methods to detect the presence of HNSCC at early stages and use of personalised therapy can help in improving HNSCC treatment outcomes and this can be possible by the use of new biomarkers for this type of cancer (22). Biomarkers can be biological molecules found in body fluids including blood, cerebrospinal fluid, saliva, urine or tissues, which can indicate normal or abnormal conditions, disease state or assess the body's response to a certain treatment (23).
Current diagnosis and management of HNSCC is challenging as currently there is no single modern tool available that can accurately detect and predict the progression of this type of cancer (24). Identifying mechanisms of tumorigenesis and potential biomarkers will aid in early diagnosis, assist in selecting appropriate therapies for patients with HNSCC, and predict clinical outcomes.
The present review addressed significant gaps in the existing literature on the molecular mechanisms, evolution and novel biomarkers of HNSCC. Despite numerous studies, there is a noticeable lack of comprehensive reviews that integrate molecular approaches to the evolution of HNSCC and utilise multi-omics platforms for an in-depth analysis. Current literature tends to focus on specific aspects, such as genetic mutations or individual biomarkers. The present review aimed to take a broader perspective by integrating cancer evolution, molecular mechanisms and omics approaches to provide a comprehensive understanding of the mechanisms underlying the initiation, development and progression of HNSCC.
HNSCC tumorigenesis mechanisms; premalignant squamous evolution
Cancer often arises from a cell line that undergoes multiple mutations in cancer-related genes, such as tumour suppressor genes, oncogenes, or multiple epigenetic changes (25). Premalignant describes a condition that can turn into cancer. By definition, pre-malignancy is the presence of abnormal tissue that is considered a precursor to cancer and is linked with high risk of develop to cancer (26). HNSCC lesions containing driver mutations are often associated with clonal expansion and are observed more frequently in precancerous and malignant lesions than expected from the normal background mutation frequencies. The exact role of each driver mutation in the early initiation and development of cancer remains under investigation (27).
Early lesions in development in HNSCC are associated with oncogenic changes that generally lead to tumorigenesis, these lesions may be due to two main factors; firstly, transformation by a key initiating agent such as high-risk HPV (hrHPVs). HPVs enter through micro-abrasions or other breaches in the epithelial barrier, exposing the basal layer and primarily infect the basal epithelial cells by attaching to heparan sulphate proteoglycans, that induce conformational changes in the viral capsid proteins (L1 and L2), exposing additional receptor binding sites (such as α6 integrin), which aids in further binding and internalisation. After HPV enters the host cell through endocytosis, the L2 protein assists in the transport of the viral DNA to the nucleus, where the viral genome can be maintained episomally (as an independent circular DNA) or can be integrated into the genome (markedly more common in cancer). The viral genome replicates using the host's DNA replication machinery, with the assistance of viral proteins E1, E2, E6 and E7 disrupting the normal cell cycle regulation by interacting with tumour suppressor proteins p53, CDKN2A and pRb, preventing apoptosis and promoting cell proliferation and transformation (28). Secondly, transformation of conventional HNSCC (HPV-negative). In non-HPV HNSCC, driver mutations can be caused by smoking, alcohol consumption and other environmental carcinogens, which are important risk factors. Exposure to these carcinogens causes DNA damage, genetic mutations and epigenetic changes that lead to early lesions such as leukoplakia and erythroplakia. Signs associated with these early lesions may include genetic changes in tumour protein P53 (TP53), CDKN2A, DNA methylation and dysplasia-expressing cell markers (29).
Normal tissue architecture and the pre-malignant microenvironment have a complex relationship that plays out a key role in natural selection and cancer evolution. Surrounding changes, such as fibroblast activation and remodelling of the immune microenvironment, can alter the strength of epigenetic changes caused by damage, by driver and passenger mutations. Environmental pressure on tumour cell transformation can be facilitated by the presence of growth factors or the introduction of normal cells that inhibit and force them to prevent invasion by creating an extracellular matrix. This microenvironment also regulates self-renewal in the epithelium (30).
Therefore, the evolution of HNSCC follows histologically ordered stages starting with pre-malignant epithelial cell hyperplasia, then dysplasia ranging from mild, moderate and severe, followed by carcinoma in situ, and finally to metastasis. In numerous cases, cancerous evolution from normal cells or the progenitor cell candidates, results in cancer stem cells (CSCs) with ability of self-renewing and proliferative properties after cancerous transformation (31).
In the following section, the potential biomarkers for HNSCC were discussed, which play roles in therapeutic approach, diagnosis and predicting of the progression of premalignant stages of conventional squamous cancer cells, such as progression of cancers from pre-malignant to metastasis. Biomarkers of HNSCC are classified into the following categories based on the aetiology of the disease; HPV-associated oro-pharyngeal carcinoma biomarkers and conventional squamous carcinoma biomarkers.
HPV-associated oro-pharyngeal carcinoma biomarkers
These biomarkers are specifically associated with oropharyngeal carcinomas caused by HPV infection, particularly types 16 and 18. HPV is a non-enveloped double-stranded DNA virus with a circular genome of ~8,000 base pairs (32). It is a common sexually transmitted infection that can cause various diseases, including HNSCC. In particular, HPV primarily infects and replicates in epithelial cells, predominantly in the oropharynx (33). HPV can persist in the basal cells in a latent state without any symptoms for numerous years. The viral genome is maintained episomally and can reactivate under certain conditions. In numerous cases, the host immune system successfully clears the infection, often within 1–2 years. However, persistent infection with hrHPV types (such as HPV-16 and HPV-18) can lead to the integration of the viral DNA into the host genome, disruption of normal cellular functions and the development of malignancies. This makes HPV a prominent independent marker for HNSCC development (34). HPV 16 and 18 strains have recently become a powerful and widely used biomarker with excellent clinical outcomes as they are highly present in most HPV-related HNSCC cases (35). The oncoproteins from this virus play a significant role in cellular transformation and disruption of human cell cycle control (36). These viral proteins induce the degradation of key regulators such as TP53 and RB Transcriptional Corepressor 1 (RB1) proteins, resulting in the cell cycle's dysregulation and suppression of normal apoptosis (37). Evidence of viral DNA integration into cancer cells is crucial for the assessment of HPV status in oropharyngeal carcinomas (38).
In most HNSCCs from hrHPV-16 strains which can be used as biomarkers, NFE2L2 expression is reduced which in turn increases sensitivity to chemotherapy and radiotherapy (39).
Diagnostic implications of p16 expression and HPV testing in patients with HNSCC
P16, a cell cycle regulatory protein of CDKN2A gene, is always overexpressed in HPV-positive HNSCC. Currently, p16 is a commonly used immunohistochemical biomarker test for patients with HNSCC. In HPV-positive oral squamous cell carcinoma (OSCC), p16 overexpression results from the inactivation of the tumour suppressor protein retinoblastoma 1 (RB1) by the hrHPV E7 oncoprotein (40). Occasionally, p16 overexpression can also result from HPV-negative cases (41).
Analysis of overexpression of p16 protein through immunohistochemistry is widely used in determination of HPV infection. In some cases, most of patients with overexpression of p16 (P16+) are HPV-negative and also some of patients without p16 (16-negative) are HPV-positive. Therefore, for improved practice, both p16 IHC and HPV testing is often used to ensure accurate classification of HPV-associated HNSCC (33).
Detection of P16 can be reliably used as high validity prognostic biomarker for HNSCC which is caused by HPV (42). In a research study where the prognostic relevance of the best biomarkers was compared to assess the status of HPV infection in HNSCC, it was found that CDKN2A, cyclin D1 (CCND1) and RB1 protein levels were significantly higher in HPV+ cases compared with negative controls as confirmed by PCR (42). HPV detection has potential in diagnosis and prognosis of cancer cells appearing in the head and neck area (39,43–45). Therefore, detecting HPV+ (45) and transcriptional products of E6 and E7, can serve as valuable diagnostic biomarkers for accurately identifying HPV-related HNSCC and will allow appropriate management and treatment of patients (33,39,42,44,46).
Conventional squamous carcinoma biomarkers
Conventional squamous carcinoma biomarkers that are associated with HNSCC are discussed in the following section.
TP53
Mutation on the TP53 tumour suppressor gene is a key factor in cancer development. Normally, when human cells are exposed to internal or external stresses (47), TP53 triggers either cell cycle arrest, cellular senescence or DNA repair and if DNA repair mechanism fails, cells undergo apoptosis (48). Determination of the role of the mutant TP53 in oral cancer using RNA sequencing (RNA-seq) showed that activation of endogenous TP53 through gain-of-function mutations in carcinogen-induced epithelial cells promotes resistance to anti-CD274 prevention immunity associated with increased IL17 signalling and a lack of exhausted CD8 T cells in the tumour microenvironment. These results support the use of TP53 as a biomarker to predict CD274 inhibitions in order to prevent HNSCC development (49).
In most cases, HNSCC with mutation in TP53 is strongly associated with invasive carcinomas and poor survival (50). Several studies have assessed these abnormalities of TP53 and it was found that TP53 has prognostic significance, indicating its potential as a marker for predicting outcomes in this type of cancer (51).
In an analysis of mutations in TP53 using next generation sequencing technology on samples of patients diagnosed with HNSCC of the pharynx, oral cavity and larynx, it was discovered that TP53 was the most commonly altered gene, indicating that TP53 has prognostic value for this type of cancer (52). In a comprehensive analysis of patients with HNSCC at different stages of the disease, at least one significant mutation of TP53 was identified in all tumour samples, consistent with previous studies that identified a somatic mutation of TP53 genes in smoking-related HNSCC (53).
Mutations in the TP53 gene disrupt its function, allowing cells with damaged DNA to survive and potentially become cancerous (47). This abnormality was frequently observed in HNSCC (54), and is associated with reduced survival (55). Hence, presence of mutations in TP53 can be used as a prognostic biomarker to provide valuable information for HNSCC (10,50,53,56).
RAS family of genes
The RAS family of genes comprises three highly conserved genes: KRAS proto-oncogene (GTPase), NRAS gene NRAS proto-oncogene (GTPase) and HRas proto-oncogene (GTPase), that were also known as Kirsten rat sarcoma viral oncogene homolog (KRAS), neuroblastoma RAS viral (v-ras) oncogene homolog (NRAS) and Harvey rat sarcoma viral oncogene homolog (HRAS), respectively (57). These genes encode proteins that act as molecular switches, cycling between an active GTP-bound state and an inactive GDP-bound state. RAS proteins can bind to and activate downstream effector proteins in their active state, initiating a signalling cascade that promotes cell proliferation and division.
Abnormal activation of RAS genes typically occurs from mutations and epigenetic alteration preventing RAS proteins from functioning properly and cause uncontrolled cell proliferation, a hallmark of HNSCC (57,58). In HNSCC it was found that in samples with the KRAS variant, there may be an altered immune status as compared with those who do not have this mutation (59).
DNA from 180 HNSCC samples was analysed to determine whether or not oncogenes HRAS are associated with drug resistance; it was identified that when HRAS proto-oncogene, GTPase signalling is mutated, it triggers an increase of ERK signalling, which in turn renders cancer cells resistant to cetuximab treatment (60).
Utilizing large-scale genomic sequencing databases, it was revealed that HNSCC from various anatomical subsites may have genetic variations that disrupt RAS signalling in malignant cells (61). From The Cancer Genome Atlas database it was found that HRAS gene alterations-mutations are more frequently found in primary untreated HNSCC tumours compared with KRAS and NRAS genes' mutations. Additionally, it was demonstrated that most HRAS alterations can lead to aggressive form of cancer (61).
Detection of HRAS mutations can be used as a valuable biomarker in HNSCC and help in providing prognostic information (58). Most of the tumours with HRAS mutations have a high chance of recurrence (61). Therefore, detection of RAS mutations can used in clinical settings and serves as a diagnostic and prognostic biomarker and will help to guide treatment choices which can improve the treatment outcomes in patients with HNSCC (62–64).
Phosphatase and tensin homolog (PTEN)
PTEN is a tumour suppressor gene that plays a significant role in molecular biology by regulating cellular survival and proliferation (65). PTEN acts as a phosphatase that dephosphorylates phosphatidylinositol 3,4,5-trisphosphate (PIP3), a signalling molecule that can activate the PI3K/AKT/mTOR signalling. By dephosphorylating PIP3 and PDK1, PTEN antagonises the PI3K/AKT signalling pathway and prevents its oncogenic effects (66).
The deactivation of PTEN in cancer can lead to activation of the PI3K/AKT pathway, which increases cell proliferation, inhibits apoptosis, activates cell cycle progression, survival, invasion and metastasis (67). In addition, PTEN can also lead to activation of other signalling pathways, such as the mTOR and NF-κB pathways, which further contribute to tumorigenesis, oxidative stress and genomic instability in tumorigenesis development (68,69).
In experiments where human cell lines were used to model different stages of OSCC progression, PTEN levels in cancer were found to be lower compared with normal cells. The decrease of PTEN expression level was consistent with the predicted downward trend of PTEN expression in successive stages of OSCC progression (68).
In silico analysis on the involvement of the PTEN in HNSCC found high target score for PTEN. PTEN was involved in proliferation and chemotherapy resistance, which were increased due to PTEN loss of function. These in silico studies revealed that PTEN could help as prognostic and predictive biomarker in HNSCC (70).
Loss of function mutation of PTEN expression is invariably linked with PI3K activation sustaining an important role in HNSCC tumorigenesis. This provides a framework for PTEN as a biomarker for targeted therapy (71). Therefore, alterations of PTEN can be used as prognostic biomarkers in cancer (68,69,72,73).
MKI67
MKI67 (Ki-67 enhancer marker) is a protein-coding gene located on human chromosome 10 (10q26.2) (74). MKI67 keeps specific mitotic chromosomes separated in the cytoplasm after nuclear envelope breakdown. MKI67 prevents chromosomes from collapsing into clumps of chromatin (75,76). MKi-67 expression normally is low in the G1 and early S phases but gradually increases to a maximum during mitosis (77). A study on the development of leukoplakia found that an increase in the MKI67 index can indicate a poor prognosis in leukoplakia (78).
MKI67 analysis in predicting HNSCC metastasis showed that MKI67 was an independent predictor of metastatic, recurrent disease and poor patient outcomes. This indicates that MKI67 has prognostic value regardless of the size of the primary tumour (79). MKI67 gene expression by quantitative PCR (qPCR) was performed in HNSCC samples in comparison with non-cancer control samples. The results of the experiment revealed that MKI67 was highly upregulated in HNSCC samples compared with control tissues. MKI67 upregulation was observed in all advanced tumour stages and lymph node status compared with early-stage HNSCC. Therefore, MKI67 expression can be used as a prognostic biomarker (80).
Higher MKi-67 index in premalignant lesions might indicate a higher risk of progression. A study evaluating the prediction value of MKI67 expression in patients with nasopharyngeal carcinoma (NPC) using immunohistochemistry found that MKI67 level was higher in tumours than in normal tissue. Thus, MKI67 can be employed as a predictive marker of HNSCC in a clinical environment, as its overexpression has been linked to poor overall survival in patients with NPC (81).
CCND1
CCND1 is a gene located on chromosome 11q13 and codes for protein that plays a crucial role in cell cycle progression (82). This protein belongings to the cyclin family of proteins that can bind and activate cyclin-dependent kinases (CDKs). CCND1 specifically binds to CDK4 and CDK6, forming active CDK4/6-CCND1 complexes that phosphorylate the retinoblastoma protein (pRb) a tumour suppressor that prevents cell cycle progression from the G1 to S phase (83). Phosphorylation of pRb inactivates its tumour suppressor function, allowing cells to enter S phase and replicate their DNA (84).
CCND1 overexpression is mostly observed in HNSCC and affects cell migration, angiogenesis and uncontrolled cell proliferation (85). CCND1 promotes tumorigenesis, which is associated with more aggressive form of tumour phenotype, poor prognosis and metastasis (86). OSCC with CCND1 overexpression was correlated with lymph node metastasis; thus, CCND1 is a prognostic biomarker for this type of cancer (82).
A cohort study analysed CCND1 in HNSCC as a prognostic biomarker and found that the CCND1 molecule is involved in cell cycle control. CCND1 overexpression was linked with several clinical and pathological stages of HNSCC tumour development and can be used as predictive biomarker (87).
CCND1 protein is often overexpressed in HNSCC; its elevated levels in premalignant lesions may indicate higher risk of progression to cancer. This was observed more frequently in numerous cases of HNSCC (85). By detecting the level of CCND1 protein in tissues, valuable prognostic information on HNSCC can be extracted (83,84,86,87).
CD274
CD274 cell surface protein, also commonly referred as PD-L1, is essential for controlling the immune response. It interacts with the immune system's PDCD1, also known as PD-1 receptor, causing immune system suppression and enabling cancer cells to avoid immune identification and elimination (88).
One way in which cancer cells resist the antitumour treatment is by overexpressing CD274 immune system reaction. In HNSCC, transmembrane protein CD274 is expressed in both haematopoietic and non-haematopoietic cancer cells. The intratumorally lymphocyte response is suppressed by the interaction of CD274 molecules with the PDCD1 receptor on T cells (89). Analysis of HNSCC tumour microenvironment found that tumour cells can utilise the PDCD1/CD274 axis to inhibit immune system. Furthermore, higher CD274 expression is linked with cancer metastasis (23).
An immunological analysis of CD274 role in a prospective cohort of oral precancer samples from 188 patients has shown that patients with oral precancer may exhibit characteristics of immune resistance. Hence, CD274 is a predictive biomarker for patients with oral precancer (90).
Analysis of CD274 and PDL2 expression in two well-controlled and independent cohorts of the primary head and neck tumour, reported that CD274 was observed as highly expressed mainly at the membrane surface and in the cytoplasm of the HNSCC tumour cells. CD274 proved to be useful as a valuable predictive biomarker for the overall survival rate (91).
The immunosuppressive impact of the PDCD1 receptor and its ligand CD274 was investigated using qPCR and western blot analysis of tissue from patients with HNSCC versus healthy individuals. The results revealed considerably greater mRNA expression of PDCD1 and CD274 in the tissues of patients with HNSCC compared with the healthy group. Increased expression of PDCD1 and CD274 could be a prognostic in HNSCC (92). Patients with HNSCC who exhibit high levels of CD274 expression typically have a poor prognosis and more aggressive form of disease (23,93).
CD274 is a potential theragnostic biomarker because it can predict therapy responsiveness in HNSCC disease such as the response to ICIs that target the CD274 pathway (50,91).
Notch Receptor 1 (NOTCH1)
NOTCH1 is membrane receptor protein that belongs to the NOTCH receptor family (NOTCH 1–4) (94), is one of the important evolutionarily conserved signalling in the process of development of an embryo from zygote and tissue homeostasis. The NOTCH receptor signalling pathway involves complex steps that end up in regulation of important cellular functions, including proliferation, differentiation (95), cell-cell interactions, apoptosis and stem cell maintenance (96).
NOTCH1 regulates gene expression when its ligands (DLL1, DLL3, DLL4, JAG1 and JAG2) bind its receptor. This triggers arrangement of intracellular flagging pathways that lead to cleavage of the NOTCH1 intracellular space (NICD) (94). The NICD enter nucleus, where it works on transcriptional co-activator to regulate the expression of numerous NOTCH1-specific target genes, such as HES1 and HEY1 (97).
NOTCH1 analysis by exome sequencing from HNSCC samples found that the most frequently mutated gene was NOTCH1, which is mostly affected by copy number changes. In the same results, loss of heterozygosity at the NOTCH1 gene locus was detected in two of three tumours with mutations that were analysed for copy number variations. These data from the sequencing experiment support the idea that NOTCH1 functions as a tumour suppressor gene in HNSCC. Moreover, those results emphasised the importance of evaluating the role of the NOTCH1 in cancer as a prognostic biomarker (98). NOTCH1 mutations are common and present in the majority of HNSCC; these changes may affect the normal function of NOTCH1, causing a loss of its tumour suppressor activities (95,97).
Single-cell RNA-seq was employed for analysis of specimens from patients with thyroid cancer who were undergoing a lobectomy of the thyroid compared with the non-malignant thyroid specimens collected from patients undergoing normal surgery (control). NOTCH1 was the key target gene of SATB2, and the activation of the SATB2/NOTCH1 pathway was one of the reasons for the highly aggressive carcinogenicity of this subgroup of HNSCC (99). Alterations in NOTCH1 signalling or mutational status may provide prognostic information and potentially guide treatment decisions (95–97,99).
Telomerase reverse transcriptase (TERT)
TERT is the catalytic subunit of telomerase (a special enzyme responsible for maintaining telomeres). In normal cells, the telomere length is shortened after each cell division, and this shortening is considered the biological unit that limits the lifespan of cells (100). In cancer cells, TERT enzyme is considered to counteract this shortening by adding DNA repeats to telomeres, allowing cancer cells to maintain their reproductive potential (101). TERT can perform reverse transcription, where it synthesises DNA from an RNA template called the enzyme internal RNA component (TERC), by adding TTAGGG repeats to the end of telomeres (101).
Upregulating TERT expression and telomerase activity, resulting in unlimited replication of cancer cells similar to embryonic or stem cells, has been shown to overcome replicative senescence in the vast majority of head and neck tumours (100). Pathways affecting telomerase reactivation are mutations in the DNA sequence controlling TERT gene expression and alternative telomere lengthening which use non-homologous recombination (101). TERT mutations lead to continuous cell division which can cause development of HNSCC with aggressive features, poor clinical outcomes and shorter survival (102,103).
Telomerase activity evaluation from patients with HNSCC as a biomarker for evaluation and identification of recurrence during follow-up revealed that telomerase activity turned negative following surgical removal of cancerous tissues. In the same study, follow-up of most of the patients with HNSCC revealed a conversion of telomerase activity from negative to positive after a subsequent recurrence of HNSCC (104). Comparison of the rate of telomerase activity between HNSCC and healthy individuals revealed to be significantly higher in patients with cancer compared with controls (104).
In a meta-analysis study of TERT to assess its prognostic use in HNSCC it was found that most of cancer sample express high amount of TERT that is linked with treatments response. Therefore, TERT is a promising biomarker for prognosis of patients with HNSCC (103).
Epidermal growth factor receptor (EGFR)
The human EGFR belongs to a family of transmembrane receptors called tyrosine kinases. Normally, EGFR responds to the binding of its natural ligands EGF and TGFA, which are present in the extracellular environment (105). EGFR initiates a signalling cascade within the cell, activating downstream effectors that are involved in cell proliferation (106,107).
In HNSCC, EGFR is often dysregulated, resulting in its overexpression or constitutive activation (108). This dysregulation of EGFR can occur through various mechanisms, including gene mutations and hormone signalling (107). Human EGFR alterations or mutations can be used as prognostic indicators for disease and therapeutic responses because they are directly correlated with worse clinical outcomes (109). Overexpression of EGFR was linked to reduced overall survival rate in patients with cancer (110). The EGFR-related cancer patient survival rate ranges from low to high as it was observed by numerous different research studies (111–113). Thyroid cancer cells consistently overexpress EGFR, which is associated with continuing cellular proliferation. These high expressions of EGFR in cancer cells appeared to have potential as a prognostic biomarker in HNSCC (114).
A meta-analysis has provided evidence that EGFR overexpression is common oncogenic mechanism in oral oncogenesis and metastasis. It was suggested that immunohistochemical analysis of EGFR should be integrated in routine prognostic evaluation in HNSCC (115).
Real-time analysis of EGFR in fluorescence-labelled fresh tissue from fresh biopsies of the oral cavity using a specific molecular contrast agent, revealed that EGFR is a biomarker for diagnosis of oral cancer, as it was found that >50% of patients with oral cancer overexpress EGFR in their cells. During carcinogenesis, the basal cells do not differentiate and continue to proliferate constantly in the epithelium. As a result, EGFR is mostly expressed in the cancerous epithelium and cells in the lining of an organ with abnormal changes are more likely to become cancerous (116).
Most of HNSCC cases exhibit EGFR overexpression which is linked with a poor cancer prognosis, increased tumour growth and therapy resistance (109). Therefore, EGFR can be widely used as a prognostic for HNSCC (108–110,117).
Vascular endothelial growth factor A (VEGFA)
VEGFA is an angiogenic protein factor that plays a crucial role in increasing vascular permeability. Abnormal regulation of VEGFA can lead to initiation and development of different human malignancies (118). Downregulation of VEGFA stimulates the cell migration and proliferation of endothelial cells and leads to cancer development (119).
A research study conducted on patients with laryngeal squamous cell carcinoma (LSCC) shows overexpression of VEGFA in cancer cells was strongly linked with re-occurrence of cancer and metastasis (120). Thus, VEGF angiogenesis biomarkers can be used in identifying high-risk patients specifically in the early stage of development of HNSCC (120,121).
Increased VEGF level is linked with more advanced tumour stages and invasiveness. Specifically in laryngeal lesions, VEGFA expression has been observed to rise progressively from pre-malignant to metastasis stages (122). Another study suggests that VEGFA may serve as a significant predictive biomarker in laryngeal squamous cell cancer (123). VEGFA can be used in prognostic and diagnostic of HNSCC, aiding in prognosis and treatment decision-making for patients (119,121,124).
Squamous cell carcinoma antigen (SCCA)
SCCA is a tumour-specific antigen which has two homologous isoforms, SCCA1 and SCCA2; they are coded by two genes, SERPINB3 and SERPINB4, respectively, located on chromosome 18 (18q21.3). SCCA belongs to the family of serine protease inhibitors. SCCA is frequently elevated in HNSCC and is used as a marker of tumour severity and recurrence (125).
A research study was conducted to determine the association of SCCA with clinical features in patients with HNSCC and analyse the clinical utility of serum SCCA in treatment. It was found that blood levels of SCCA were higher in HNSCC samples compared with normal controls (126).
In a follow-up analysis of patients with HNSCC, an SCCA test was performed during the examination and their results indicated that SCCA was higher in patients with cancer; therefore, SCCA could be a suitable predictive biomarker in cancer (127).
HNSCC samples were subjected to fine needle aspiration to determine the additional diagnostic value of SCCA. The experimental results revealed that the SCCA level was high in the nodes of HNSCC compared with control patients. Therefore, presence of SCCA in patients with cancer can be used as prognostic biomarker (126,128).
Lactate
Lactate is a product of glycolysis from pyruvate, especially without oxygen. Recently, it was found that lactate is a crucial regulator of the glycolytic phenotype and has been observed to promote tumour growth and metastatic spread in HNSCC (129).
Elevated lactic acid levels in HNSCC may indicate dependence on this altered metabolic pathway for energy production (130). Study of HNSCC biopsy from the primary site was conducted for quantification of lactate concentration using the luciferase bioluminescence technique. Most tumour lactate concentrations were found to be associated with subsequent metastasis HNSCC, thus rendering lactate a favourable prognostic biomarker (131).
Analysis of serum metabolites from salivary gland tumours by using the mass spectrometry method was compared with healthy controls. It was found that lactic acid levels were consistently higher in HNSCC samples than control samples (132).
An ex vivo study performed with the Proton HR-MAS MR method on oral squamous HNSCC tissues for metabolic abnormalities and signs of inflammation in OSCC showed a high level of lactic acid in OSCC compared with the controls; thus, lactic acid can be used as indicative biomarker for HNSCC (133).
CD44
CD44 is the surface glycoprotein of a cell, and has numerous functions including cell to cell adhesion, migration and signalling (134). A previous study has highlighted the involvement of a small subset of cancer cells called CSCs in HNSCC tumour initiation, development and resistance to treatments (135). CSCs in HNSCC are characterised by the presence of certain surface markers, such as CD44, PROM1, ALDH1A1 and ABCG2; their overexpression is generally linked with an unfavourable prognosis (136).
CD44 is involved in the Wnt/β-catenin pathway, which functions in cell proliferation, differentiation and stem cell maintenance. Activation of this pathway may promote CSC properties and contribute to tumour growth and invasion in HNSCC (137).
An in silico study on HNSCC found that CD44 was a frequently observed CSC marker and is implicated in various tumour-related process including proliferation and metastasis (138). CD44 expression in patients with HNSCC appears to differ among different subsequent of squamous cancer on the oral cavity, pharynx or larynx (134).
A study on untreated patients with laryngeal cancer found that CD44 level is associated with lymph node metastases and resistance to treatment (139). Therefore, CD44 holds promise as a valuable biomarker on diagnosis and prognosis for HNSCC (138–141).
Epstein-Barr virus (EBV)
EBV is a commonly occurring herpesvirus infecting humans and is mostly transmitted through contact with infected bodily fluids such as saliva or blood. The EBV genome is a linear dsDNA, ~172 kbp long (32,37). EBV causes cancer through expression of two important proteins, EBV-induced nuclear antigen 1 and latent membrane protein 1 (LMP1) that facilitate cell proliferation, hinder apoptosis and disrupt the normal functioning of the immune system (142).
Research conducted on determination of EBV status in HNSCC using an in situ hybridisation method found that infection with EBV can disrupt several intracellular signalling processes involved in cell cycle control. In the same study, it was reported that EBV LMP1 can inhibit CDKN2A expression and inhibit downstream effectors, including the transcription factors E2F4 and E2F5, and can promote cell proliferation. In addition, it was found that LMP1 was expressed in >50% of EBV-positive tumours with aggressive features (143).
In a case-control study, EBV DNA positivity detected by qPCR was higher in the laryngeal cancer samples suggesting that individuals who tested positive for EBV DNA virus in the laryngeal area had increased risk of developing laryngeal HNSCC cancer. Furthermore, in the same case-control study, it was concluded that EBV DNA is positively linked with anti-apoptosis by BCL2 expression in their tissues (144).
A nomogram analysis incorporated EBV DNA as a new immunotropic marker to predict the survival rate in nasopharyngeal cancer. The results of nomogram analysis found that EBV can be recognised as a potential prognostic factor for HNSCC (145).
A meta-analysis on HNSCC found that EBV+ patients had higher probability of developing laryngeal cancer compared with healthy controls (146). Therefore, the presence of EBV can be utilised as a prognostic marker of HNSCC (147–149).
Non-coding RNA (ncRNA) biomarkers of HNSCC
In this section, both long non-coding RNA (lncRNA) and small RNA molecules that are not translated into a protein but have regulatory functions are discussed. lncRNAs are a varied class of ncRNAs that play important roles in biological processes and are linked to cancer formation and progression. Their presence in body fluids and link with tumour characteristics make them promising non-invasive biomarkers for cancer diagnosis and therapy (150–152). MicroRNAs (miRNAs or miRs) are short non-protein coding RNAs whose sequences are ~18-22 nucleotides in length. A wide range of miRNAs have been identified in humans, and numerous of them have the ability to target and regulate numerous genes. Abnormal expression of miRNAs has been observed in HNSCC, suggesting their involvement in the initiation and progression of this type of cancer (153). In this section, specific ncRNAs which have been identified as potential drivers of HNSCC and can be used as diagnostic, prognostic or predictive biomarkers, are included.
HOXA transcript at the distal tip (HOTTIP)
HOTTIP is a lncRNA that exhibits a unique genomic arrangement where it is brought in close proximity to the HOXA genes through chromosomal looping (154). By binding to the WDR5 protein, HOTTIP forms a complex with the histone methyltransferase protein KMT2A (previously known as MLL). This complex, known as the WDR5-MLL complex, specifically targets the HOXA region, leading to H3K4 methylation and subsequent transcriptional activation of the HOXA locus. HOTTIP has been identified to have a key role in various cancers, including HNSCC (155,156).
Analysis of HOTTIP as a prognostic biomarker of HNSCC integrated with computational analysis, have consistently demonstrated elevated HOTTIP expression in tissues of individuals who developed HNSCC compared with those who did not have cancer (157).
HOTTIP expression was discovered to be related with various stages of cancer as well as patient survival, establishing it as an important prognostic marker. This indicates that high HOTTIP levels contribute to cancer growth, proliferation and progression. HOTTIP has a favourable correlation with survival and can be employed as a unique and important diagnostic and prognostic biomarker in HNSCC (158).
The involvement of HOTTIP in the initiation and drug resistance in oesophageal cancer was determined by using MTT assay. HOTTIP levels in the serum of patients with oesophageal cancer were detected by reverse transcription (RT) PCR. Drug resistance was determined after treating Eca109 cells with various concentrations of Adriamycin for 24 h; the IC50 was measured using MTT assay. The results revealed that serum levels of HOTTIP are higher in patients with HNSCC compared with controls; HOTTIP can activate the ABCG2 axis to cause drug resistance in cancer. The expression profile of HOTTIP can be documented as a prognostic marker in monitoring the response in treatment for patients with HNSCC (159).
Analysis of HOTTIP levels in patients with NPC using RT-qPCR combined with western blotting demonstrated that HOTTIP expression was extremely high in HNSCC cell lines. Knockdown of HOTTIP resulted in decreased expression of HOXA13, which ended up in the inhibition of the proliferation of cells and metastasis (154).
LncRNA HOTTIP can facilitate HNSCC formation by modulating the expression of HOXA13 in cancer cells. Therefore, it can be suggested that measuring the expression levels of HOTTIP could potentially be useful as biomarker in prognosis of HNSCC (154,157,160).
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
MALAT1 is a lncRNA that is upregulated in cancer. This ncRNA can interact with chromatin-modifying complexes and regulate expression of proto-oncogenes, genes involved in cell cycle progression and apoptosis. MALAT1 binds to miRNAs that regulate gene expression and by binding them, it indirectly regulates the expression of miRNA for tumour suppressor genes (161). MALAT1 is overexpressed in HNSCC tumours influencing the expression of genes associated with cell proliferation, angiogenesis and cancer invasion (162,163). MALAT1 levels were found to be elevated in tongue squamous cell carcinoma (TSCC), particularly in those with metastasis stages (164). Experimental studies involving the knockdown of MALAT1 in TSCC cells, demonstrated impaired cancer cell migration and proliferation abilities in vitro, as well as few metastases occurred in vivo (165).
PCR was used to investigate MALAT1 expression levels as a potential therapeutic target in HNSCC samples and healthy adjacent tissues. The Transwell cell migration assay and the MTT assay were used to assess cell proliferation, migration or invasion capabilities and apoptosis. MALAT1 was shown to be overexpressed in cancerous tissues relative to cells of healthy adjacent tissues. This high level of expression was connected to a lower survival rate in HNSCC. In the same study, MALAT1 knockdown resulted in a corresponding reduction in cancer cell proliferation, increased apoptosis and cell cycle arrest (166).
In HNSCC, MALAT1 is highly expressed in tumour cells and can be measured using simple methods such as blood tests. Overexpression of MALAT1 is associated with angiogenesis and invasion (162,163). Therefore, determination of levels of MALAT1 in HNSCC could be used as a theragnostic biomarker (164,167,168).
HOX transcript antisense RNA (HOTAIR)
HOTAIR is a type of lncRNA that plays a role in regulating gene expression by interacting with the polycomb repressive complex 2 and lysine-specific demethylase 1 (169). HOTAIR promotes cancer by recruiting EZH2 and H3K27me3 to regulate CDH1, inducing cancer proliferation, invasion and lower level of apoptosis (170). It is considered that HOTAIR can also induce cancer development by suppressing the expression of tumour suppressor genes (171).
HOTAIR level of expression is constantly increased in LSCC tissues compared with control ones (172). This upregulation is associated with advanced tumour stage, poor differentiation, metastasis and worse clinical outcomes (163). A meta-analysis discovered that patients with HNSCC have high levels of HOTAIR, which can speed up tumour growth and lead to the development of malignancies. HOTAIR expression, particularly in LSCC tissues, is strongly linked to the T phase, histopathological grades and the probability of lymphatic metastasis (170). To determine the role of HOTAIR in LSCC, PCR was used to assess its activity in vitro by using Hep-2 and Tu212 LSCC cell lines transfected by pHOTAIR or si-HOTAIR, and then survival and apoptosis were measured during the experiment. The results of the experiment revealed that HOTAIR gene expression was enhanced by pHOTAIR and oppositely suppressed by si-HOTAIR. In the same experiment, knockdown of HOTAIR resulted in significant inhibitory effect on cell division observed in Hep-2 and Tu212 cells; on the other hand, the opposite effect was observed in HOTAIR-overexpressing cells. Therefore, HOTAIR promotes proliferation and inhibits cell senescence of LSCC cells (173). HOTAIR can be employed as a potential prognostic biomarker for assessing the clinicopathological characteristics and prognosis of HNSCC (169,174).
Urothelial carcinoma-associated 1 (UCA1)
UCA1 is a lncRNA that has been recently found to be upregulated in HNSCC (175). Its upregulation is associated with promoting tumour growth, invasion and metastasis through regulating proliferation genes, angiogenesis and epithelial-mesenchymal transition. Overexpression of UCA1 can increase the level of WNT6 and trigger subsequent responses of the WNT signalling pathway (176). UCA1 is also employed in regulating the level of the WNT/β-catenin signalling pathway (177).
The levels of UCA1 were found higher in TSCC tissues, and there was a notable positive correlation with lymph node metastasis (178). HSCC specimens were examined and compared with adjacent normal tissues acquired from patients with proven HNSCC illness. UCA1 was overexpressed in all HSCC tumour cells. RT-qPCR was used to determine the comparative expression amounts of UCA1 in all pairs of HNSCC and their surrounding (normal) non-tumour cells. The overall level of UCA1 in HSCC tissues was markedly higher than in paired neighbouring normal tissues, showing that UCA1 is involved in the development of HNSCC (179).
Overexpression of UCA1 in HNSCC tissues exhibits correlation with tumour stage and prognosis, and has the capability to differentiate between HNSCC and benign lesions. UCA1 is easily accessible through blood, saliva and other bodily fluids. These characteristics indicate that UCA1 is used in prognosis and treatment determination of HNSCC (178,180).
Myocardial infarction-associated transcript (MIAT)
MIAT is a lncRNA located on chromosome 22q12 (181). Initially identified for its involvement in myocardial infarction, MIAT has been found to play significant roles in development of various cancers (182). MIAT is involved in initiation and progression of cancers such as thyroid cancer by binding to miR-324-3p and promoting PI3K/AKT signalling (183). MIAT was reported in other studies consistently supporting the notion that MIAT functions as an oncogene in HNSCC (184,185). A study focusing on LSCC discovered that MIAT expression level was notably elevated in almost all of LSCC samples (185).
To validate MIAT's role in the progression of papillary thyroid cancer, HNSCC tissues and cell lines were analysed using RT-qPCR. The investigation revealed that the expression level of MIAT was significantly higher in cancer tissue than in normal surrounding tissues. The same approach was used to assess the degree of MIAT expression in papillary thyroid cancer cell lines, which revealed that MIAT was overexpressed in all cancer cell lines as well. On the other hand, knocking down MIAT had the effect of decreased cancer cell progression and invasion (186). Thus, MIAT could be used in diagnostic or prognostic tests for HNSCC because it is expression level can be measured with simple blood tests (187,188).
miR-21
miR-21 is a highly researched miRNA, particularly because it is consistently found to be overexpressed in numerous types of cancers (189). In numerous experiments, miR-21 level is constantly high in tumour tissues, serum, plasma, saliva and in blood of patients with HNSCC compared with healthy individuals (190,191). miRNA-21 overexpression in HNSCC is linked to oncogenic properties as it targets several tumour-suppressor genes including PTEN, TP53, TP63 and PDCD4. This miRNA plays a crucial role in cancer proliferation, invasion, metastasis and apoptosis (192). Elevated levels of miR-21 expression are considered as prognostic marker in HNSCC. It was also proved that its expression levels tend to decrease or become low after surgery (removal of cancer tissue) in patients with HNSCC with a favourable prognosis (193).
A pilot study that looked at salivary and serum miRNAs' association with OSCC development was conducted, where saliva and matched serum miRNAs were collected from patients with OSCC and healthy volunteers, with an equal proportion of smokers and non-smokers in each group. It was identified that miR-21 in saliva was among the most commonly overexpressed miRNAs in OSCC compared with healthy controls (194). Furthermore, miR-21 was shown to be statistically considerably more abundant in the OSCC saliva of smokers than non-smokers (controls) (194).
Meta-analysis research on the miR-21 level in patients with HNSCC found that miRNA can be used in clinical setting to evaluate the risk to develop head and neck tumours (195). Higher levels of miR-21 in patients are more likely linked to progression of HNSCC with worse clinical outcomes. Implementation of the method that can detect levels of miR-21 is crucial because it demonstrates significant promise in diagnosis and prognosis biomarker for HNSCC (193–196).
Let-7
Let-7 is a highly conserved family of miRNAs that usually regulate expression of genes by binding to the 3′ untranslated region of target mRNAs, resulting in their degradation or repression of translation (197). Let-7 is often considered a tumour suppressor miRNA and its under-expression (decreased levels) is associated with the loss of its tumour suppressor functions, allowing uncontrolled cell proliferation and avoidance of apoptosis (198).
In HNSCC, Let-7 miRNAs function as tumour suppressors by targeting important oncogenes such as KRAS and HMGA2 (199,200). It has been found that the Let-7 family has decreased levels in tumour tissues, serum and saliva of patients with HNSCC relative to healthy individuals. Furthermore, Let-7a expression levels have been associated to different clinical phases of the disease, with advanced laryngeal carcinoma exhibiting lower Let-7a expression than early-stage data. These data suggest that Let-7 miRNAs could be useful prognostic indicators for HNSCC (197).
Levels of the Let-7 miRNA family and CD274 in HNSCC were compared with controls. The results showed that Let-7 family miRNAs are lower in HNSCC and have a negative correlation with CD274 expression (201). In vivo, the analysis demonstrated that overexpression of Let-7a/b enhanced anticancer immunotherapy by CTLA4 blockade. Those findings indicate targeting the Let-7 family as a viable way to improve immune checkpoint treatment for HNSCC (201). Let-7 is a promising biomarker for HNSCC; it is considered as a stable biomarker that has the potential to revolutionise early detection and prognosis of the disease (198,202).
miR-375
miR-375 is a type of miRNA that works as a regulator of numerous cellular processes in organisms like cell differentiation, proliferation and apoptosis (203). Moreover, miR-375 targets and regulates several genes that control cell proliferation and survival, including Janus kinase 2 and insulin-like growth factor 1 receptor (204).
In HNSCC, miR-375 has been found to be downregulated, and this indicates its potential role and involvement in tumour development and progression. These low levels of miR-375 expression are linked with unfavourable prognosis, increased invasion of head and neck tumours, and higher chances of cancer metastasis (153,204).
A meta-analysis on HNSCC showed that miR-375 is a potential biomarker for HNSCC because the results from clinical samples revealed that miR-375 in plasma of HNSCC was significantly different when compared with controls, but did not differ by sex or age. Based on the analysis, it was suggested that miR-375 may be a diagnostic marker in HNSCC (205).
A research study on well-characterised oral squamous cell lines revealed a positive association between miR-375 and rearranged L-myc fusion transcription factor expression in chemo-resistant oral cancers. It was indicated that miR-375 influenced chemotherapy resistance (206). Therefore, miR-375 can be identified as a potential biomarker for HNSCCs. Measuring its gene expression level can help in detection, prognosis and therapy decision in HNSCC (205,206).
Epigenetic biomarkers
Epigenetic modifications are heritable, reversible changes in the way gene is expressed that do not result from changes in the sequence of DNA bases. Therefore, epigenetic processes change the phenotype without disturbing the DNA sequence (207). Epigenetic processes include DNA methylation, histone post-translational modification, modification in chromatin structure and some effects from gene regulators such as ncRNA (208).
A meta-analysis demonstrated that methylation is involved in cancer initiation and progression. Methylation of repetitive sequences such as Long Interspersed Elements-1 (LINE), Alu and Sat-α can be used as prognostic biomarkers. Based on their findings, it is supported that patients with cancer with global hypomethylation in all of the populations studied have poor outcomes. From the point of view of clinicians, hypomethylation benchmark can be used, and pyrosequencing to detect LINE-1 in combination with other detectors of methylation levels in samples from a patient may be a useful way of predicting their patients' outcome. If low methylation is detected, a strengthening treatment, such as the use of postoperative radiotherapy, chemotherapy or some type of intervention treatment can be applied. Therefore, the level of global hypomethylation of DNA is an integral predictor of cancer (209). A research study showed that epigenetic mechanisms play a crucial activity in the initiation, progression and treatment of cancer. Methylation of repetitive sequences such as LINE-1 or tumour suppressors including DAPK, RASSF1 and ECA is very important in tumour progression. Furthermore, epigenetic changes associated with histone modification and chromatin remodelling can lead to open chromatin structure and transcriptional activation of HNSCC-related factors. Therefore, epigenetics modifications can serve as predictive and prognostic biomarkers for HNSCC (207).
Liquid biomarkers
Liquid biopsies are methods to detect and manage cancer through non-invasive procedures by analysing readily available body fluids including saliva, blood, spinal fluid and urine. In general, those fluids may contain various molecules released by tumour cells that contain important cancer information (210). This section will discuss the most important circulating molecules used in liquid biopsy technology and their role in patients with HNSCC.
Circulating tumour cells (CTCs)
CTCs are represented by transient cancer cells from primary tumours or metastatic tissue that can enter nearby blood vessels and spread to distant sites. Their presence can alert presence of cancer and the possibility of metastases (211).
Currently, numerous tumour cell detection platforms and biochemical systems are widely accepted for CTC isolation, and the CellSearch system is the first FDA-approved platform (212).
Circulating tumour DNA (ctDNA)
ctDNA is represented by cell-free DNA that is released into the blood by the degeneration of cancer cells through apoptosis and necrosis (213). Mutations in ctDNA can reflect the genetic makeup of the tumour, dead cells or the release of DNA fragments during cell division (214). Therefore, detection of ctDNA from samples of patients with cancer can provide valuable information on malignant transformation state and holds promise as the easy way to provide both predictive and prognostic information in patients with HNSCC (215).
Exosomes
Exosomes are a subset of extracellular membrane-bound vesicles that have a certain size range (30–150 nm); they are released by cells and can be detected in numerous body fluids (216). Exosomes are composed of a lipid bilayer containing cancer proteins, cancer miRNA and DNA that enable cell-to-cell communication. Moreover, the layer of lipid contributes to the tumour microenvironment and affects immune response in HNSCC (217). The exosomes in HNSCC may be a potential drug delivery vehicle for targeted therapy (218).
miRNAs
Circulating miRNA is a liquid biomarker in HNSCC including plasma-free miRNA, mRNA and lncRNA (219). miRNA expression in tumour cells can help to identify therapeutic targets or prognostic markers (220). A study to determine miRNA cycle stability in blood samples showed that plasma miR-196a and mi-196b expression was higher in HNSCC precancerous lesions when compared with normal controls (221). Therefore, measurement of tumour-derived miRNA in serum or plasma is a useful method for cancer diagnosis and treatment decision (219).
Overall potential of liquid biomarkers in HNSCC management
Liquid biopsies are an important tool for early detection of HNSCC, which can lead to improved patient outcomes and monitoring of treatment response and disease progression over time. Furthermore, liquid biomarker helps in the decision of using treatment strategies based on the specific genetic profile of the HNSCC detected by liquid biomarkers (222).
Current status of liquid-based biomarkers in HNSCC
Advancements in science and technology in this century have been made to adapt and use liquid biopsy as new alternative and non-invasive method for molecular characterisation of HNSCC. Liquid biopsies offer a promising method for monitoring patients with HNSCC and may guide treatment decisions. However, the development of liquid biopsies continues, and more studies are needed to confirm their accuracy and improve sensitivity, as well as to improve the special requirements that must be met before these platforms are recognised as alternative or complementary tests to the standard based on tissue biopsy (223).
Multi-omics platforms for patients with HNSCC
Currently, the standard method for the diagnosis of HNSCC is based on biopsy samples and histopathological interpretation. However, the current method does not describe all the characteristics of the cancer microenvironment and carcinogenesis processes. Multi-omics analysis is a comprehensive and integrated analysis of combined data derived from different omics techniques including proteomics, genomics, transcriptomics and metabolomics (224). Multi-omics analysis can provide a large database compared with a single analysis and provide important information about the pathogenesis of complex events, which will greatly contribute to the development of HNSCC diagnosis and treatment (225). The following are the currently available omics platform that can be used in HNSCC diagnosis and disease management (Fig. 2).
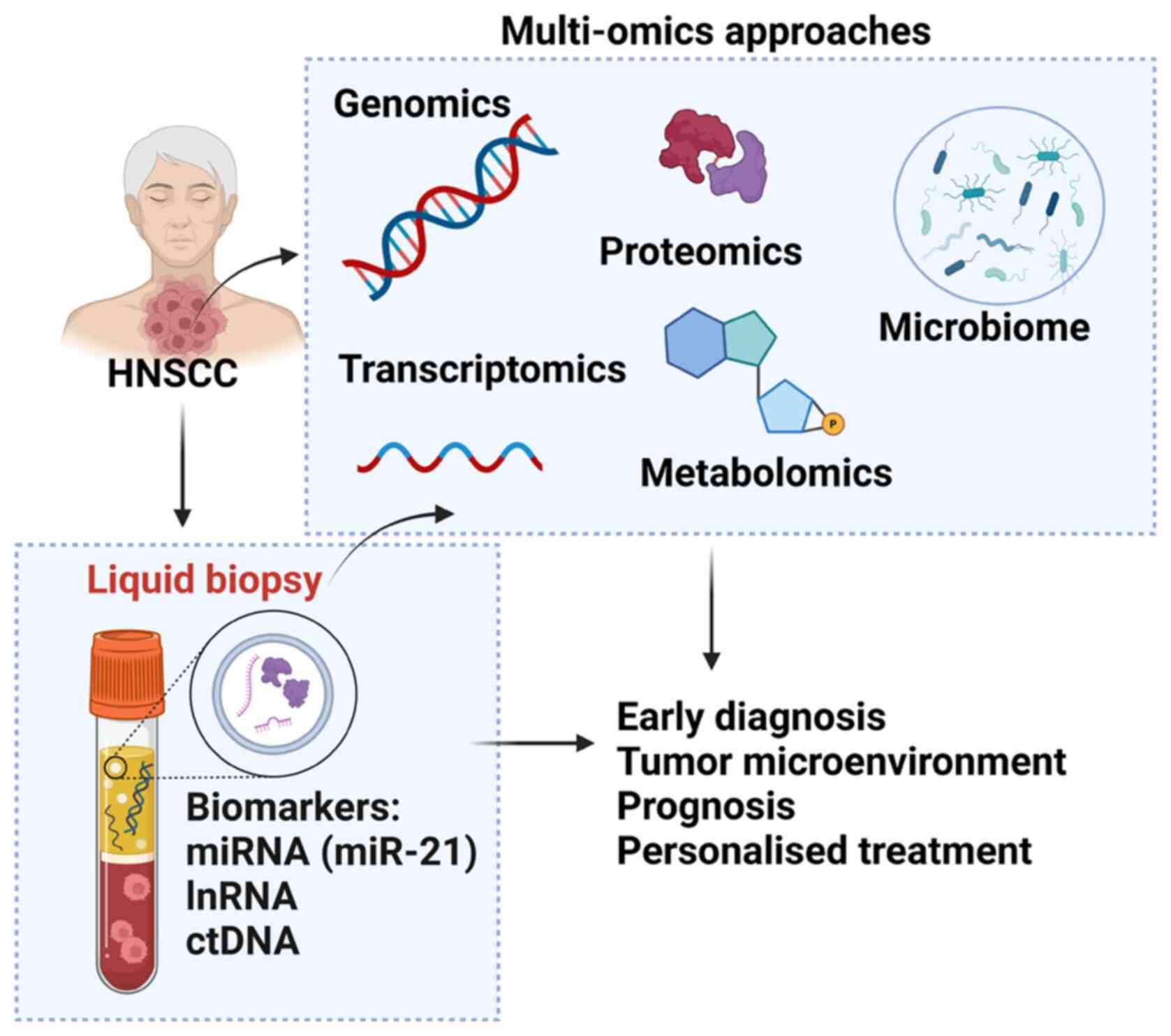 |
Figure 2.
Multi-omics approaches available for patients with HNSCC. Genomics includes examining DNA sequences to detect genetic mutations, copy number variations and structural variations that contribute to tumorigenesis. The Transcriptomics approach focuses on the analysis of RNA sequences to explore gene expression patterns, alternative splicing events, and non-coding RNA profiles, offering insights into the functional consequences of genetic alterations. Proteomics entails studying proteins, their structures and functions to uncover post-translational modifications and protein-protein interactions that play crucial roles in cancer pathways. Epigenomics studies modifications such as DNA methylation and histone modifications, which control gene expression without changing the DNA sequence and play a crucial role in the development and progression of HNSCC. Metabolomics analyses all metabolites in the biological sample of HNSCC, offering insight into the metabolic state and changes in metabolic pathways related to this cancer. Microbiomics involves the study of all microbiomes associated with HNSCC, investigating their influence on cancer development and responses to treatment. HNSCC, head and neck squamous cell carcinoma.
|
Genomic platforms: Genomics is the most studied and used field of omics. For patients with HNSCC it can involve whole genome sequencing, whole-exome sequencing, gene panels and copy number variations. The most common genomic indicators in initiation of HNSCC are caused by changes in CDKN2A, TP53, SMAD4, PIK3CA and KDM5C (226). In addition, activation of PIK3CA through mutations enhances the PI3K/AKT signalling pathway and mutations on NOTCH1, MET, STAT3, AKT, etc (95,227,228).
Epigenomic platforms: Nowadays, there are high-throughput and high-resolution DNA methylation analysis methods such as pyrosequencing and luminometric methylation assays that can analyse global DNA methylation levels in different HNSCC tumours (229). The most commonly detected indictors in epigenetics are DNA methylation and chromatin immunoprecipitation-sequencing for histone modifications. This epigenetic changes can lead to the silencing of tumour suppressor genes and the activation of oncogenes, facilitating tumorigenesis (230).
Transcription platforms: In HNSCC, transcriptomic analysis helps to find the differentially expressed genes between tumour and neighbouring normal tissues, revealing biomarkers and therapeutic targets. Available transcriptomic technologies include RNA-Seq and microarrays (231). Transcriptomic findings include altered RNA expression patterns of coding and non-coding genes, as well as alternative splicing events, offering insights into the functional consequences of these factors in HNSCC, for example, the detection of changes in miRNA and lncRNA levels.
Proteomics platforms: Generally, proteomics is used to quantify peptide abundance, modifications and interactions. Modern techniques such as mass spectrometry have revolutionised protein analysis and quantification and have recently been adapted for high-throughput analysis of thousands of proteins found in cells or body fluids (232). To efficiently investigate new protein biomarkers in HNSCC, a high amplification method is required for expression profiling and combining multiple biomarkers to increase discriminatory power (233). Example of protein indicators for initiation and development of HNSCC are changes in E-cadherin, EGFR, CD44, CyclinD1, PD-L1, PAI-1, XRCC1, RB, raptor proteins, cytokines and matrix metalloproteinases (234–237).
Metabolomics platforms involve the simultaneous evaluation of several types of small molecules including amino acids, fatty acids, carbohydrates or other products of cellular metabolism. Metabolic levels and ratios reflect metabolic function, and abnormal frequency disturbances often indicate disease (132). High-throughput LC-MS/MS analysis was performed on patients with HNSCC of all TNM to check the metabolic pathways of ESCC stages; the results provided evidence that cancer cell metabolites can be used to provide useful prognostic information about HNSCC development (238).
Microbiomics platforms: Microbes from oral cavity have been associated with development of HNSCC. Microbiome analysis platforms such as 16S rRNA-seq and metagenomic sequencing can be used to analyse disturbances in the human oral microbiome that may lead to imbalances and possible dysbiosis which are employed in the aetiology of HNSCC (239). It has been found that OSCC is associated with numerous oral bacteria such as Porphyromonas gingivalis, Fusobacterium nucleatum and some yeast types (240). The microbiome can be oncogenic, enhancing mucosal inflammation, immune effects and changes in cancer resistance or preventing the effectiveness of cancer treatment (241). Therefore, analysis of different microbial taxonomic groups may help to explore the role of the oral microbiome in initiation and progression of HNSCC (242). The presence of dysbiosis in HNSCC can be identified by conducting metagenomic sequencing of 16s RNA and shotgun sequencing. These bacteria can have an oncogenic effect on the mucosa, leading to enhanced tumor staging and grading, as well as impacting the response to chemotherapy treatments (241). The development of gingival squamous cell carcinoma might be influenced by the combined impact of Porphyromonas gingivalis, Fusibacterium nucleatum and Prevotella intermedia within the oral microbiome. This combination can trigger NF-κB-mediated immune responses, potentially providing support for cancer survival, activating oncogenic pathways, and enabling cancer cell migration and invasion (242).
Clinical trials evaluating biomarkers for HNSCC in diagnosis, prognosis and treatment
Diagnosis, prognosis and treatment of HNSCC are particularly difficult due to complicated disease heterogeneity and a severe absence of validated novel biological markers with high clinical value, that may enhance patient quality of life and therapeutic strategies.
In the present review, clinical trials in which HNSCC biomarkers are the primary focus of several in vitro and in vivo studies aimed at determining the diagnostic, prognostic, or therapeutic value of biomarkers in HNSCC, were also included (Table I).
 |
Table I.
Clinical trials evaluating biomarkers for HNSCC in diagnosis, prognosis and treatment.
|
Table I.
Clinical trials evaluating biomarkers for HNSCC in diagnosis, prognosis and treatment.
| Identifier |
Sample type |
Biomarkers, Targeted |
Purpose/Objectives |
| NCT05708209 |
Saliva |
MALAT1, miR-124 |
The clinical trial aimed to determine useful and diagnostic accuracy of lncRNA MALAT1 as a potential salivary biomarker of oral squamous cell carcinoma as well as assessment of the salivary expression level of miRNA 124 which is targeted by MALAT1. |
| NCT02748707 |
Tumour tissues |
TP53, PTGS2 (COX-2), EGFR |
This clinical trial's objective is to study the effect of COX-2 inhibitor Celecoxib and EGFR tyrosine kinase inhibitor Erlotinib alone or in combination on molecular markers of apoptosis and angiogenesis. |
| NCT04305366 |
Saliva, blood, tissue |
miRs |
The main objective of this clinical trial study is to examine presence of miRNA markers in saliva, blood, FNA and tissue specimens in HNSCC patients and control. They evaluate whether these miRNA markers can provide prognostic or diagnostic clinical significance in management of this type of cancer. |
| NCT01487733 |
Tissue |
HPV |
The main objective of this clinical trial study is to analyse the association between biomarkers (HPV tumour status and ERCC1 expression) in patients with metastasis. |
| NCT03469544 |
Blood |
HOTAIR |
The main objective of this trial is quantitation of long non-coding RNA HOTAIR by Real time polymerase chain reaction in peripheral blood. |
| NCT03412058 |
Biopsy |
CD274 (PD-L1) |
To identify predictive factors of response to PD-1 and PD-L1 antagonists authorised for use in France in treatment of melanoma, NSCLC, or HNSCC. |
| NCT05904327 |
Plasma samples |
HPV |
The main objective is to investigate about circulating tumour HPV-DNA as a biomarker for HPV positive HNSCC. |
| NCT04085900 |
Blood and saliva |
EBV |
Objective of the study was to identify earlier detection biomarkers by test all participants for EBV associated biomarkers, including EBV-induced nuclear antigen 1-IgA, VCA-IgA, BNLF2b total antibodies (P85-Ab) |
Current challenges in HNSCC and future directions
Currently, there is a deficiency in comprehensive omics research for HNSCC. While numerous individual omics studies have offered valuable insights, there is a lack of comprehensive omics studies that integrate data across numerous aspects, such as DNA, RNA, proteins, epigenomics, microbiomics and metabolites. Collaborative studies of this nature are crucial for gaining a full understanding of the complex biology of HNSCC.
Currently, massive validation of biomarkers is limited, despite the identification of numerous potential biomarkers, as only a few have been confirmed in large, independent cohorts. There is a need for comprehensive validation studies to establish the clinical usefulness of these biomarkers for early detection, prognosis and treatment response in HNSCC (243).
A great unresolved challenge in HNSCC is the significant heterogeneity in the cancer microenvironment and multidrug resistance mechanisms, contributing to variability in treatment response and resistance. It is essential to comprehend the underlying mechanisms of this heterogeneity and resistance to develop more personalised and effective treatment strategies (244). At present, the clinical and molecular data are not currently well integrated, therefore there is a requirement for research that combines clinical information with molecular discoveries to create algorithms that can assist with clinical decision-making for patients with HNSCC. Classification of biomarkers and their clinical significance in HNSCC is included in Table II.
 |
Table II.
Classification of biomarkers and their clinical significance in HNSCC.
|
Table II.
Classification of biomarkers and their clinical significance in HNSCC.
| First author/s, year |
Marker |
Category |
Clinical application in HNSCC |
(Refs.) |
| Solomon et al, 2020; |
TP53 |
Predictive |
Mutant Tp53 can predict individuals at high risk of |
(50,245) |
| Li et al, 2023 |
|
|
developing HNSCC |
|
| Kovesi and Szende, 2003 |
MKI67 |
Predictive, |
Presence of elevated MKi-67 index may indicate an |
(78) |
| |
|
prognostic |
unfavourable prognosis |
|
| Jasphin et al, 2016; |
PTEN |
Prognostic, |
PTEN level can be used as both prognostic and |
(72,246) |
| Snietura et al, 2012 |
|
predictive |
predictive marker after radiotherapy for high-risk HNSCC |
|
| Gioacchini et al, 2016 |
CCND1 |
Prognostic |
The level of CCND1 expression is positively correlated |
(84) |
| |
|
|
with different stages of cancer |
|
| Murphrey et al, 2023; |
EGFR |
Predictive, |
Determining the level of EGFR as predictive and |
(106,247) |
| Bossi et al, 2012 |
|
prognostic |
prognostic role may help select the patients who can |
|
| |
|
|
mostly benefit from accelerated treatment or personalised |
|
| |
|
|
medicine |
|
| Li et al, 2019; |
CD274 |
Prognostic, |
The level of CD274 expression is directly related to the |
(88,248) |
| Qiao et al, 2020 |
(PD-L1) |
therapeutic |
clinical behaviour of HNSCC |
|
| Gisterek et al, 2006; |
VEGFA |
Prognostic, |
VEGFA expression is usually correlated with stage and |
(123,249) |
| Edirisinghe et al, 2023 |
|
therapeutic |
state of the tumour. This makes it a favourable biomarker |
|
| |
|
|
in prognosis of HNSCC |
|
| Zhu, 2022 |
SCC-Ag |
Prognostic |
Squamous cell carcinoma antigen is always elevated in |
(125) |
| |
|
|
HNSCC compared with healthy cells |
|
| Kavitha et al, 2023; |
CD44 |
Diagnostic, |
CD44 is upregulated and correlated with |
(134,135) |
| Gomez et al, 2020 |
|
prognostic |
clinicopathological characteristics of HNSCC |
|
| Roman and Aragones, 2021; |
HPV |
Prognostic, |
HPV infection causes cancer through its |
(34,250) |
| Augustin et al, 2020 |
|
theragnostic |
oncoproteins E6 and E7. HPV-positive HNSCC |
|
| |
|
|
has more favourable prognosis |
|
| Tan et al, 2017; |
EBV |
Diagnostic, |
Presence of EBV infection is closely associated |
(251,252) |
| Yoshizaki et al, 2023 |
|
prognostic |
with pathological characteristics of nasopharyngeal |
|
| |
|
|
carcinoma |
|
| Yin et al, 2021; |
HOTTIP |
Prognostic |
The level of HOTTIP in clinical samples can be |
(157,160) |
| Shen, et al, 2019 |
|
|
implemented as indicative biomarker in HNSCC |
|
| Yang and Deng, 2014; |
MALAT1 |
Prognostic, |
MALAT1 promotes HNSCC progression, its level |
(151,152,253) |
| Luo et al, 2018; |
|
theragnostic |
in samples may be used in diagnosis and treatment |
|
| Ye et al, 2021 |
|
|
|
|
| Cossu et al, 2019; |
HOTAIR |
Diagnostic, |
In HNSCC, HOTAIR interactions interfere with |
(163,254) |
| Cantile et al, 2021 |
|
prognostic |
different cellular processes during cancer evolution |
|
| Cossu et al, 2019; |
UCA1 |
Prognostic |
Level of UCA1 may play an important role in different |
(163,255) |
| Wang et al, 2020 |
|
|
stages of cancer, as its level in metastasis stages is |
|
| |
|
|
higher than that in the primary tumour |
|
| Song et al, 2019; |
MIAT |
Prognostic |
The level of MIAT in clinical samples can indicate |
(183,184) |
| Liu et al, 2019 |
|
|
progression of HNSCC |
|
| Shah et al, 2016; |
miR-21 |
Theragnostic |
Presence of miR-21 can be applied as theragnostic |
(191,256) |
| Mohammadi et al, 2021 |
|
|
biomarkers for predicting of chemotherapy and |
|
| |
|
|
radiotherapy response in HNSCC |
|
| Thomaidou et al, 2022 |
miR-200 |
Prognostic |
The level of miR-200 family has been found to have the |
(153) |
| |
|
|
potential role of prognosis in patients with HNSCC |
|
| Yu et al, 2019; |
let-7 |
Prognostic, |
Determining the level of Let-7 miRNAs in |
(201,257) |
| Ma et al, 2021 |
|
theragnostic |
clinical samples can help in prognosis and treatment |
|
| |
|
|
planning because Let-7 is involved in tumour growth, |
|
| |
|
|
differentiation and regenerative potential |
|
Future directions
Generally, the future direction for HNSCC should focus on large-scale multi-omics, biomarker discovery and validations, use of new technologies for diagnosis and treatment development of targeted therapy and personalised treatment of patients with HNSCC.
Conclusions
In conclusion, the complex process of tumorigenesis in HNSCC was described and the important role of genetic mutations, epigenetic modifications, dysregulated signalling pathways and biomarkers was highlighted. HNSCC development involves a series of stages from pre-malignant evolution that starts at the early stages of lesion development to the advanced stages of cancer invasion and metastasis. This complex interplay involves molecular events resulting in the transforming of normal epithelial cells into highly malignant phenotypes.
Identifying biomarkers for HNSCC is a dynamic and continuously evolving field in science, with promising implications for patient care. Biomarkers found in tissues and body fluids can indicate the presence of HNSCC, offering the potential for earlier and more accurate diagnosis. Furthermore, these biomarkers can provide prognostic information that could guide treatment decisions and predict patient outcomes.
To enhance HNSCC cancer management and personalised treatment in this new era of science and technology, it is crucial to discover more reliable novel biomarkers and incorporate them into clinical use. Multi-omics analysis can provide a large database compared with a single analysis and offer comprehensive insights into the heterogeneity of pathogenesis, significantly contributing to the development of new diagnostic tools and targeted therapies for HNSCC. By integrating multi-omics data, clinicians can improve understanding of the molecular underpinnings of HNSCC, leading to more effective treatment plans and improved patient management strategies.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
CAS performed literature review, wrote the original draft, and wrote, reviewed and edited the manuscript. CB, DAS, AKEN and GGP conceptualized the study, and wrote, reviewed and edited the manuscript. IM conceptualized and supervised the study, wrote the original draft, and wrote, reviewed and edited the manuscript. All authors read and approved the final version of the manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor‑in‑Chief for the journal, but had no personal involvement in the reviewing process, or any influence in terms of adjudicating on the final decision, for this article.
Glossary
Abbreviations
Abbreviations:
|
CSC
|
cancer stem cells
|
|
EBV
|
Epstein-Barr virus
|
|
HNSCC
|
head and neck squamous cell carcinoma
|
|
HPV
|
human papillomavirus
|
|
lncRNA
|
long non-coding RNA
|
|
LSCC
|
laryngeal squamous cell carcinoma
|
|
miR or miRNA
|
microRNA
|
|
NPC
|
nasopharyngeal carcinoma
|
|
OSCC
|
oral squamous cell carcinoma
|
|
TNM
|
tumour, lymph node and metastasis
|
|
TSCC
|
tongue squamous cell carcinoma
|
References
|
1
|
Brown JS, Amend SR, Austin RH, Gatenby RA, Hammarlund EU and Pienta KJ: Updating the definition of cancer. Mol Cancer Res. 21:1142–1147. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Szyfter K: Genetics and molecular biology of head and neck cancer. Biomolecules. 11:12932021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jerjes WK, Upile T, Wong BJ, Betz CS, Sterenborg HJ, Witjes MJ, Berg K, van Veen R, Biel MA, El-Naggar AK, et al: The future of medical diagnostics: review paper. Head Neck Oncol. 3:382011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ruffin AT, Li H, Vujanovic L, Zandberg DP, Ferris RL and Bruno TC: Improving head and neck cancer therapies by immunomodulation of the tumour microenvironment. Nat Rev Cancer. 23:173–188. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Langer CJ: Exploring biomarkers in head and neck cancer. Cancer. 118:3882–3892. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Head and Neck Cancer Study Group (HNCSG), . Monden N, Asakage T, Kiyota N, Homma A, Matsuura K, Hanai N, Kodaira T, Zenda S, Fujii H, et al: A review of head and neck cancer staging system in the TNM classification of malignant tumors (eighth edition). Jpn J Clin Oncol. 49:589–595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Field JK, Zoumpourlis V, Spandidos DA and Jones AS: p53 expression and mutations in squamous cell carcinoma of the head and neck: Expression correlates with the patients' use of tobacco and alcohol. Cancer Detect Prev. 18:197–208. 1994.PubMed/NCBI
|
|
8
|
Barsouk A, Aluru JS, Rawla P, Saginala K and Barsouk A: Epidemiology, risk factors, and prevention of head and neck squamous cell carcinoma. Med Sci (Basel). 11:422023.PubMed/NCBI
|
|
9
|
Watling DL, Gown AM and Coltrera MD: Overexpression of p53 in head and neck cancer. Head Neck. 14:437–444. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Field JK, Spandidos DA, Malliri A, Gosney JR, Yiagnisis M and Stell PM: Elevated P53 expression correlates with a history of heavy smoking in squamous cell carcinoma of the head and neck. Br J Cancer. 64:573–577. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Svider PF, Blasco MA, Raza SN, Shkoukani M, Sukari A, Yoo GH, Folbe AJ, Lin HS and Fribley AM: Head and neck cancer. Otolaryngol Head Neck Surg. 156:10–13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Field JK, Spandidos DA and Stell PM: Overexpression of p53 gene in head-and-neck cancer, linked with heavy smoking and drinking. Lancet. 339:502–503. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Steuer CE, El-Deiry M, Parks JR, Higgins KA and Saba NF: An update on larynx cancer. CA Cancer J Clin. 67:31–50. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Anderson G, Ebadi M, Vo K, Novak J, Govindarajan A and Amini A: An updated review on head and neck cancer treatment with radiation therapy. Cancers (Basel). 13:49122021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang Z, Wu C, Zhao Y, Zhan Q, Wang K and Li Y: Global research trends in immunotherapy for head and neck neoplasms: A scientometric study. Heliyon. 9:e153092023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pereira D, Martins D and Mendes F: Immunotherapy in head and neck cancer when, how, and why? Biomedicines. 10:21512022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kleszcz R: Advantages of the combinatorial molecular targeted therapy of head and neck cancer-A step before anakoinosis-based personalized treatment. Cancers (Basel). 15:42472023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hijazi MA, Gessner A and El-Najjar N: Repurposing of chronically used drugs in cancer therapy: A chance to grasp. Cancers (Basel). 15:31992023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xia Y, Sun M, Huang H and Jin WL: Drug repurposing for cancer therapy. Signal Transduct Target Ther. 9:922024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Johnson FM, Janku F, Gouda MA, Tran HT, Kawedia JD, Schmitz D, Streefkerk H, Lee JJ, Andersen CR, Deng D, et al: Inhibition of the phosphatidylinositol-3 kinase pathway using bimiralisib in loss-of-function NOTCH1-Mutant head and neck cancer. Oncologist. 27:1004–e1926. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li C, Geng H, Ji L, Ma X, Yin Q and Xiong H: ESM-1: A novel tumor biomaker and its research advances. Anticancer Agents Med Chem. 19:1687–1694. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wurzba S, Salo TA and Coletta RD: Editorial: Prognostic biomarkers for oral cancer. Front Oral Health. 3:9943872022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang HC, Yeh TJ, Chan LP, Hsu CM and Cho SF: Exploration of feasible immune biomarkers for immune checkpoint inhibitors in head and neck squamous cell carcinoma treatment in real World clinical practice. Int J Mol Sci. 21:76212020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
El-Naggar A: Pathobiology of head and neck squamous tumorigenesis. Curr Cancer Drug Targets. 7:606–612. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes. Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Curtius K, Wright NA and Graham TA: Evolution of premalignant disease. Cold Spring Harb Perspect Med. 7:a0265422017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES and Getz G: Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 505:495–501. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Leshchiner I, Mroz EA, Cha J, Rosebrock D, Spiro O, Bonilla-Velez J, Faquin WC, Lefranc-Torres A, Lin DT, Michaud WA, et al: Inferring early genetic progression in cancers with unobtainable premalignant disease. Nat Cancer. 4:550–563. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE and Grandis JR: Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 6:922020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mbeunkui F and Johann DJ Jr: Cancer and the tumor microenvironment: A review of an essential relationship. Cancer Chemother Pharmacol. 63:571–582. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Krishnamurthy S, Dong Z, Vodopyanov D, Imai A, Helman JI, Prince ME, Wicha MS and Nör JE: Endothelial cell-initiated signaling promotes the survival and self-renewal of cancer stem cells. Cancer Res. 70:9969–9978. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vashisht S, Mishra H, Mishra PK, Ekielski A and Talegaonkar S: Structure, genome, infection cycle and clinical manifestations associated with human papillomavirus. Curr Pharm Biotechnol. 20:1260–1280. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mehanna H, Taberna M, von Buchwald C, Tous S, Brooks J, Mena M, Morey F, Grønhøj C, Rasmussen JH, Garset-Zamani M, et al: Prognostic implications of p16 and HPV discordance in oropharyngeal cancer (HNCIG-EPIC-OPC): A multicentre, multinational, individual patient data analysis. Lancet Oncol. 24:239–251. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Roman BR and Aragones A: Epidemiology and incidence of HPV-related cancers of the head and neck. J Surg Oncol. 124:920–922. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dokianakis DN, Papaefthimiou M, Tsiveleka A and Spandidos DA: High prevalence of HPV18 in correlation with ras gene mutations and clinicopathological parameters in cervical cancer studied from stained cytological smears. Oncol Rep. 6:1327–1331. 1999.PubMed/NCBI
|
|
36
|
Galati L, Chiocca S, Duca D, Tagliabue M, Simoens C, Gheit T, Arbyn M and Tommasino M: HPV and head and neck cancers: Towards early diagnosis and prevention. Tumour Virus Res. 14:2002452022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lechner M, Liu J, Masterson L and Fenton TR: HPV-associated oropharyngeal cancer: Epidemiology, molecular biology and clinical management. Nat Rev Clin Oncol. 19:306–327. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
El-Naggar AK and Westra WH: p16 expression as a surrogate marker for HPV-related oropharyngeal carcinoma: A guide for interpretative relevance and consistency. Head Neck. 34:459–461. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ramesh PS, Bovilla VR, Swamy VH, Manoli NN, Dasegowda KB, Siddegowda SM, Chandrashekarappa S, Somasundara VM, Kabekkodu SP, Rajesh R, et al: Human papillomavirus-driven repression of NRF2 signalling confers chemo-radio sensitivity and predicts prognosis in head and neck squamous cell carcinoma. Free Radic Biol Med. 205:234–243. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Holzinger D, Flechtenmacher C, Henfling N, Kaden I, Grabe N, Lahrmann B, Schmitt M, Hess J, Pawlita M and Bosch FX: Identification of oropharyngeal squamous cell carcinomas with active HPV16 involvement by immunohistochemical analysis of the retinoblastoma protein pathway. Int J Cancer. 133:1389–1399. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lechner M, Chakravarthy AR, Walter V, Masterson L, Feber A, Jay A, Weinberger PM, McIndoe RA, Forde CT, Chester K, et al: Frequent HPV-independent p16/INK4A overexpression in head and neck cancer. Oral Oncol. 83:32–37. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kuhn JP, Schmid W, Korner S, Bochen F, Wemmert S, Rimbach H, Smola S, Radosa JC, Wagner M, Morris LGT, et al: HPV status as prognostic biomarker in head and neck cancer-which method fits the best for outcome prediction? Cancers (Basel). 13:47302021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sánchez Barrueco A, González Galán F, Villacampa Aubá JM, Díaz Tapia G, Fernández Hernández S, Martín-Arriscado Arroba C, Cenjor Español C and Almodóvar Álvarez C: p16 influence on laryngeal squamous cell carcinoma relapse and survival. Otolaryngol Head Neck Surg. 160:1042–1047. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chung CH, Zhang Q, Kong CS, Harris J, Fertig EJ, Harari PM, Wang D, Redmond KP, Shenouda G, Trotti A, et al: p16 protein expression and human papillomavirus status as prognostic biomarkers of nonoropharyngeal head and neck squamous cell carcinoma. J Clin Oncol. 32:3930–3938. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kori M and Arga KY: HPV16 status predicts potential protein biomarkers and therapeutics in head and neck squamous cell carcinoma. Virology. 582:90–99. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wise-Draper TM, Bahig H, Tonneau M, Karivedu V and Burtness B: Current therapy for metastatic head and neck cancer: Evidence, opportunities, and challenges. Am Soc Clin Oncol Educ Book. 42:1–14. 2022.PubMed/NCBI
|
|
47
|
Liloglou T, Scholes AG, Spandidos DA, Vaughan ED, Jones AS and Field JK: p53 mutations in squamous cell carcinoma of the head and neck predominate in a subgroup of former and present smokers with a low frequency of genetic instability. Cancer Res. 57:4070–4074. 1997.PubMed/NCBI
|
|
48
|
Carpov D, Buiga R and Nagy VM: DNA damage response and potential biomarkers of radiosensitivity in head and neck cancers: clinical implications. Rom J Morphol Embryol. 64:5–13. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang J, Hu Y, Escamilla-Rivera V, Gonzalez CL, Tang L, Wang B, El-Naggar AK, Myers JN and Caulin C: Epithelial mutant p53 promotes resistance to Anti-PD-1-Mediated oral cancer immunoprevention in carcinogen-induced mouse models. Cancers (Basel). 13:14712021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Solomon B, Young RJ and Rischin D: Head and neck squamous cell carcinoma: Genomics and emerging biomarkers for immunomodulatory cancer treatments. Semin Cancer Biol. 52((Pt 2)): 228–240. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Amenabar JM, Da Silva BM and Punyadeera C: Salivary protein biomarkers for head and neck cancer. Expert Rev Mol Diagn. 20:305–313. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kampel L, Feldstein S, Tsuriel S, Hannes V, Carmel Neiderman NN, Horowitz G, Warshavsky A, Leider-Trejo L, Hershkovitz D and Muhanna N: Mutated TP53 in circulating tumor DNA as a risk level biomarker in head and neck squamous cell carcinoma patients. Biomolecules. 13:14182023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yu R, Sun T, Zhang X, Li Z, Xu Y, Liu K, Shi Y, Wu X, Shao Y and Kong L: TP53 Co-Mutational Features and NGS-Calibrated immunohistochemistry threshold in gastric cancer. Onco Targets Ther. 14:4967–4978. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Peltonen JK, Helppi HM, Paakko P, Turpeenniemi-Hujanen T and Vahakangas KH: p53 in head and neck cancer: functional consequences and environmental implications of TP53 mutations. Head Neck Oncol. 2:362010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Field J, Malliri A, Butt S, Gosney J, Phillips D, Spandidos D and Jones A: P53 expression in end-stage squamous-cell carcinoma of the head and neck prior to chemotherapy treatment-expression correlates with a very poor clinical outcome. Int J Oncol. 3:431–435. 1993.PubMed/NCBI
|
|
56
|
Field J, Malliri A, Jones A and Spandidos D: Mutations in the p53 gene at codon 249 are rare in squamous-cell carcinoma of the head and neck. Int J Oncol. 1:253–256. 1992.PubMed/NCBI
|
|
57
|
Jagadeeshan S, Novoplansky OZ, Cohen O, Kurth I, Hess J, Rosenberg AJ, Grandis JR and Elkabets M: New insights into RAS in head and neck cancer. Biochim Biophys Acta Rev Cancer. 1878:1889632023. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Novoplansky O, Jagadeeshan S, Regev O, Menashe I and Elkabets M: Worldwide prevalence and clinical characteristics of RAS mutations in head and neck cancer: A systematic review and meta-analysis. Front Oncol. 12:8389112022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Weidhaas JB, Harris J, Schaue D, Chen AM, Chin R, Axelrod R, El-Naggar AK, Singh AK, Galloway TJ, Raben D, et al: The KRAS-Variant and cetuximab response in head and neck squamous cell cancer: A secondary analysis of a Randomized clinical trial. JAMA Oncol. 3:483–491. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Rampias T, Giagini A, Siolos S, Matsuzaki H, Sasaki C, Scorilas A and Psyrri A: RAS/PI3K crosstalk and cetuximab resistance in head and neck squamous cell carcinoma. Clin Cancer Res. 20:2933–2946. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang J, Al-Majid D, Brenner JC and Smith JD: Mutant HRas signaling and rationale for use of farnesyltransferase inhibitors in head and neck squamous cell carcinoma. Target Oncol. 18:643–655. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Coleman N, Marcelo KL, Hopkins JF, Khan NI, Du R, Hong L, Park E, Balsara B, Leoni M, Pickering C, et al: HRAS mutations define a distinct subgroup in head and neck squamous cell carcinoma. JCO Precis Oncol. 7:e22002112023. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sasaki E, Masago K, Fujita S, Hanai N and Yatabe Y: Frequent KRAS and HRAS mutations in squamous cell papillomas of the head and neck. J Pathol Clin Res. 6:154–159. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Papanikolaou V, Chrysovergis A, Mastronikolis S, Tsiambas E, Ragos V, Peschos D, Spyropoulou D, Pantos P, Niotis A, Mastronikolis N and Kyrodimos E: Impact of K-Ras Over-expression in laryngeal squamous cell carcinoma. In Vivo. 35:1611–1615. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wei F, Wu Y, Wang Z, Li Y, Wang J, Shao G, Yang Y and Shi B: Diagnostic significance of DNA methylation of PTEN and DAPK in thyroid tumors. Clin Endocrinol (Oxf). 93:187–195. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Qiu R, Wang W, Li J and Wang Y: Roles of PTEN inactivation and PD-1/PD-L1 activation in esophageal squamous cell carcinoma. Mol Biol Rep. 49:6633–6645. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kim B, Kang SY, Kim D, Heo YJ and Kim KM: PTEN Protein Loss and Loss-of-Function mutations in gastric cancers: The relationship with microsatellite instability, EBV, HER2, and PD-L1 expression. Cancers (Basel). 12:17242020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kerdjoudj M, De La Torre RA and Arnouk H: Characterization of DJ-1, PTEN, and p-Akt as prognostic biomarkers in the progression of oral squamous cell carcinoma. Cureus. 15:e344362023.PubMed/NCBI
|
|
69
|
Squarize CH, Castilho RM, Abrahao AC, Molinolo A, Lingen MW and Gutkind JS: PTEN deficiency contributes to the development and progression of head and neck cancer. Neoplasia. 15:461–471. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Blessy S, Sankar S and Girija AS: In silico analysis of key miRNAs and gene-network analysis of pten gene involved in head and neck cancer. J Surv Fish Sci. 10:198–204. 2023.
|
|
71
|
Saintigny P, Mitani Y, Pytynia KB, Ferrarotto R, Roberts DB, Weber RS, Kies MS, Maity SN, Lin SH and El-Naggar AK: Frequent PTEN loss and differential HER2/PI3K signaling pathway alterations in salivary duct carcinoma: Implications for targeted therapy. Cancer. 124:3693–3705. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Jasphin SS, Desai D, Pandit S, Gonsalves NM, Nayak PB and Iype A: Immunohistochemical expression of phosphatase and tensin homolog in histologic gradings of oral squamous cell carcinoma. Contemp Clin Dent. 7:524–528. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Vidotto T, Melo CM, Lautert-Dutra W, Chaves LP, Reis RB and Squire JA: Pan-cancer genomic analysis shows hemizygous PTEN loss tumors are associated with immune evasion and poor outcome. Sci Rep. 13:50492023. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, et al: The GeneCards Suite: From gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 54:1.30.1–1.30.33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Cuylen S, Blaukopf C, Politi AZ, Müller-Reichert T, Neumann B, Poser I, Ellenberg J, Hyman AA and Gerlich DW: Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature. 535:308–312. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
MacCallum DE and Hall PA: The biochemical characterization of the DNA binding activity of pKi67. J Pathol. 191:286–298. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Urruticoechea A, Smith IE and Dowsett M: Proliferation marker Ki-67 in early breast cancer. J Clin Oncol. 23:7212–7220. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Kovesi G and Szende B: Changes in apoptosis and mitotic index, p53 and Ki67 expression in various types of oral leukoplakia. Oncology. 65:331–336. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Hellgren LS, Stenman A, Paulsson JO, Höög A, Larsson C, Zedenius J and Juhlin CC: Prognostic Utility of the Ki-67 labeling index in follicular thyroid tumors: A 20-Year experience from a tertiary thyroid center. Endocr Pathol. 33:231–242. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ahmed MW, Kayani MA, Shabbir G, Ali SM, Shinwari WU and Mahjabeen I: Expression of PTEN and its correlation with proliferation marker Ki-67 in head and neck cancer. Int J Biol Markers. 31:e193–e203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
He QY, Jin F, Li YY, Wu WL, Long JH, Luo XL, Gong XY, Chen XX, Bi T, Li ZL, et al: Prognostic significance of downregulated BMAL1 and upregulated Ki-67 proteins in nasopharyngeal carcinoma. Chronobiol Int. 35:348–357. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Huang SF, Cheng SD, Chuang WY, Chen IH, Liao CT, Wang HM and Hsieh LL: Cyclin D1 overexpression and poor clinical outcomes in Taiwanese oral cavity squamous cell carcinoma. World J Surg Oncol. 10:402012. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Rauf M, Azmat H, Shahab S, Ahmad A, Khadija S and Firidi J: Immunohistochemical expression of cyclind1 in conventional squamous cell carcinomaof oral cavity. J Ayub Med Coll Abbottabad. 35:11–16. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Gioacchini FM, Alicandri-Ciufelli M, Kaleci S, Magliulo G, Presutti L and Re M: The prognostic value of cyclin D1 expression in head and neck squamous cell carcinoma. Eur Arch Otorhinolaryngol. 273:801–809. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yang K, Zhu G, Sun Y, Hu Y, Lv Y, Li Y, Pan J, Chen F, Zhou Y and Zhang J: Prognostic significance of cyclin D1 expression pattern in HPV-negative oral and oropharyngeal carcinoma: A deep-learning approach. J Oral Pathol Med. 52:919–929. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wang CC, Lu DD, Shen MH, Chen RL, Zhang ZH and Lv JH: Clinical value of Cyclin D1 and P21 in the differential diagnosis of papillary thyroid carcinoma. Diagn Pathol. 18:1232023. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
John RR, Sam N and Chandrasekaran B: Prognostic significance of proliferative markers: Cyclin D1 and CENPF in oral squamous cell carcinoma patients-A cohort study. J Maxillofac Oral Surg. 22:734–740. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Li C, Li C, Zhi C, Liang W, Wang X, Chen X, Lv T, Shen Q, Song Y, Lin D and Liu H: Clinical significance of PD-L1 expression in serum-derived exosomes in NSCLC patients. J Transl Med. 17:3552019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Oliva M, Spreafico A, Taberna M, Alemany L, Coburn B, Mesia R and Siu LL: Immune biomarkers of response to immune-checkpoint inhibitors in head and neck squamous cell carcinoma. Ann Oncol. 30:57–67. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
William WN Jr, Zhang J, Zhao X, Parra ER, Uraoka N, Lin HY, Peng SA, El-Naggar AK, Rodriguez-Canales J, Song J, et al: Spatial PD-L1, immune-cell microenvironment, and genomic copy-number alteration patterns and drivers of invasive-disease transition in prospective oral precancer cohort. Cancer. 129:714–727. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Müller T, Braun M, Dietrich D, Aktekin S, Höft S, Kristiansen G, Göke F, Schröck A, Brägelmann J, Held SAE, et al: PD-L1: A novel prognostic biomarker in head and neck squamous cell carcinoma. Oncotarget. 8:52889–52900. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Malinowska K, Kowalski A, Merecz-Sadowska A, Paprocka-Zjawiona M, Sitarek P, Kowalczyk T and Zielińska-Bliźniewska H: PD-1 and PD-L1 expression levels as a potential biomarker of chronic Rhinosinusitis and head and neck cancers. J Clin Med. 12:20332023. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Jiang D, Song Q, Wang H, Huang J, Wang H, Hou J, Li X, Xu Y, Sujie A, Zeng H, et al: Independent prognostic role of PD-L1expression in patients with esophageal squamous cell carcinoma. Oncotarget. 8:8315–8329. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Shah PA, Huang C, Li Q, Kazi SA, Byers LA, Wang J, Johnson FM and Frederick MJ: NOTCH1 signaling in head and neck squamous cell carcinoma. Cells. 9:26772020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Sadeghi ES, Nematpour FS, Mohtasham N and Mohajertehran F: The oncogenic role of NOTCH1 as biomarker in oral squamous cell carcinoma and oral lichen planus. Dent Res J (Isfahan). 20:1022023. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Lee SH, Do SI, Lee HJ, Kang HJ, Koo BS and Lim YC: Notch1 signaling contributes to stemness in head and neck squamous cell carcinoma. Lab Invest. 96:508–516. 2016.(In Amharic, English). View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Fukusumi T and Califano JA: The NOTCH pathway in head and neck squamous cell carcinoma. J Dent Res. 97:645–653. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et al: Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 333:1154–1157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Zhang W, Wang J, Huang D, Liu Z, Lu T, Cui C and Li Z: Single-cell sequencing reveals SATB2/NOTCH1 signaling promotes the progression of malignancy of epithelial cells from papillary thyroid cancer. Mol Carcinog. 63:22–33. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Yeh TJ, Luo CW, Du JS, Huang CT, Wang MH, Chuang TM, Gau YC, Cho SF, Liu YC, Hsiao HH, et al: Deciphering the functions of telomerase reverse transcriptase in head and neck cancer. Biomedicines. 11:6912023. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Leão R, Apolónio JD, Lee D, Figueiredo A, Tabori U and Castelo-Branco P: Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: Clinical impacts in cancer. J Biomed Sci. 25:222018. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Arantes LMRB, Cruvinel-Carloni A, de Carvalho AC, Sorroche BP, Carvalho AL, Scapulatempo-Neto C and Reis RM: TERT Promoter Mutation C228T increases risk for tumor recurrence and death in head and neck cancer patients. Front Oncol. 10:12752020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Boscolo-Rizzo P, Tirelli G, Polesel J, Sia E, Phillips V, Borsetto D, De Rossi A and Giunco S: TERT promoter mutations in head and neck squamous cell carcinoma: A systematic review and meta-analysis on prevalence and prognostic significance. Oral Oncol. 140:1063982023. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Ganesh MS, Narayanan GS and Kumar R: Change of telomerase activity in peripheral blood of patients with head and neck squamous cell carcinoma pre and post curative treatment. Rep Pract Oncol Radiother. 25:28–34. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Xu Y, Quan Z, Zhan Y, Wang H, Luo J, Wang W and Fan S: SSTR2 positively associates with EGFR and predicts poor prognosis in nasopharyngeal carcinoma. J Clin Pathol. Sep 27–2023.(Epub ahead of print). View Article : Google Scholar
|
|
106
|
Murphrey MB, Quaim L and Varacallo M: Biochemistry, Epidermal Growth Factor Receptor. Disclosure: Lamisa Quaim declares no relevant financial relationships with ineligible companies. Disclosure: Matthew Varacallo declares no relevant financial relationships with ineligible companies. StatPearls; Treasure Island, FL: 2023
|
|
107
|
Zimmermann M, Zouhair A, Azria D and Ozsahin M: The epidermal growth factor receptor (EGFR) in head and neck cancer: its role and treatment implications. Radiat Oncol. 1:112006. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Boccellino M, De Rosa A and Di Domenico M: An ELISA test able to predict the development of oral cancer: The significance of the interplay between steroid receptors and the EGF receptor for early diagnosis. Diagnostics (Basel). 13:20012023. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Nair S, Bonner JA and Bredel M: EGFR mutations in head and neck squamous cell carcinoma. Int J Mol Sci. 23:38182022. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Sivarajah S, Kostiuk M, Lindsay C, Puttagunta L, O'Connell DA, Harris J, Seikaly H and Biron VL: EGFR as a biomarker of smoking status and survival in oropharyngeal squamous cell carcinoma. J Otolaryngol Head Neck Surg. 48:12019. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Rehmani HS and Issaeva N: EGFR in head and neck squamous cell carcinoma: Exploring possibilities of novel drug combinations. Ann Transl Med. 8:8132020. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Licitra L, Perrone F, Tamborini E, Bertola L, Ghirelli C, Negri T, Orsenigo M, Filipazzi P, Pastore E, Pompilio M, et al: Role of EGFR family receptors in proliferation of squamous carcinoma cells induced by wound healing fluids of head and neck cancer patients. Ann Oncol. 22:1886–1893. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Numico G, Russi EG, Colantonio I, Lantermo RA, Silvestris N, Vitiello R, Comino A, Abrate M, Zavattero C, Melano A and Merlano M: EGFR status and prognosis of patients with locally advanced head and neck cancer treated with chemoradiotherapy. Anticancer Res. 30:671–676. 2010.PubMed/NCBI
|
|
114
|
Schiff BA, McMurphy AB, Jasser SA, Younes MN, Doan D, Yigitbasi OG, Kim S, Zhou G, Mandal M, Bekele BN, et al: Epidermal growth factor receptor (EGFR) is overexpressed in anaplastic thyroid cancer, and the EGFR inhibitor gefitinib inhibits the growth of anaplastic thyroid cancer. Clin Cancer Res. 10:8594–8602. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Cívico-Ortega JL, González-Ruiz I, Ramos-García P, Cruz-Granados D, Samayoa-Descamps V and González-Moles MÁ: Prognostic and clinicopathological significance of epidermal growth factor receptor (EGFR) expression in oral squamous cell carcinoma: Systematic Review and meta-analysis. Int J Mol Sci. 24:118882023. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Hsu ER, Gillenwater AM, Hasan MQ, Williams MD, El-Naggar AK and Richards-Kortum RR: Real-time detection of epidermal growth factor receptor expression in fresh oral cavity biopsies using a molecular-specific contrast agent. Int J Cancer. 118:3062–3071. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Chen X, Liang R and Zhu X: Anti-EGFR therapies in nasopharyngeal carcinoma. Biomed Pharmacother. 131:1106492020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Neufeld G, Cohen T, Gengrinovitch S and Poltorak Z: Vascular endothelial growth factor (VEGF) and its receptors. FASEB J. 13:9–22. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Bouzoubaa SM, Benlahfid M, Sidqui M, Aboussaouira T, Rifki C, Brouillet S, Traboulsi W, Alfaidy N and Benharouga M: Vascular endothelial growth factor (VEGF) and Endocrine gland-VEGF (EG-VEGF) are down regulated in head and neck cancer. Clin Otolaryngol. 45:788–795. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Schlüter A, Weller P, Kanaan O, Nel I, Heusgen L, Höing B, Haßkamp P, Zander S, Mandapathil M, Dominas N, et al: CD31 and VEGF are prognostic biomarkers in early-stage, but not in late-stage, laryngeal squamous cell carcinoma. BMC Cancer. 18:2722018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Dumitru CS and Raica M: Vascular endothelial growth factor family and head and neck squamous cell carcinoma. Anticancer Res. 43:4315–4326. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Patel SA, Nilsson MB, Le X, Cascone T, Jain RK and Heymach JV: Molecular mechanisms and future implications of VEGF/VEGFR in cancer therapy. Clin Cancer Res. 29:30–39. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Gisterek I and Kornafel J: Vascular endothelial growth factor in head and neck cancer. Pol Merkur Lekarski. 20:242–244. 2006.(In Polish). PubMed/NCBI
|
|
124
|
Edirisinghe ST, Weerasekera M, De Silva DK, Devmini MT, Pathmaperuma S, Wijesinghe GK, Nisansala T, Maddumage A, Huzaini H, Rich AM, et al: Vascular endothelial growth factor A (VEGF-A) and vascular endothelial growth factor receptor 2 (VEGFR-2) as potential biomarkers for oral squamous cell carcinoma: A Sri Lankan study. Asian Pac J Cancer Prev. 24:267–274. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Zhu H: Squamous cell carcinoma antigen: Clinical application and research status. Diagnostics (Basel). 12:10652022. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Yasumatsu R, Nakano T, Hashimoto K, Kogo R, Wakasaki T and Nakagawa T: The clinical value of serum squamous cell carcinoma antigens 1 and 2 in head and neck squamous cell carcinoma. Auris Nasus Larynx. 46:135–140. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Schepens EJA, Al-Mamgani A, Karssemakers LHE, van den Broek D, van den Brekel MWM and Lopez-Yurda M: Squamous cell carcinoma antigen in the follow-up of patients with head and neck cancer. Otolaryngol Head Neck Surg. 170:422–430. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
van Schaik JE, Muller Kobold AC, van der Laan B, van der Vegt B, van Hemel BM and Plaat BEC: Squamous cell carcinoma antigen concentration in fine needle aspiration samples: A new method to detect cervical lymph node metastases of head and neck squamous cell carcinoma. Head Neck. 41:2561–2565. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Souza SP, Serra MGDS, Oliveira NDS, Oliveira MC, Junior JB and Reis TG: Arterial lactate as a predictor of postoperative complications in head and neck squamous cell carcinoma. Braz J Otorhinolaryngol. 88 (Suppl 1):S97–S101. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Blatt S, Voelxen N, Sagheb K, Pabst AM, Walenta S, Schroeder T, Mueller-Klieser W and Ziebart T: Lactate as a predictive marker for tumor recurrence in patients with head and neck squamous cell carcinoma (HNSCC) post radiation: A prospective study over 15 years. Clin Oral Investig. 20:2097–2104. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Brizel DM, Schroeder T, Scher RL, Walenta S, Clough RW, Dewhirst MW and Mueller-Klieser W: Elevated tumor lactate concentrations predict for an increased risk of metastases in head-and-neck cancer. Int J Radiat Oncol Biol Phys. 251:349–353. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Wu M, Li B, Zhang X and Sun G: Serum metabolomics reveals an innovative diagnostic model for salivary gland tumors. Anal Biochem. 655:1148532022. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Srivastava S, Roy R, Gupta V, Tiwari A, Srivastava AN and Sonkar AA: Proton HR-MAS MR spectroscopy of oral squamous cell carcinoma tissues: An ex vivo study to identify malignancy induced metabolic fingerprints. Metabolomics. 7:278–288. 2011. View Article : Google Scholar
|
|
134
|
Kavitha L, Vijayashree Priyadharsini J, Kattula D, Rao UKM, Balaji Srikanth R, Kuzhalmozhi M and Ranganathan K: Expression of CD44 in head and neck squamous cell carcinoma-an in-silico study. Glob Med Genet. 10:221–228. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Gomez KE, Wu F, Keysar SB, Morton JJ, Miller B, Chimed TS, Le PN, Nieto C, Chowdhury FN, Tyagi A, et al: Cancer Cell CD44 mediates macrophage/monocyte-driven regulation of head and neck cancer stem cells. Cancer Res. 80:4185–4198. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Yu SS and Cirillo N: The molecular markers of cancer stem cells in head and neck tumors. J Cell Physiol. 235:65–73. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Chen J, Zhou J, Lu J, Xiong H, Shi X and Gong L: Significance of CD44 expression in head and neck cancer: A systemic review and meta-analysis. BMC Cancer. 14:152014. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Xu H, Niu M, Yuan X, Wu K and Liu A: CD44 as a tumor biomarker and therapeutic target. Exp Hematol Oncol. 9:362020. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Puzzo L, Bianco MR, Salvatorelli L, Tinnirello G, Occhiuzzi F, Latella D and Allegra E: CD44, PDL1, and ATG7 expression in laryngeal squamous cell carcinomas with tissue microarray (TMA) Technique: Evaluation of the potential prognostic and predictive roles. Cancers (Basel). 15:24612023. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Franzmann EJ, Reategui EP, Pedroso F, Pernas FG, Karakullukcu BM, Carraway KL, Hamilton K, Singal R and Goodwin WJ: Soluble CD44 is a potential marker for the early detection of head and neck cancer. Cancer Epidemiol Biomarkers Prev. 16:1348–1355. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Ali F, Siddiqui TA, Sukhia RH, Maqsood A and Ghandhi D: Diagnostic and prognostic role of cancer stem cell biomarkers in oral squamous cell carcinoma; A systematic review. J Pak Med Assoc. 73 (Suppl 1):S32–S39. 2023. View Article : Google Scholar
|
|
142
|
Vasudevan HN and Yom SS: Nasopharyngeal carcinoma and its association with epstein-barr virus. Hematol Oncol Clin North Am. 35:963–971. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Jiang W, Chamberlain PD, Garden AS, Kim BY, Ma D, Lo EJ, Bell D, Gunn GB, Fuller CD, Rosenthal DI, et al: Prognostic value of p16 expression in Epstein-Barr virus-positive nasopharyngeal carcinomas. Head Neck. 38 (Suppl 1):E1459–E1466. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Lee LA, Fang TJ, Li HY, Chuang HH, Kang CJ, Chang KP, Liao CT, Chen TC, Huang CG and Yen TC: Effects of Epstein-Barr virus infection on the risk and prognosis of primary laryngeal squamous cell carcinoma: A hospital-based case-control study in Taiwan. Cancers (Basel). 13:17412021. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Wu S, Yuan X, Huang H, Li Y, Cui L, Lin D, Lu W, Feng H, Chen Z, Liu X, et al: Nomogram incorporating Epstein-Barr virus DNA and a novel immune-nutritional marker for survival prediction in nasopharyngeal carcinoma. BMC Cancer. 23:12172023. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
de Lima MAP, Silva ADL, do Nascimento Filho ACS, Cordeiro TL, Bezerra JPS, Rocha MAB, Pinheiro SFL, Pinheiro Junior RFF, Gadelha MDSV and da Silva CGL: Epstein-Barr virus-associated carcinoma of the larynx: A systematic review with meta-analysis. Pathogens. 10:14292021. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Goldenberg D, Benoit NE, Begum S, Westra WH, Cohen Y, Koch WM, Sidransky D and Califano JA: Epstein-Barr virus in head and neck cancer assessed by quantitative polymerase chain reaction. Laryngoscope. 114:1027–1031. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Javadirad E, Yekta AM, Lorestani RC and Azimivaghar J: A survey of human papillomavirus and epstein-barr virus immunohistochemical status in patients with head and neck squamous cell carcinoma (HNSCC). Head Neck Pathol. 17:325–330. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Wuerdemann N, Joosse S, Klasen C, Prinz J, Demers I, George J, Speel EM, Wagner S and Klußmann JP: ctHPV-DNA based precision oncology for patients with oropharyngeal cancer-Where are we? Laryngorhinootologie. 102:728–734. 2023.(In German). PubMed/NCBI
|
|
150
|
Yan H and Bu P: Non-coding RNA in cancer. Essays Biochem. 65:625–639. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Yang QQ and Deng YF: Long non-coding RNAs as novel biomarkers and therapeutic targets in head and neck cancers. Int J Clin Exp Pathol. 7:1286–1292. 2014.PubMed/NCBI
|
|
152
|
Luo X, Qiu Y, Jiang Y, Chen F, Jiang L, Zhou Y, Dan H, Zeng X, Lei YL and Chen Q: Long non-coding RNA implicated in the invasion and metastasis of head and neck cancer: Possible function and mechanisms. Mol Cancer. 17:142018. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Thomaidou AC, Batsaki P, Adamaki M, Goulielmaki M, Baxevanis CN, Zoumpourlis V and Fortis SP: Promising biomarkers in head and neck cancer: The most clinically important miRNAs. Int J Mol Sci. 23:82572022. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Feng H, Zhao F, Luo J, Xu S, Liang Z, Xu W, Bao Y and Qin G: Long non-coding RNA HOTTIP exerts an oncogenic function by regulating HOXA13 in nasopharyngeal carcinoma. Mol Biol Rep. 50:6807–6818. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Fan Y, Yan T, Chai Y, Jiang Y and Zhu X: Long noncoding RNA HOTTIP as an independent prognostic marker in cancer. Clin Chim Acta. 482:224–230. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Jiang H, Zhou L, Shen N, Ning X, Wu D, Jiang K and Huang X: M1 macrophage-derived exosomes and their key molecule lncRNA HOTTIP suppress head and neck squamous cell carcinoma progression by upregulating the TLR5/NF-κB pathway. Cell Death Dis. 13:1832022. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Yin X, Yang W, Xie J, Wei Z, Tang C, Song C, Wang Y, Cai Y, Xu W and Han W: HOTTIP functions as a key candidate biomarker in head and neck squamous cell carcinoma by integrated bioinformatic analysis. Biomed Res Int. 2019:54506172019. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Kozlowska J, Kolenda T, Poter P, Sobocińska J, Guglas K, Stasiak M, Bliźniak R, Teresiak A and Lamperska K: Long intergenic non-coding RNAs in HNSCC: From ‘Junk DNA’ to important prognostic factor. Cancers (Basel). 13:29492021. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Li Y, Liu Y, Chen G, Liu H, Wu Y, Liu J and Zhang Z: HOTTIP is upregulated in esophageal cancer and triggers the drug resistance. J BUON. 26:1056–1061. 2021.PubMed/NCBI
|
|
160
|
Shen M, Li M and Liu J: Long Noncoding RNA HOTTIP promotes nasopharyngeal cancer cell proliferation, migration, and invasion by inhibiting miR-4301. Med Sci Monit. 25:778–785. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Hussein MA, Valinezhad K, Adel E and Munirathinam G: MALAT-1 is a key regulator of epithelial-mesenchymal transition in cancer: A potential therapeutic target for metastasis. Cancers (Basel). 16:2342024. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Masrour M, Khanmohammadi S, Fallahtafti P and Rezaei N: Long non-coding RNA as a potential diagnostic biomarker in head and neck squamous cell carcinoma: A systematic review and meta-analysis. PLoS One. 18:e02919212023. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Cossu AM, Mosca L, Zappavigna S, Misso G, Bocchetti M, De Micco F, Quagliuolo L, Porcelli M, Caraglia M and Boccellino M: Long Non-coding RNAs as important biomarkers in laryngeal cancer and other head and neck tumours. Int J Mol Sci. 20:34442019. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Han X, Xu Z, Tian G, Tang Z, Gao J, Wei Y and Xu X: Suppression of the long non-coding RNA MALAT-1 impairs the growth and migration of human tongue squamous cell carcinoma SCC4 cells. Arch Med Sci. 15:992–1000. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Kangboonruang K, Wongtrakoongate P, Lertsuwan K, Khachonkham S, Changkaew P, Tangboonduangjit P, Siripoon T, Ngamphaiboon N and Chairoungdua A: MALAT1 decreases the sensitivity of head and neck squamous cell carcinoma cells to radiation and cisplatin. Anticancer Res. 40:2645–2655. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Yao W, Bai Y, Li Y, Guo L, Zeng P, Wang Y, Qi B, Liu S, Qin X, Li Y and Zhao B: Upregulation of MALAT-1 and its association with survival rate and the effect on cell cycle and migration in patients with esophageal squamous cell carcinoma. Tumour Biol. 37:4305–4312. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Liu X, Zhao W and Wang X: Inhibition of long non-coding RNA MALAT1 elevates microRNA-429 to suppress the progression of hypopharyngeal squamous cell carcinoma by reducing ZEB1. Life Sci. 262:1184802020. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Duan Y, Yue K, Ye B, Chen P, Zhang J, He Q, Wu Y, Lai Q, Li H, Wu Y, et al: LncRNA MALAT1 promotes growth and metastasis of head and neck squamous cell carcinoma by repressing VHL through a non-canonical function of EZH2. Cell Death Dis. 14:1492023. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Ghafouri-Fard S, Dashti S, Farsi M and Taheri M: HOX transcript antisense RNA: An oncogenic lncRNA in diverse malignancies. Exp Mol Pathol. 118:1045782021. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Xiang Y and Hua Q: The role and mechanism of long non-coding RNA HOTAIR in the oncogenesis, diagnosis, and treatment of head and neck squamous cell carcinoma. Clin Med Insights Oncol. 17:117955492311690992023. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Shi L, Zhang D, Han H, Zhang L, Li S, Yang F and He C: HOTAIR knockdown impairs metastasis of cervical cancer cells by down-regulating metastasis-related genes. J Obstet Gynaecol. 43:21810602023. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Hu D and Messadi DV: Immune-Related long non-coding RNA signatures for tongue squamous cell carcinoma. Curr Oncol. 30:4817–4832. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Yuan X, Shen Q and Ma W: Long Noncoding RNA Hotair promotes the progression and immune escape in laryngeal squamous cell carcinoma through MicroRNA-30a/GRP78/PD-L1 Axis. J Immunol Res. 2022:51414262022. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Yang G, Huang T, Wu Y, Wang J, Pan Z, Chen Y, Pan F and Wang Y: Clinicopathological and prognostic significance of the long non-coding RNA HOTAIR high expression in head and neck squamous cell carcinoma: A systematic review and meta-analysis. Transl Cancer Res. 11:2536–2552. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Li F and Hu CP: Long Non-Coding RNA urothelial carcinoma associated 1 (UCA1): Insight into Its role in human diseases. Crit Rev Eukaryot Gene Expr. 25:191–197. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Wang X, Sun M, Gao Z, Yin L, Pu Y, Zhu Y, Wang X and Liu R: N-nitrosamines-mediated downregulation of LncRNA-UCA1 induces carcinogenesis of esophageal squamous by regulating the alternative splicing of FGFR2. Sci Total Environ. 855:1589182023. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Sun S, Gong C and Yuan K: LncRNA UCA1 promotes cell proliferation, invasion and migration of laryngeal squamous cell carcinoma cells by activating Wnt/β-catenin signaling pathway. Exp Ther Med. 17:1182–1189. 2019.PubMed/NCBI
|
|
178
|
Huang HH, You GR, Tang SJ, Chang JT and Cheng AJ: Molecular signature of long non-coding RNA associated with areca nut-induced head and neck cancer. Cells. 12:8732023. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Qian Y, Liu D, Cao S, Tao Y, Wei D, Li W, Li G, Pan X and Lei D: Upregulation of the long noncoding RNA UCA1 affects the proliferation, invasion, and survival of hypopharyngeal carcinoma. Mol Cancer. 16:682017. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Liu C, Jin J, Shi J, Wang L, Gao Z, Guo T and He Y: Long noncoding RNA UCA1 as a novel biomarker of lymph node metastasis and prognosis in human cancer: A meta-analysis. Biosci Rep. 39:BSR201809952019. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Zhang C, Xie L, Fu Y, Yang J and Cui Y: lncRNA MIAT promotes esophageal squamous cell carcinoma progression by regulating miR-1301-3p/INCENP axis and interacting with SOX2. J Cell Physiol. 235:7933–7944. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Da CM, Gong CY, Nan W, Zhou KS, Wu ZL and Zhang HH: The role of long non-coding RNA MIAT in cancers. Biomed Pharmacother. 129:1103592020. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Song F, Yang Y and Liu J: Long non-coding RNA MIAT promotes the proliferation and invasion of laryngeal squamous cell carcinoma cells by sponging microRNA-613. Exp Ther Med. 21:2322021. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Liu W, Wang Z, Wang C and Ai Z: Long non-coding RNA MIAT promotes papillary thyroid cancer progression through upregulating LASP1. Cancer Cell Int. 19:1942019. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Zhong W, Xu Z, Wen S, Xie T, Wang F, Wang Q and Chen J: Long non-coding RNA myocardial infarction associated transcript promotes epithelial-mesenchymal transition and is an independent risk factor for poor prognosis of tongue squamous cell carcinoma. J Oral Pathol Med. 48:720–727. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Wang R, Zhao L, Ji L, Bai L and Wen Q: Myocardial infarction associated transcript (MIAT) promotes papillary thyroid cancer progression via sponging miR-212. Biomed Pharmacother. 118:1092982019. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Guo K, Qian K, Shi Y, Sun T and Wang Z: LncRNA-MIAT promotes thyroid cancer progression and function as ceRNA to target EZH2 by sponging miR-150-5p. Cell Death Dis. 12:10972021. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Dey Ghosh R and Guha Majumder S: Circulating long non-coding RNAs could be the potential prognostic biomarker for liquid biopsy for the clinical management of oral squamous cell carcinoma. Cancers (Basel). 14:55902022. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Wang H, Zhou Z, Lin W, Qian Y, He S and Wang J: MicroRNA-21 promotes head and neck squamous cell carcinoma (HNSCC) induced transition of bone marrow mesenchymal stem cells to cancer-associated fibroblasts. BMC Cancer. 23:11352023. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Doukas SG, Vageli DP, Lazopoulos G, Spandidos DA, Sasaki CT and Tsatsakis A: The Effect of NNK, A tobacco smoke carcinogen, on the miRNA and Mismatch DNA repair expression profiles in lung and head and neck squamous cancer cells. Cells. 9:10312020. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Shah S, Jadhav K, Shah V, Gupta N and Dagrus K: miRNA 21: Diagnostic prognostic and therapeutic marker for oral cancer. Microrna. 5:175–179. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Arantes LM, Laus AC, Melendez ME, de Carvalho AC, Sorroche BP, De Marchi PR, Evangelista AF, Scapulatempo-Neto C, de Souza Viana L and Carvalho AL: MiR-21 as prognostic biomarker in head and neck squamous cell carcinoma patients undergoing an organ preservation protocol. Oncotarget. 8:9911–9921. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Ishinaga H, Okugawa Y, Hou B, He F, Yin C, Murata M, Toiyama Y and Takeuchi K: The role of miR-21 as a predictive biomarker and a potential target to improve the effects of chemoradiotherapy against head and neck squamous cell carcinoma. J Radiat Res. 64:668–676. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Vageli DP, Doukas PG, Shah R, Boyi T, Liu C and Judson BL: A novel saliva and serum miRNA panel as a potential useful index for oral cancer and the association of miR-21 with smoking history: A pilot study. Cancer Prev Res (Phila). 16:653–659. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Irimie-Aghiorghiesei AI, Pop-Bica C, Pintea S, Braicu C, Cojocneanu R, Zimța AA, Gulei D, Slabý O and Berindan-Neagoe I: Prognostic Value of MiR-21: An updated meta-analysis in head and neck squamous cell carcinoma (HNSCC). J Clin Med. 8:20412019. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Sheng S, Su W, Mao D, Li C, Hu X, Deng W, Yao Y and Ji Y: MicroRNA-21 induces cisplatin resistance in head and neck squamous cell carcinoma. PLoS One. 17:e02670172022. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Kabzinski J, Maczynska M and Majsterek I: MicroRNA as a novel biomarker in the diagnosis of head and neck cancer. Biomolecules. 11:8442021. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Luo C, Zhang J, Zhang Y, Zhang X, Chen Y and Fan W: Low expression of miR-let-7a promotes cell growth and invasion through the regulation of c-Myc in oral squamous cell carcinoma. Cell Cycle. 19:1983–1993. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Ulusan M, Sen S, Yilmazer R, Dalay N and Demokan S: The let-7 microRNA binding site variant in KRAS as a predictive biomarker for head and neck cancer patients with lymph node metastasis. Pathol Res Pract. 239:1541472022. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Lee YS and Dutta A: The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 21:1025–1030. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Yu D, Liu X, Han G, Liu Y, Zhao X, Wang D, Bian X, Gu T and Wen L: The let-7 family of microRNAs suppresses immune evasion in head and neck squamous cell carcinoma by promoting PD-L1 degradation. Cell Commun Signal. 17:1732019. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Chirshev E, Oberg KC, Ioffe YJ and Unternaehrer JJ: Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin Transl Med. 8:242019. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Kinoshita T, Hanazawa T, Nohata N, Okamoto Y and Seki N: The functional significance of microRNA-375 in human squamous cell carcinoma: Aberrant expression and effects on cancer pathways. J Hum Genet. 57:556–563. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Mazumder S, Datta S, Ray JG, Chaudhuri K and Chatterjee R: Liquid biopsy: miRNA as a potential biomarker in oral cancer. Cancer Epidemiol. 58:137–145. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Chen J, Cai Z, Hu J, Zhou L, Zhang P and Xu X: MicroRNA-375 in extracellular vesicles-novel marker for esophageal cancer diagnosis. Medicine (Baltimore). 102:e328262023. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Huni KC, Cheung J, Sullivan M, Robison WT, Howard KM and Kingsley K: Chemotherapeutic drug resistance associated with differential miRNA expression of miR-375 and miR-27 among oral cancer cell lines. Int J Mol Sci. 24:12442023. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Gazdzicka J, Golabek K, Strzelczyk JK and Ostrowska Z: Epigenetic modifications in head and neck cancer. Biochem Genet. 58:213–244. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Castilho RM, Squarize CH and Almeida LO: Epigenetic modifications and head and neck cancer: Implications for tumor progression and resistance to therapy. Int J Mol Sci. 18:15062017. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Li J, Huang Q, Zeng F, Li W, He Z, Chen W, Zhu W and Zhang B: The prognostic value of global DNA hypomethylation in cancer: A meta-analysis. PLoS One. 9:e1062902014. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Temilola DO, Adeola HA, Grobbelaar J and Chetty M: Liquid biopsy in head and neck cancer: Its present state and future role in Africa. Cells. 12:26632023. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Liu HE, Triboulet M, Zia A, Vuppalapaty M, Kidess-Sigal E, Coller J, Natu VS, Shokoohi V, Che J, Renier C, et al: Workflow optimization of whole genome amplification and targeted panel sequencing for CTC mutation detection. NPJ Genom Med. 2:342017. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Kong L and Birkeland AC: Liquid biopsies in head and neck cancer: Current state and future challenges. Cancers (Basel). 13:18742021. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Huang X, Duijf PHG, Sriram S, Perera G, Vasani S, Kenny L, Leo P and Punyadeera C: Circulating tumour DNA alterations: emerging biomarker in head and neck squamous cell carcinoma. J Biomed Sci. 30:652023. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Thierry AR, El Messaoudi S, Gahan PB, Anker P and Stroun M: Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 35:347–376. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Silvoniemi A, Laine J, Aro K, Nissi L, Bäck L, Schildt J, Hirvonen J, Hagström J, Irjala H, Aaltonen LM, et al: Circulating tumor DNA in head and neck squamous cell carcinoma: Association with metabolic tumor burden determined with FDG-PET/CT. Cancers (Basel). 15:39702023. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Allevato MM, Smith JD, Brenner MJ and Chinn SB: Tumor-Derived exosomes and the role of liquid biopsy in human papillomavirus oropharyngeal squamous cell carcinoma. Cancer J. 29:230–237. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, et al: Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 24:766–769. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Li S, Yi M, Dong B, Tan X, Luo S and Wu K: The role of exosomes in liquid biopsy for cancer diagnosis and prognosis prediction. Int J Cancer. 148:2640–2651. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al: Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Kabzinski J, Kucharska-Lusina A and Majsterek I: RNA-Based liquid biopsy in head and neck cancer. Cells. 12:19162023. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Lu YC, Chang JT, Huang YC, Huang CC, Chen WH, Lee LY, Huang BS, Chen YJ, Li HF and Cheng AJ: Combined determination of circulating miR-196a and miR-196b levels produces high sensitivity and specificity for early detection of oral cancer. Clin Biochem. 48:115–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Kumar P, Gupta S and Das BC: Saliva as a potential non-invasive liquid biopsy for early and easy diagnosis/prognosis of head and neck cancer. Transl Oncol. 40:1018272024. View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Mishra V, Singh A, Chen X, Rosenberg AJ, Pearson AT, Zhavoronkov A, Savage PA, Lingen MW, Agrawal N and Izumchenko E: Application of liquid biopsy as multi-functional biomarkers in head and neck cancer. Br J Cancer. 126:361–370. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Wang X and Li BB: Deep learning in head and neck tumor multiomics diagnosis and analysis: Review of the literature. Front Genet. 12:6248202021. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Haider M, Jagal J, Bajbouj K, Sharaf BM, Sahnoon L, Okendo J, Semreen MH, Hamda M and Soares NC: Integrated multi-omics analysis reveals unique signatures of paclitaxel-loaded poly(lactide-co-glycolide) nanoparticles treatment of head and neck cancer cells. Proteomics. 23:e22003802023. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Eichberger J, Froschhammer D, Schulz D, Scholz KJ, Federlin M, Ebensberger H, Reichert TE, Ettl T and Bauer RJ: BMSC-HNC Interaction: Exploring effects on bone integrity and head and neck cancer progression. Int J Mol Sci. 24:144172023. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Borkowska EM, Barańska M, Kowalczyk M and Pietruszewska W: Detection of PIK3CA gene mutation in head and neck squamous cell carcinoma using droplet digital PCR and RT-qPCR. Biomolecules. 11:8182021. View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Khedkar HN, Chen LC, Kuo YC, Wu ATH and Huang HS: Multi-Omics identification of genetic alterations in head and neck squamous cell carcinoma and therapeutic efficacy of HNC018 as a novel multi-target agent for c-MET/STAT3/AKT signaling axis. Int J Mol Sci. 24:102472023. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Ehrlich M: DNA hypomethylation in cancer cells. Epigenomics. 1:239–259. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Guo Z, Liu W, Yang Y, Zhang S, Li C and Yang W: DNA methylation in the genesis, progress and prognosis of head and neck cancer. Holist Integ Oncol. 2:232023. View Article : Google Scholar
|
|
231
|
Capkova M, Sachova J, Strnad H, Kolář M, Hroudová M, Chovanec M, Čada Z, Šteffl M, Valach J, Kastner J, et al: Microarray analysis of serum mRNA in patients with head and neck squamous cell carcinoma at whole-genome scale. Biomed Res Int. 2014:4086832014. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Li Y, Porta-Pardo E, Tokheim C, Bailey MH, Yaron TM, Stathias V, Geffen Y, Imbach KJ, Cao S, Anand S, et al: Pan-cancer proteogenomics connects oncogenic drivers to functional states. Cell. 186:3921–3944.e25. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Assarsson E, Lundberg M, Holmquist G, Björkesten J, Thorsen SB, Ekman D, Eriksson A, Rennel Dickens E, Ohlsson S, Edfeldt G, et al: Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 9:e951922014. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Pawlicka M, Gumbarewicz E, Blaszczak E and Stepulak A: Transcription factors and markers related to epithelial-mesenchymal transition and their role in resistance to therapies in head and neck cancers. Cancers (Basel). 16:13542024. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Fornieles G, Núñez MI and Expósito J: Matrix metalloproteinases and their inhibitors as potential prognostic biomarkers in head and neck cancer after radiotherapy. Int J Mol Sci. 25:5272023. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Liao C, An J, Tan Z, Xu F, Liu J and Wang Q: Changes in protein glycosylation in head and neck squamous cell carcinoma. J Cancer. 12:1455–1466. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
237
|
Zhao X, Liu X and Cui L: Development of a five-protein signature for predicting the prognosis of head and neck squamous cell carcinoma. Aging (Albany NY). 12:19740–19755. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
238
|
Yang T, Hui R, Nouws J, Sauler M, Zeng T and Wu Q: Untargeted metabolomics analysis of esophageal squamous cell cancer progression. J Transl Med. 20:1272022. View Article : Google Scholar : PubMed/NCBI
|
|
239
|
Wu Z, Han Y, Wan Y, Hua X, Chill SS, Teshome K, Zhou W, Liu J, Wu D, Hutchinson A, et al: Oral microbiome and risk of incident head and neck cancer: A nested case-control study. Oral Oncol. 137:1063052023. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Stasiewicz M and Karpinski TM: The oral microbiota and its role in carcinogenesis. Semin Cancer Biol. 86:633–642. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Dorobisz K, Dorobisz T and Zatonski T: The Microbiome's influence on head and neck cancers. Curr Oncol Rep. 25:163–171. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Benjamin WJ, Wang K, Zarins K, Bellile E, Blostein F, Argirion I, Taylor JMG, D'Silva NJ, Chinn SB, Rifkin S, et al: Oral microbiome community composition in head and neck squamous cell carcinoma. Cancers (Basel). 15:25492023. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Kunhabdulla H, Manas R, Shettihalli AK, Reddy CRM, Mustak MS, Jetti R, Abdulla R, Sirigiri DR, Ramdan D and Ammarullah MI: Identifying biomarkers and therapeutic targets by multiomic analysis for HNSCC: Precision medicine and healthcare management. ACS Omega. 9:12602–12610. 2024.PubMed/NCBI
|
|
244
|
Xiao M, Zhang X, Zhang D, Deng S, Zheng A, Du F, Shen J, Yue L, Yi T, Xiao Z and Zhao Y: Complex interaction and heterogeneity among cancer stem cells in head and neck squamous cell carcinoma revealed by single-cell sequencing. Front Immunol. 13:10509512022. View Article : Google Scholar : PubMed/NCBI
|
|
245
|
Li M, Sun D, Song N, Chen X, Zhang X, Zheng W, Yu Y and Han C: Mutant p53 in head and neck squamous cell carcinoma: Molecular mechanism of gain-of-function and targeting therapy (Review). Oncol Rep. 50:1622023. View Article : Google Scholar : PubMed/NCBI
|
|
246
|
Snietura M, Jaworska M, Mlynarczyk-Liszka J, Goraj-Zajac A, Piglowski W, Lange D, Wozniak G, Nowara E and Suwinski R: PTEN as a prognostic and predictive marker in postoperative radiotherapy for squamous cell cancer of the head and neck. PLoS One. 7:e333962012. View Article : Google Scholar : PubMed/NCBI
|
|
247
|
Bossi P, Resteghini C, Paielli N, Licitra L, Pilotti S and Perrone F: Prognostic and predictive value of EGFR in head and neck squamous cell carcinoma. Oncotarget. 7:74362–74379. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
248
|
Qiao XW, Jiang J, Pang X, Huang MC, Tang YJ, Liang XH and Tang YL: The Evolving Landscape of PD-1/PD-L1 pathway in head and neck cancer. Front Immunol. 11:17212020. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Edirisinghe S, Weerasekera M, De Silva D, Devmini MT, Pathmaperuma S, Wijesinghe GK, Nisansala T, Maddumage A, Huzaini H, Rich AM, et al: Vascular endothelial growth factor A (VEGF-A) and vascular endothelial growth factor receptor 2 (VEGFR-2) as potential biomarkers for oral squamous cell carcinoma: A Sri Lankan study. Asian Pac J Cancer Prev. 24:267–274. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
250
|
Augustin JG, Lepine C, Morini A, Brunet A, Veyer D, Brochard C, Mirghani H, Péré H and Badoual C: HPV detection in head and neck squamous cell carcinomas: What is the issue? Front Oncol. 10:17512020. View Article : Google Scholar : PubMed/NCBI
|
|
251
|
Tan D, Wu Y, Hu L, He P, Xiong G, Bai Y and Yang K: Long noncoding RNA H19 is up-regulated in esophageal squamous cell carcinoma and promotes cell proliferation and metastasis. Dis Esophagus. 30:1–9. 2017.
|
|
252
|
Yoshizaki T, Kondo S, Dochi H, Kobayashi E, Mizokami H, Komura S and Endo K: Recent advances in assessing the clinical implications of epstein-barr virus infection and their application to the diagnosis and treatment of nasopharyngeal carcinoma. Microorganisms. 12:142023. View Article : Google Scholar : PubMed/NCBI
|
|
253
|
Ye D, Deng Y and Shen Z: The role and mechanism of MALAT1 Long Non-Coding RNA in the diagnosis and treatment of head and neck squamous cell carcinoma. Onco Targets Ther. 14:4127–4136. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
254
|
Cantile M, Di Bonito M, Tracey De Bellis M and Botti G: Functional Interaction among lncRNA HOTAIR and MicroRNAs in cancer and other human diseases. Cancers (Basel). 13:5702021. View Article : Google Scholar : PubMed/NCBI
|
|
255
|
Wang Y, Wang S, Ren Y and Zhou X: The Role of lncRNA Crosstalk in leading cancer metastasis of head and neck squamous cell carcinoma. Front Oncol. 10:5618332020. View Article : Google Scholar : PubMed/NCBI
|
|
256
|
Mohammadi C, Gholamzadeh Khoei S, Fayazi N, Mohammadi Y and Najafi R: miRNA as promising theragnostic biomarkers for predicting radioresistance in cancer: A systematic review and meta-analysis. Crit Rev Oncol Hematol. 157:1031832021. View Article : Google Scholar : PubMed/NCBI
|
|
257
|
Ma Y, Shen N, Wicha MS and Luo M: The Roles of the Let-7 Family of MicroRNAs in the regulation of cancer stemness. Cells. 10:24152021. View Article : Google Scholar : PubMed/NCBI
|
















