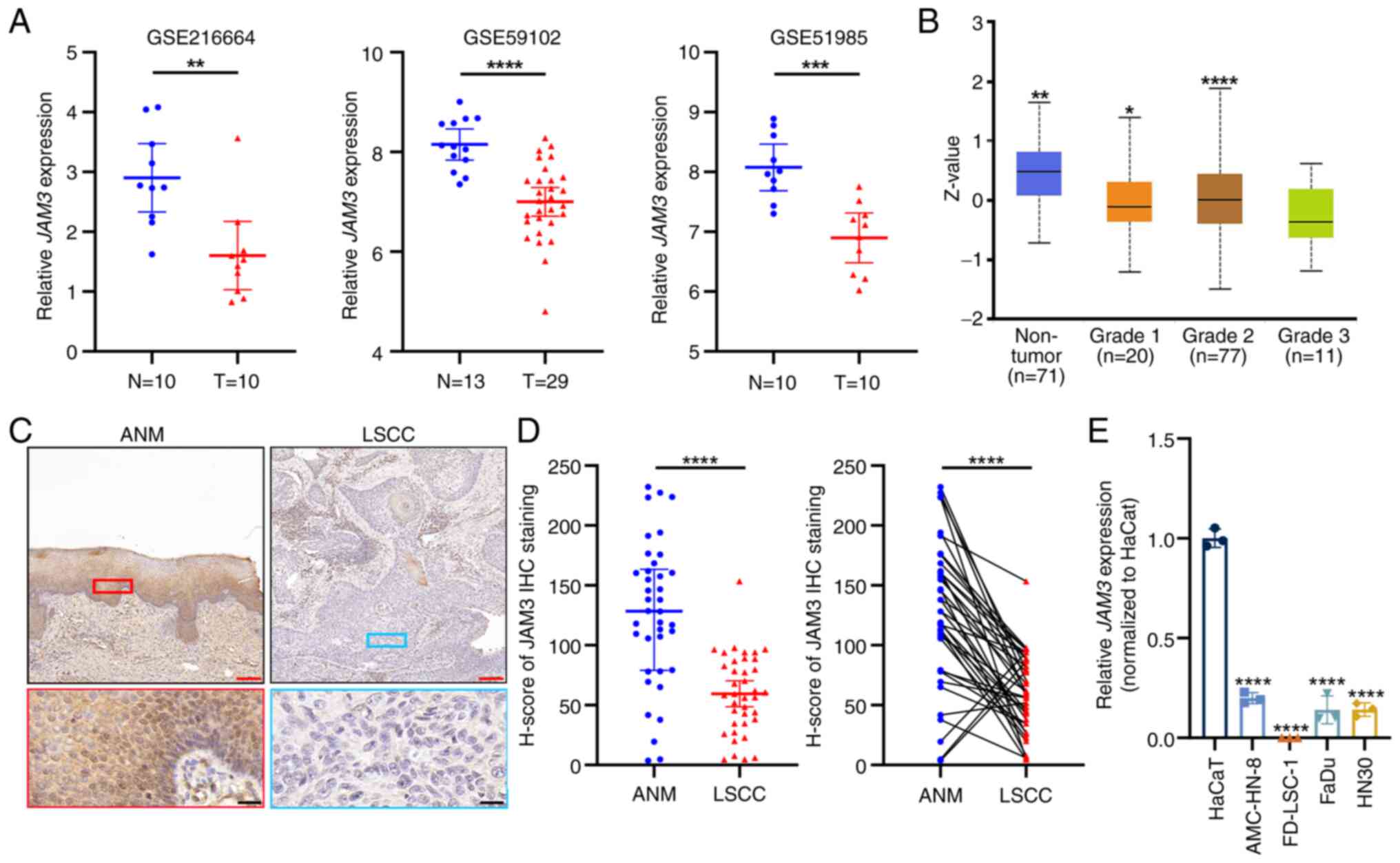
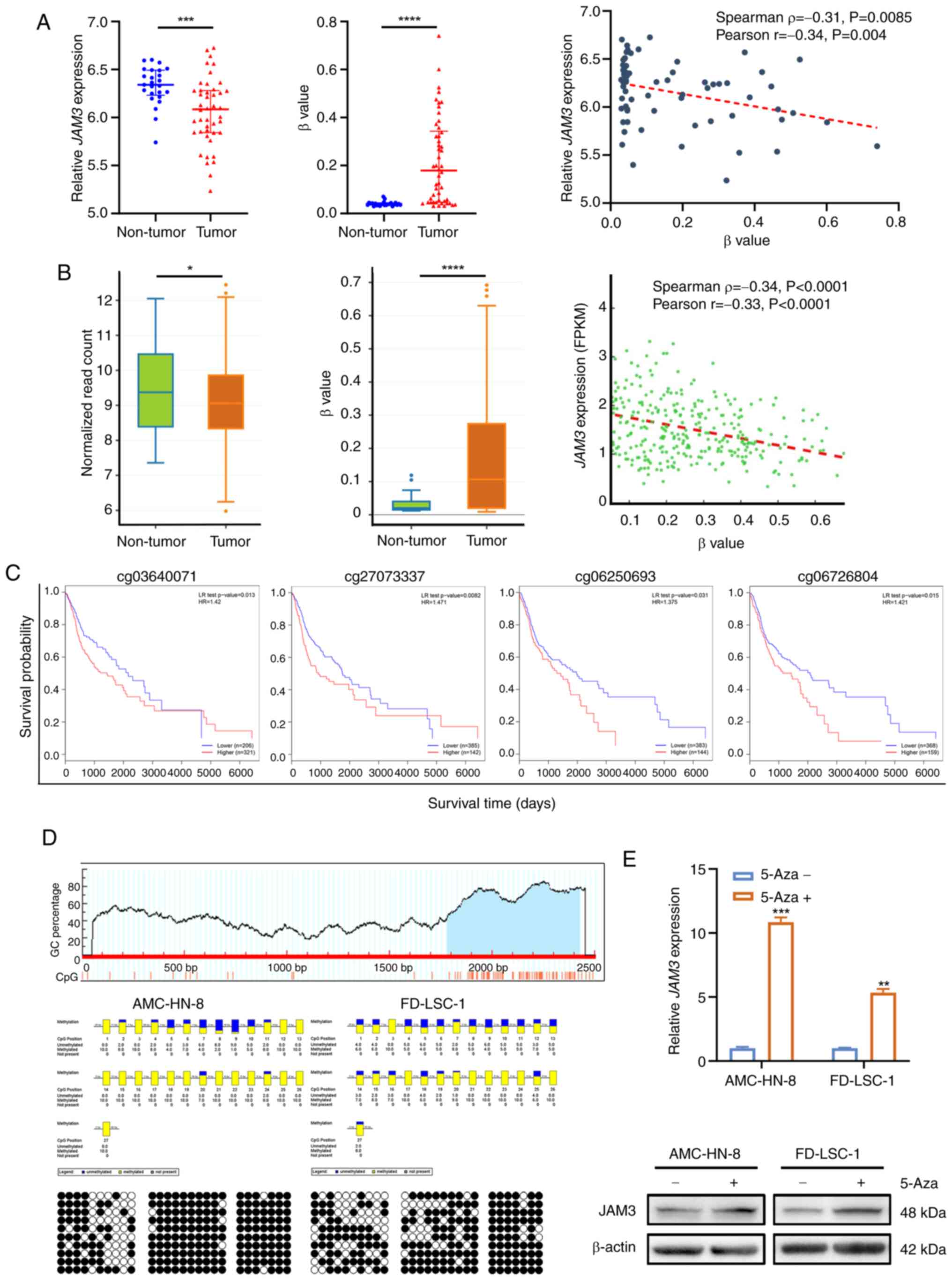
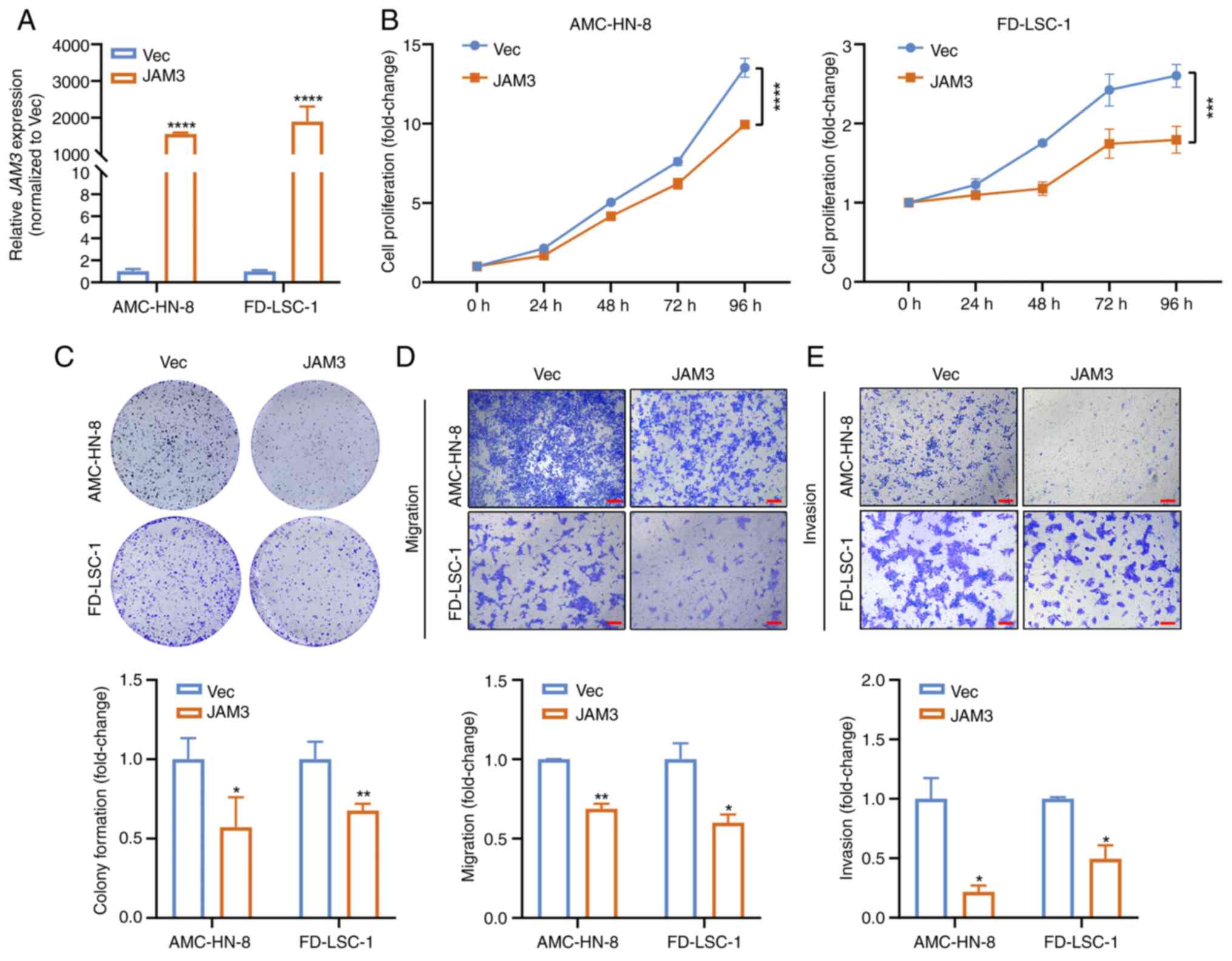
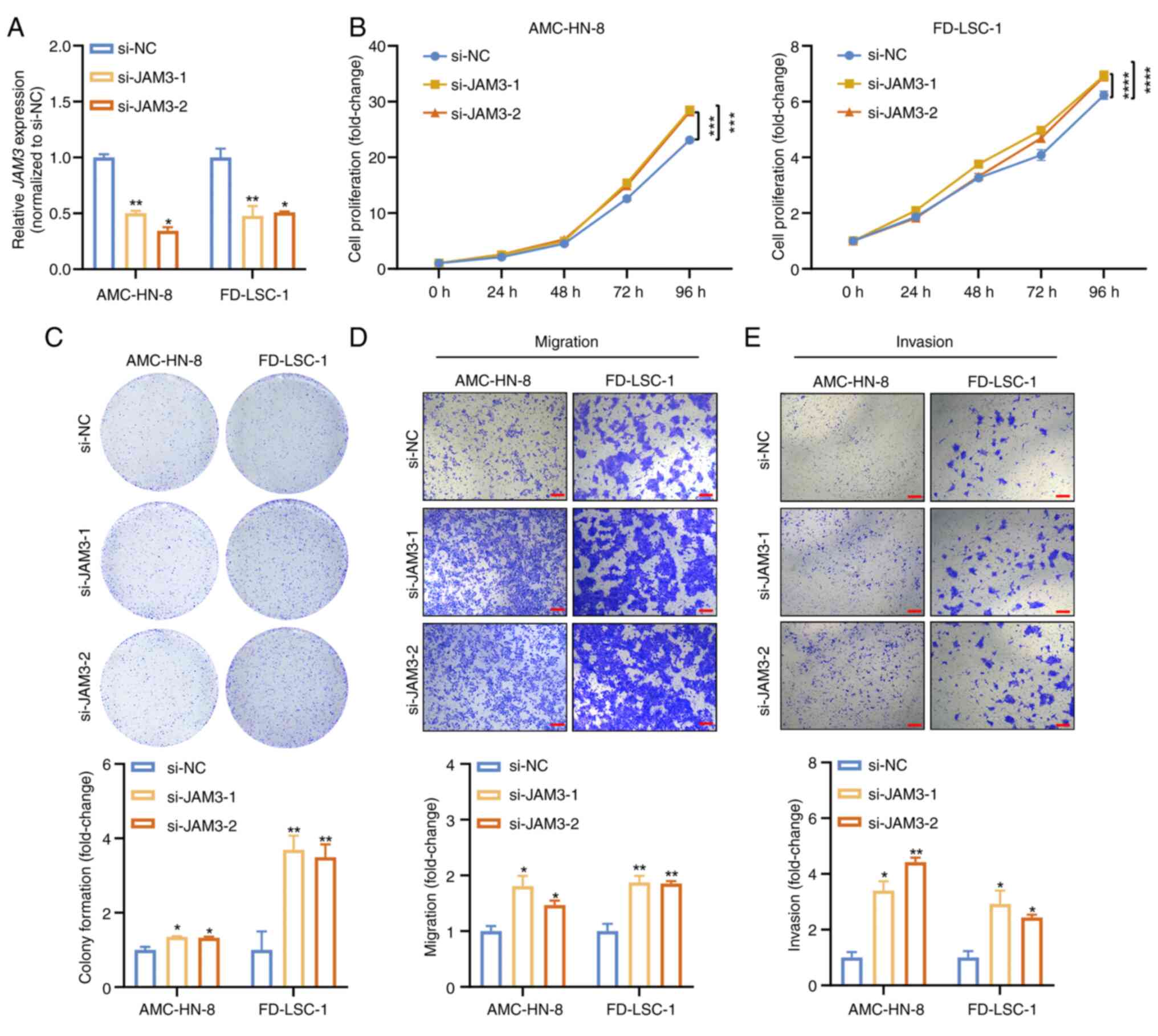
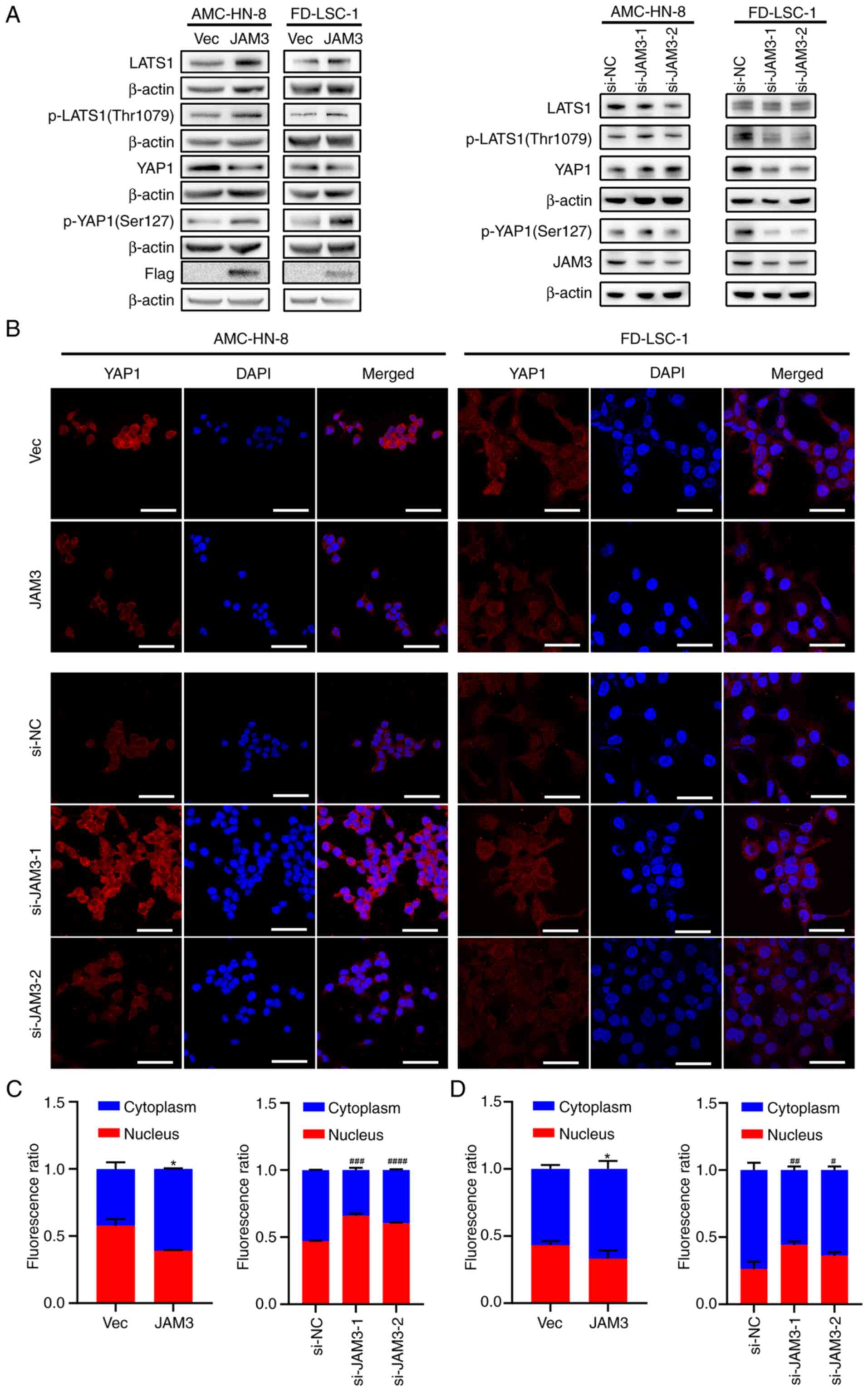
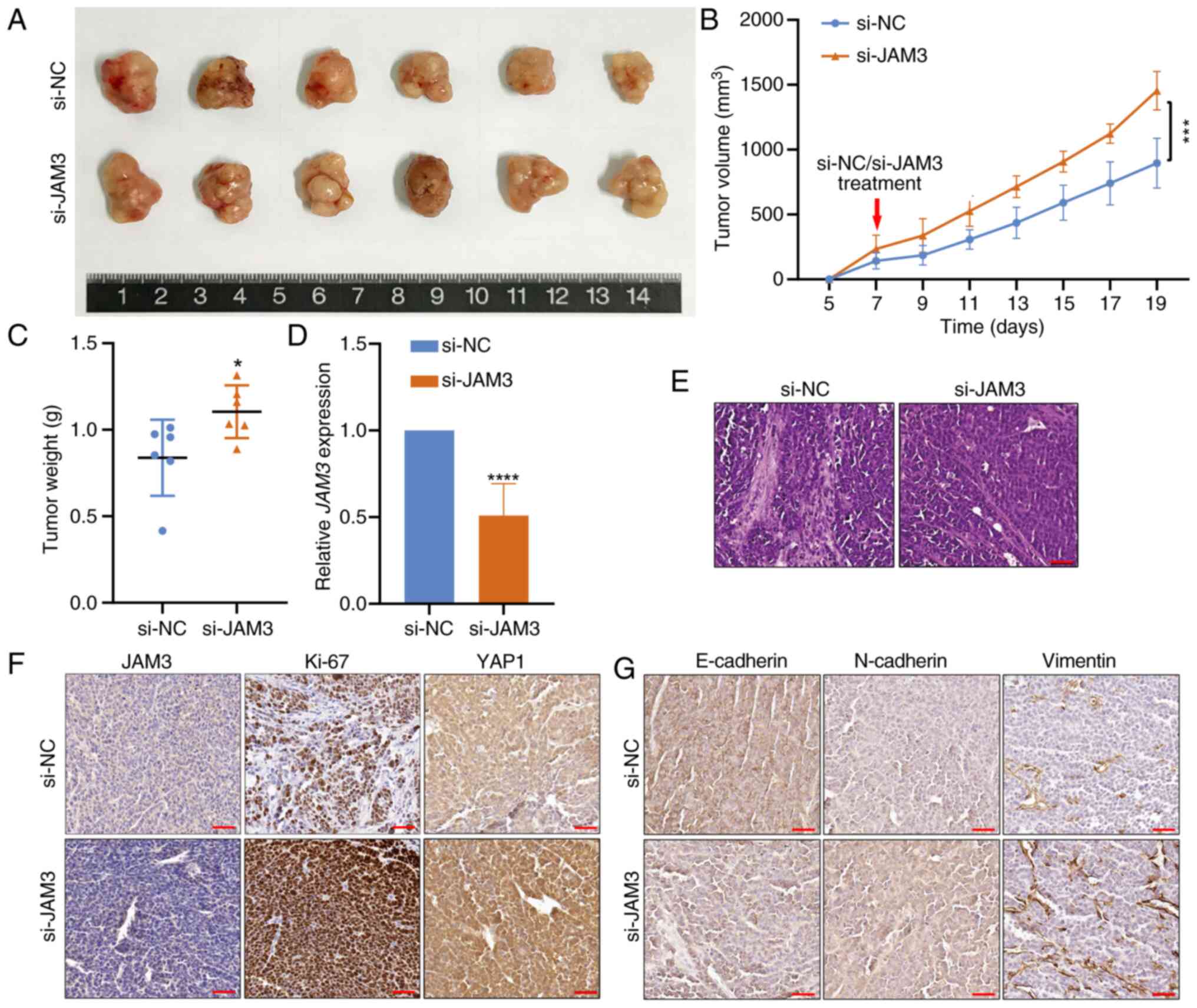
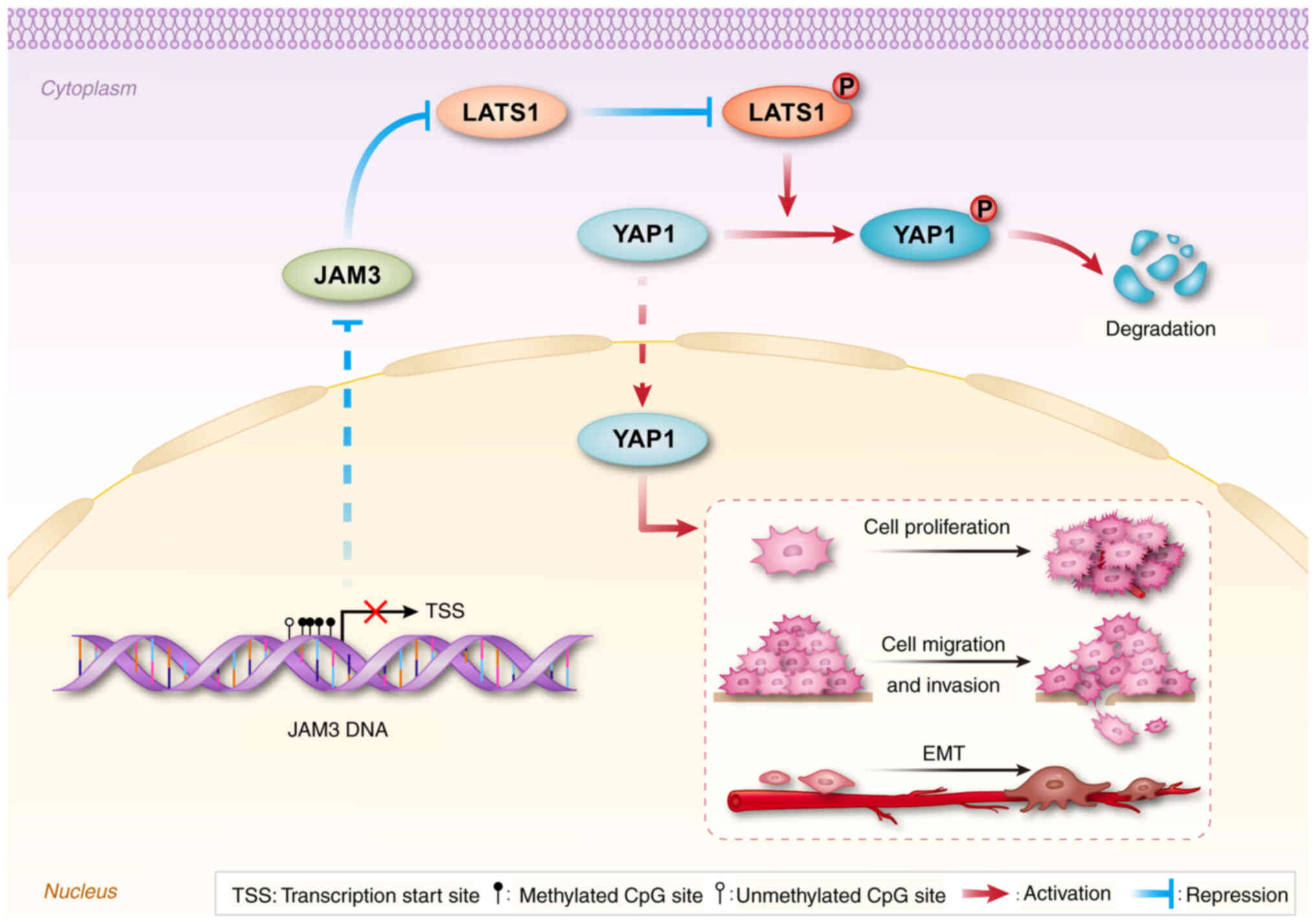
|
1
|
Chow LQM: Head and neck cancer. N Engl J
Med. 382:60–72. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Johnson DE, Burtness B, Leemans CR, Lui
VWY, Bauman JE and Grandis JR: Head and neck squamous cell
carcinoma. Nat Rev Dis Primer. 6:922020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bhat AA, Yousuf P, Wani NA, Rizwan A,
Chauhan SS, Siddiqi MA, Bedognetti D, El-Rifai W, Frenneaux MP,
Batra SK, et al: Tumor microenvironment: An evil nexus promoting
aggressive head and neck squamous cell carcinoma and avenue for
targeted therapy. Signal Transduct Target Ther. 6:122021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bray F, Laversanne M, Sung H, Ferlay J,
Siegel RL, Soerjomataram I and Jemal A: Global cancer statistics
2022: GLOBOCAN estimates of incidence and mortality worldwide for
36 cancers in 185 countries. CA Cancer J Clin. 74:229–263. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cossu AM, Mosca L, Zappavigna S, Misso G,
Bocchetti M, De Micco F, Quagliuolo L, Porcelli M, Caraglia M and
Boccellino M: Long non-coding RNAs as important biomarkers in
laryngeal cancer and other head and neck tumours. Int J Mol Sci.
20:34442019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Verro B, Saraniti C, Carlisi D,
Chiesa-Estomba C, Maniaci A, Lechien JR, Mayo M, Fakhry N and
Lauricella M: Biomarkers in laryngeal squamous cell carcinoma: The
literature review. Cancers (Basel). 15:50962023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lyu H, Huang J, He Z and Liu B: Epigenetic
mechanism of Survivin dysregulation in human cancer. Sci China Life
Sci. 61:808–814. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Inbar-Feigenberg M, Choufani S, Butcher
DT, Roifman M and Weksberg R: Basic concepts of epigenetics. Fertil
Steril. 99:607–615. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Moore LD, Le T and Fan G: DNA methylation
and its basic function. Neuropsychopharmacology. 38:23–38. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mattei AL, Bailly N and Meissner A: DNA
methylation: A historical perspective. Trends Genet. 38:676–707.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li D, Da L, Tang H, Li T and Zhao M: CpG
methylation plays a vital role in determining tissue- and
cell-specific expression of the human cell-death-inducing
DFF45-like effector A gene through the regulation of Sp1/Sp3
binding. Nucleic Acids Res. 36:330–341. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bird A: DNA methylation patterns and
epigenetic memory. Genes Dev. 16:6–21. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Smith J, Sen S, Weeks RJ, Eccles MR and
Chatterjee A: Promoter DNA hypermethylation and paradoxical gene
activation. Trends Cancer. 6:392–406. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schübeler D: Epigenomics: Methylation
matters. Nature. 462:296–297. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Martìn-Padura I, Lostaglio S, Schneemann
M, Williams L, Romano M, Fruscella P, Panzeri C, Stoppacciaro A,
Ruco L, Villa A, et al: Junctional adhesion molecule, a novel
member of the immunoglobulin superfamily that distributes at
intercellular junctions and modulates monocyte transmigration. J
Cell Biol. 142:117–127. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li X, Yin A, Zhang W, Zhao F, Lv J, Lv J
and Sun J: Jam3 promotes migration and suppresses apoptosis of
renal carcinoma cell lines. Int J Mol Med. 42:2923–2929.
2018.PubMed/NCBI
|
|
17
|
Zhou D, Tang W, Zhang Y and An HX: JAM3
functions as a novel tumor suppressor and is inactivated by DNA
methylation in colorectal cancer. Cancer Manag Res. 11:2457–2470.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi Y, Feng X, Zhang Y, Gao J, Bao W, Wang
J and Bai JF: Downregulation of JAM3 occurs in cholangiocarcinoma
by hypermethylation: A potential molecular marker for diagnosis and
prognosis. J Cell Mol Med. 28:e180382024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu CP, Zhou L, Gong HL, Du HD, Tian J, Sun
S and Li JY: Establishment and characterization of a novel
HPV-negative laryngeal squamous cell carcinoma cell line, FD-LSC-1,
with missense and nonsense mutations of TP53 in the DNA-binding
domain. Cancer Lett. 342:92–103. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bock C, Reither S, Mikeska T, Paulsen M,
Walter J and Lengauer T: BiQ analyzer: Visualization and quality
control for DNA methylation data from bisulfite sequencing.
Bioinformatics. 21:4067–4068. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
De Vleeschauwer SI, van de Ven M, Oudin A,
Debusschere K, Connor K, Byrne AT, Ram D, Rhebergen AM, Raeves YD,
Dahlhoff M, et al: OBSERVE: Guidelines for the refinement of rodent
cancer models. Nat Protoc. 19:2571–2596. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Detre S, Jotti GS and Dowsett M: A
‘quickscore’ method for immunohistochemical semiquantitation:
Validation for oestrogen receptor in breast carcinomas. J Clin
Pathol. 48:876–878. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Figueiredo DLA, Chao BMP and Figueiredo
FNDS: Larynx cancer: Search for molecular markers. Arch Head Neck
Surg. 48:e004320192019. View Article : Google Scholar
|
|
26
|
Lian M, Fang J, Han D, Ma H, Feng L, Wang
R and Yang F: Microarray gene expression analysis of tumorigenesis
and regional lymph node metastasis in laryngeal squamous cell
carcinoma. PLoS One. 8:e848542013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Stansfield JC, Rusay M, Shan R, Kelton C,
Gaykalova DA, Fertig EJ, Califano JA and Ochs MF: Toward
signaling-driven biomarkers immune to normal tissue contamination.
Cancer Inform. 15:15–21. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Deng M, Brägelmann J, Schultze JL and
Perner S: Web-TCGA: An online platform for integrated analysis of
molecular cancer data sets. BMC Bioinformatics. 17:722016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ding W, Chen J, Feng G, Chen G, Wu J, Guo
Y, Ni X and Shi T: DNMIVD: DNA methylation interactive
visualization database. Nucleic Acids Res. 48:D856–D862. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bibikova M, Lin Z, Zhou L, Chudin E,
Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, et al:
High-throughput DNA methylation profiling using universal bead
arrays. Genome Res. 16:383–393. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cerami1 E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Modhukur V, Iljasenko T, Metsalu T, Lokk
K, Laisk-Podar T and Vilo J: MethSurv: A web tool to perform
multivariable survival analysis using DNA methylation data.
Epigenomics. 10:277–288. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fu M, Hu Y, Lan T, Guan KL, Luo T and Luo
M: The Hippo signalling pathway and its implications in human
health and diseases. Signal Transduct Target Ther. 7:3762022.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mandell K and Parkos C: The JAM family of
proteins. Adv Drug Deliv Rev. 57:857–867. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liang TW, DeMarco RA, Mrsny RJ, Gurney A,
Gray A, Hooley J, Aaron HL, Huang A, Klassen T, Tumas DB and Fong
S: Characterization of huJAM: Evidence for involvement in cell-cell
contact and tight junction regulation. Am J Physiol Cell Physiol.
279:C1733–C1743. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mandicourt G, Iden S, Ebnet K,
Aurrand-Lions M and Imhof BA: JAM-C regulates tight junctions and
integrin-mediated cell adhesion and migration. J Biol Chem.
282:1830–1837. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lauko A, Mu Z, Gutmann DH, Naik UP and
Lathia JD: Junctional adhesion molecules in cancer: A paradigm for
the diverse functions of cell-cell interactions in tumor
progression. Cancer Res. 80:4878–4885. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jia X, Zhao C, Chen Q, Du Y, Huang L, Ye
Z, Ren X, Wang S, Lee C, Tang Z, et al: JAM-C maintains VEGR2
expression to promote retinal pigment epithelium cell survival
under oxidative stress. Thromb Haemost. 117:750–757. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sanchez-Vega F, Mina M, Armenia J, Chatila
WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia
S, et al: Oncogenic signaling pathways in the cancer genome atlas.
Cell. 173:321–337.e10. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Eun YG, Lee D, Lee YC, Sohn BH, Kim EH,
Yim SY, Kwon KH and Lee JS: Clinical significance of YAP1
activation in head and neck squamous cell carcinoma. Oncotarget.
8:111130–111143. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li Q, Wang M, Hu Y, Zhao E, Li J, Ren L,
Wang M, Xu Y, Liang Q, Zhang D, et al: MYBL2 disrupts the Hippo-YAP
pathway and confers castration resistance and metastatic potential
in prostate cancer. Theranostics. 11:5794–5812. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Huang Z, Zhang Z, Zhou C, Liu L and Huang
C: Epithelial-mesenchymal transition: The history, regulatory
mechanism, and cancer therapeutic opportunities. MedComm (2020).
3:e1442022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fan S, Smith MS, Keeney J, O'Leary MN,
Nusrat A and Parkos CA: JAM-A signals through the Hippo pathway to
regulate intestinal epithelial proliferation. iScience.
25:1043162022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Muñoz-Galván S, Felipe-Abrio B,
Verdugo-Sivianes EM, Perez M, Jiménez-García MP, Suarez-Martinez E,
Estevez-Garcia P and Carnero A: Downregulation of MYPT1 increases
tumor resistance in ovarian cancer by targeting the Hippo pathway
and increasing the stemness. Mol Cancer. 19:72020. View Article : Google Scholar : PubMed/NCBI
|