Introduction
Triple-negative breast cancer (TNBC) represents the
most aggressive subtype among all breast cancers, accounting for
~15% of breast cancer cases (1).
With respect to the fact that TNBC does not express the estrogen
receptor, progesterone receptor, or human epidermal growth factor
receptor-2 (HER-2), patients with TNBC cannot obtain benefits from
endocrine- or HER-2-targeted therapies. The treatment protocols for
TNBC encompass radiotherapy, chemotherapy and surgery procedures.
Presently, there have been some advancements in the immunotherapy
of TNBC as well (2,3). Nevertheless, TNBC has a comparatively
high vascular density. This feature enables cancer cells to
multiply swiftly and metastasize to distant tissues (4,5), which
in turn still results in the reduction of the progression-free
survival (PFS) period among patients with metastatic TNBC (6,7).
Consequently, elucidating the underlying molecular mechanisms of
angiogenesis in TNBC holds great clinical significance.
Non-coding RNAs (ncRNAs) play crucial roles in the
genetic evolution of organisms (8–10).
Piwi-interacting RNAs (piRNAs), which belong to a type of ncRNAs,
need to bind to PIWI family proteins to exert a variety of
biological effects (11). In
reproductive stem cells, piRNA/PIWI complexes maintain the
integrity of the transposon genome by silencing transposons
(12). Besides the mammalian
reproductive system, piRNAs can be expressed in numerous tissues
within the human body (13–15). Previously, it was reported that
piRNAs are associated with cancer. For instance, piR-823 can
suppress the proliferation of gastric cancer cells (16). The expression of piR-823 is elevated
in multiple myeloma cells, and it can subsequently promote their
proliferation by influencing apoptosis (17). piR-651 can impede the apoptosis of
lung cancer cells (18). In colon
cancer, the downregulation of piR-1245 expression leads cells to
undergo apoptosis (19). These
findings suggest that piRNAs have a regulatory function in the
development of cancer.
A recent study revealed that piR-31115 is aberrantly
elevated in clear cell renal cancer and it was established that the
effect of piR-31115 can enhance cancer cell invasion (20). Koduru et al (21) reported that piR-hsa-1254 (also known
as piR-31115) is upregulated in TNBC tissue samples. However, the
role of piR-31115 in TNBC has not yet been reported. In the present
study, it was discovered that piR-31115 derived from MDA-MB-231
cells promotes the proliferation and migration of HMEC-1 cells by
modulating N6-methyladenosine (m6A) modification. The results offer
a perspective for further exploration of angiogenesis induced by
TNBC.
Materials and methods
Tissue sample collection and
preparation
TNBC tissue specimens were obtained for the present
study. A total of 27 female patients, with ages ranging from 38 to
57 years old, were treated at the Department of Breast and Thyroid
Surgery in Zibo Central Hospital between January 2022 and December
2023, provided the TNBC and adjacent normal tissue samples. Both
the tumour tissues and the corresponding adjacent normal tissues
were histologically confirmed. Immediately after the surgical
procedure, the tissue specimens were placed into cryovials,
snap-frozen, and then stored in liquid nitrogen until further use.
Every participant signed a statement of informed consent. The
protocol for the utilization of patient samples was approved
(approval no. 202102005) by the Ethics Committee of Zibo Central
Hospital (Zibo, China).
Cell culture
HMEC-1 cells were kindly provided by Dr Z. Wang
(Zhongda Hospital, Affiliated Hospital of Southeast University,
Nanjing, China). Normal human breast epithelial cells (MCF-10A)
were a gift from Dr XC. Sun (Jiangsu University, Zhenjing, China).
MDA-MB-231 cells were provided by Dr H. Yang (Tai'an City Central
Hospital, Affiliated Hospital of Qingdao University, Tai'an,
China). HMEC-1 cells were cultured in DMEM/F-12 (cat. no.
SH30023.01; HyClone; Cytiva) supplemented with 10% (v/v) fetal
bovine serum (FBS; cat. no. 11012-8611; Every Green; http://www.hzsjq.com/). MDA-MB-231 cells were
maintained in Leibovitz's L-15 medium (cat. no. CM10045; MacGene;
http://www.macgene.com/) supplemented with 10%
(v/v) FBS (cat. no. 11012-8611; Every Green). MCF-10A cells were
maintained in mammary epithelial cell medium supplemented with 5%
horse serum and 1% growth medium (cat. no. ZQ-1311; Zhongqiao
Xinzhou Biotechnology Co., Ltd.). All the cells were incubated in a
thermostatic incubator at 37°C with 5% CO2 (for HMEC-1
and MCF-10A) or without CO2 (for MDA-MB-231).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
The Beyozol (cat. no. R0011; Beyotime Institute of
Biotechnology) was utilized to lyse the tissues and cells. The
total RNA was extracted using a kit (cat. no. G3607-50T; Wuhan
Servicebio Technology Co., Ltd.). cDNA was synthesized from the
total RNA by means of a kit (cat. no. TSK302S; Tsingke Biological
Technology) according to the manufacturer's instructions. The cDNA
levels were detected with an Applied Biosystems® 7500
(Thermo Fisher Scientific, Inc.) using a kit (cat. no. D7260;
Beyotime Institute of Biotechnology). The thermocycling conditions
for qPCR were as follows: Initial denaturation at 95°C for 10 min,
followed by 40 cycles of denaturation at 95°C for 15 sec, annealing
at 60°C for 30 sec, and extension at 72°C for 30 sec. The
comparative cycle threshold (Ct) value method was employed to
determine the fold-differences in expression levels in relation to
those in U6 snRNA or β-actin (22).
The sequences of primers used are presented in Table I.
 | Table I.Sequences of primers used for reverse
transcription-quantitative PCR. |
Table I.
Sequences of primers used for reverse
transcription-quantitative PCR.
| Gene name | Primer sequence
(5′-3′) |
|---|
| piR-31115 | F:
AGCCTGAGCAACATAGCGAG |
|
| R:
GTGCAGGGTCCGAGGTATTC |
| U6 | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
| METTL3 | F:
GAGTGTCGGAGGTGATT |
|
| R:
AGTACGGGTATGTTGAGC |
| METTL14 | F:
TGTACTTACAAGCCGATAT |
|
| R:
TAGCAGTGATGCCAGTT |
| WTAP | F:
AGATGACCAACGAAGAAC |
|
| R:
CTAGTCGCATTACAAGGAT |
| ALKBH5 | F:
TTTTCCCCCTTTAGTCTCC |
|
| R:
CCCTTCACCAACTCCCAT |
| FTO | F:
GGTGTCCCAAGAAATCGT |
|
| R:
CTGGTGGCAGGAAAGAGT |
| MST1 | F:
CACCGATTTACGCCAGAAAA |
|
| R:
GAAGTTCTCCTCCAGTTGTG |
| MST2 | F:
ACCTCTGGATTGTTATGGAGTACTG |
|
| R:
TCTGTGTATTTTTCTCATAAAGTGC |
| LATS1 | F:
CCACCCTACCCAAAACATCT |
|
| R:
TCACTCTCATCTTCCTTGGG |
| LATS2 | F:
GTGGACCTGTATGAATTGGG |
|
| R:
TGGTGGCTGTTGAAGGAGTT |
| YAP1 | F:
GCTACAGTGTCCCTCGAACC |
|
| R:
TCCTTCCAGTGTTCCAAGGT |
| TAZ | F:
GAGCATAGAAGGCAGGTGAGCAAC |
|
| R:
GGCAAGGGCGGTGGGTAGG |
| AREG | F:
CGGTCTCCACTCGCTCTTCC |
|
| R:
GGGCTCTCATTGGTCCTTCGC |
| CTGF | F:
GCGAGGAGTGGGTGTGTGACG |
|
| R:
TGGACCAGGCAGTTGGCTCTAATC |
| CYR61 | F:
TGAGGTGCGGCCTTGTGGA |
|
| R:
CACTCAAACATCCAGCGTAAGTAA |
Plasmids and small interfering RNA
(siRNA) transfection
Plasmids harboring METTL3, HA-Ub, Myc-YAP1 and
siRNAs targeting YAP1 (si-YAP1), METTL3 (si-METTL3), IGF2BP1
(si-IGF2BP1), IGF2BP2 (si-IGF2BP2), IGF2BP3 (si-IGF2BP3) and si-NC
were synthesized by Tsingke Biological Technology. The
concentration of nucleic acid used was 50 nM. These were
transfected into cells using TSnanofect V1 (cat. no. TSV404;
Tsingke Biological Technology) at a temperature of 37°C for a
duration of 6 h. The time interval between transfection and
subsequent experimentation was 48 h. All the siRNA sequences are
presented in Table II.
 | Table II.Sequences of si-METTL3, si-YAP1,
si-IGF2BP1, si-IGF2BP2, si-IGF2BP3 and si-NC. |
Table II.
Sequences of si-METTL3, si-YAP1,
si-IGF2BP1, si-IGF2BP2, si-IGF2BP3 and si-NC.
| Name | Sequence
(5′-3′) |
|---|
| si-METTL3 | Sense:
AGCUACAGAUCCUGAGUUAGAGA (dT)(dT) |
|
| Antisense:
UCUCUAACUCAGGAUCUGUAGCU (dT)(dT) |
| si-YAP1 | Sense:
GAGAUACUUCUUAAAUCACAUCG (dT)(dT) |
|
| Antisense:
CGAUGUGAUUUAAGAAGUAUCUC (dT)(dT) |
| si-IGF2BP1 | Sense:
CACCAUGAACAAGCUUUACAUCG (dT)(dT) |
|
| Antisense:
CGAUGUAAAGCUUGUUCAUGGUG (dT)(dT) |
| si-IGF2BP2 | Sense:
UGGAAUUGCAUGGGAAAAUCAUG (dT)(dT) |
|
| Antisense:
CAUGAUUUUCCCAUGCAAUUCCA (dT)(dT) |
| si-IGF2BP3 | Sense:
CACAAUGAACAAACUGUAUAUCG (dT)(dT) |
|
| Antisense:
CGAUAUACAGUUUGUUCAUUGUG (dT)(dT) |
| si-NC | Sense:
UUCUCCGAACGUGUCACGU (dT)(dT) |
|
| Antisense:
ACGUGACACGUUCGGAGAA (dT)(dT) |
Lentiviral transduction
The lentiviral plasmids for overexpressing piR-31115
(LV-piR-31115), knocking down piR-31115 (LV-piR-31115 inhibitor)
and the negative control (LV-NC and LV-inhibitor NC) were procured
from Shanghai GenePharma Co., Ltd. (cat. no. LV2022-7704). The
lentiviral transduction was carried out using the 2nd generation
system. The interim cell line employed in this process was 293T,
which was provided by Shanghai GenePharma Co., Ltd. For
transfection, the quantity of the lentiviral plasmid used was 4 µg
per 6-well plate, and the ratio of the lentivirus, packaging and
envelope plasmids was optimized as transfer plasmid: psPAX2:
pMD2.G=4:3:1. The transfection procedure was performed at a
temperature of 37°C, and it lasted for 6 h. After 48 h, the cell
supernatant was collected and the lentiviral particles were then
collected through ultracentrifugation. To infect the target cells,
a multiplicity of infection of 50 was applied. The transduction of
the cells of interest lasted ~6 h. Notably, there was a time
interval of 48 h between the completion of transduction and the
subsequent experimentation. Stably transduced MDA-MB-231 cell lines
(MDA-MB-231-LV-NC, MDA-MB-231-LV-piR-31115, MDA-MB-231-LV-inhibitor
NC, and MDA-MB-231-LV-piR-31115 inhibitor) were obtained via
through selection with puromycin at a concentration of 1 µg/ml
(cat. no. ST551; Beyotime Institute of Biotechnology).
Cell co-culture assay
An equal number of stably transduced MDA-MB-231
cells were seeded in 24-well plates. The wells without cells served
as the control group. Initially, the lower chamber was filled with
Leibovitz's L-15 medium when MDA-MB-231 cells were seeded. Before
the cell co-culture, the Leibovitz's L-15 medium in the lower
chamber was removed and replaced with DMEM/F-12 supplemented with
10% (v/v) fetal bovine serum. Then, a Transwell insert (cat. no.
3422; Corning, Inc.) with a pore size of 8 µm was placed.
Subsequently, 1×104 HMEC-1 cells were added to the upper
chamber of the Transwell insert, where the medium used was
DMEM/F-12 without fetal bovine serum, and cultured for 12 h at
37°C. The HEMC-1 cells that had migrated to the lower chamber were
fixed with 4% paraformaldehyde at room temperature for 30 min and
then stained with a 0.1% crystal violet solution at room
temperature for 30 min. The number of migratory cells was counted
in three randomly selected fields (at a low magnification of ×100)
using a light microscope.
Production of the conditioned medium
(CM)
Firstly, an equal quantity of stably transduced
MDA-MB-231 cells were meticulously inoculated into a culture dish
and permitted to adhere to the surface for a duration of time.
Subsequently, the original culture medium was gingerly removed, and
a serum-free DMEM/F-12 mixture was introduced into the dish for
culturing. After 24 h, the resultant CM, which now encompassed the
secreted factors, was filtered through a 0.22-µm filter (cat. no.
SLHV033RS; MilliporeSigma) to eliminate any cellular debris or
large particles. The filtered CM was then apportioned and promptly
frozen at −80°C until it was requisite for further experiments.
This painstaking process of CM generation ensured the collection of
a cell-CM that could be utilized to explore its effects on other
cell types, such as HMEC-1 cells, in subsequent assays.
Cell counting kit-8 (CCK-8) assay
First, a total of 2×103 HMEC-1 cells
(either transfected or non-transfected) per well were seeded in
96-well plates and allowed to attach. Subsequently, they were
treated with CM for 24 h. Finally, 10 µl of CCK-8 reagent (cat. no.
C0048S; Beyotime Institute of Biotechnology) was mixed with 90 µl
of DMEM/F-12 cell medium and added to one well of the 96-well
plate. The incubation with CCK-8 was performed at 37°C for 0.5 h,
and the absorbance was measured at a wavelength of 450 nm.
Transwell migration assays
Transwell inserts (cat. no. 3422; Corning, Inc.)
were employed to conduct cell migration assays. First, HMEC-1 cells
(either transfected or non-transfected) were treated with CM for 24
h. Then, 1×104 cells were resuspended in 0.1 ml of
serum-free DMEM/F-12 and seeded in the upper chamber. DMEM/F-12
containing 10% serum was added to the lower chamber. After
culturing for 12 h at 37°C, the cells that had migrated to the
lower chamber were fixed and stained with a crystal violet
solution. The number of migratory cells was counted in three
randomly selected fields (at a low magnification of ×100).
Western blotting (WB)
RIPA lysis buffer (cat. no. P0013B; Beyotime
Institute of Biotechnology) was used to extract the total cellular
protein. The proteins were separated into cytoplasmic and nuclear
fractions using a kit (cat. no. P0027; Beyotime Institute of
Biotechnology). The protein determination was performed using the
bicinchoninic acid (BCA) method. Proteins were separated by 10%
SDS-PAGE (cat. no. P0012A; Beyotime Institute of Biotechnology) and
then transferred to PVDF membranes (cat. nos. ISEQ00010 and
IPVH00010; Merck KGaA). A protein mass of 100 micrograms was loaded
per lane. The membranes were blocked with 5% defatted milk/TBST
(containing 0.1% Tween) for 1 h at room temperature, and then
incubated with primary antibodies against METTL3 (1:1,000; cat. no.
GB114688; Wuhan Servicebio Technology Co., Ltd.), PIWIL4 (1:200;
cat. no. sc-517215; Santa Cruz Biotechnology, Inc.), VEGFA
(1:5,000; cat. no. 81323-2-RR), YAP1 (1:5,000; cat. no.
66900-1-lg), IGF2BP1 (1:10,000; cat. no. 22803-1-AP), IGF2BP2
(1:2,000; cat. no. 11601-1-AP), IGF2BP3 (1:10,000; cat. no.
14642-1-Ig), proliferating cell nuclear antigen (1:10,000; cat. no.
60097-1-Ig), HA (1:10,000; cat. no. 66006-2-Ig), Myc (1:5,000; cat.
no. 66003-2-Ig; all from Proteintech Group, Inc.) and β-actin
(1:1,000; cat. no. GB15003; Wuhan Servicebio Technology Co., Ltd.)
overnight at 4°C. Next, the membranes were incubated with
horseradish peroxidase-conjugated secondary antibodies (1:10,000;
cat. nos. GB23301 and GB23303; Wuhan Servicebio Technology Co.,
Ltd.) at room temperature for 1 h. The pre-stained protein marker
was purchased from Wuhan Servicebio Technology Co., Ltd. (cat. no.
G2083). The protein bands were developed using an enhanced
chemiluminescence reagent (cat. no. P0018; Beyotime Institute of
Biotechnology).
Coimmunoprecipitation (Co-IP)
assays
Co-IP of the lysates was carried out using a kit
(cat. no. P2179; Beyotime Institute of Biotechnology). The lysis
buffer used was Lysis Buffer from the kit (containing components
and concentrations as provided by the manufacturer). For each IP
reaction, 500 µl of lysate was used. BeyoMag™ Protein
A+G magnetic beads (20 µl per 500 µl sample) were used. Protein A+G
can bind to the Fc end of the antibody specifically. After
incubation for 30 min, the Protein A+G magnetic beads-antibody
mixture (beads-Ab complex) was formed. Then the sample was added,
and the sample could be specifically recognized by the Fab end of
the antibody to form the Protein A+G magnetic
beads-antibody-antigen immune complex (beads-Ab-Ag complex). The
immunocomplex was washed to remove unbound proteins. Centrifugation
steps included centrifugation at 12,000 × g at 4°C for 5 min during
the sample preparation process. WB was performed to evaluate
protein expression in the samples obtained through
immunoprecipitation with anti-PIWIL4 antibodies (1:100; cat. no.
sc-517215; Santa Cruz Biotechnology, Inc.). The input and IgG
groups were utilized as the positive and negative controls,
respectively.
Total RNA m6A quantification
Total RNA m6A quantification was carried out using a
kit (cat. no. P-9005; EpiGentek). The cells (either transfected or
non-transfected) were treated with CM for 24 h. Subsequently, the
RNA extracted from the cells with TRIzol reagent (Thermo Fisher
Scientific, Inc.) was combined with the capture antibody. During
multiple incubations steps, colorimetric measurement of the m6A
content was performed at a wavelength of 450 nm.
Methylated RNA immunoprecipitation
(MeRIP)
MeRIP was carried out using a kit (cat. no.
Bes5203-2; Guangzhou Bersinbio Co., Ltd.). Briefly, the RNA of
cells was extracted with Beyozol reagent and fragmented by
ultrasonication. The RNA was incubated with an anti-m6A antibody at
4°C for 4 h and then with magnetic beads. The enrichment of
m6A-containing mRNAs was then analysed by RT-qPCR.
Transcriptome sequencing and pathway
enrichment analysis
The RNA from cells was extracted with Beyozol and
then used to construct an RNA library using VAHTS Universal V6
RNA-seq Library Prep Kit (cat. no. NR604; Shanghai OE Biotech Co.,
Ltd.). The quality/integrity of the processed RNA samples was
verified by Agilent 2100 Bioanalyzer. The RNA library was sequenced
on the Illumina NovaSeqTM 6000 platform by OE Biotech, Inc. with a
sequencing type of 150-bp paired end using VAHTS Universal V6
RNA-seq Library Prep Kit (cat. no. NR604; Shanghai OE Biotech Co.,
Ltd.). The loading concentration of the final library was 5 nM. The
pathway enrichment analysis was performed using the Kyoto
Encyclopedia of Genes and Genomes (KEGG) database (https://www.genome.jp/kegg/). This database was
utilized to identify the potential signaling pathways related to
the genes of interest. The analysis was based on the publicly
available data and algorithms in the KEGG database to explore the
biological functions and pathways associated with the
differentially expressed genes.
Statistical analysis
Statistical analyses were performed using the Prism
software (version 5.0; Dotmatics). In vitro data (mean ±
standard deviation) were obtained from three independent
experiments. To evaluate the statistical significance of the
differences between two separate groups, the following rules were
applied: for tissue samples, Student's t-test (paired) was used,
while for other samples, Student's t-test (unpaired) was applied.
For comparisons among more than two groups, one-way analysis of
variance (ANOVA) was utilized, followed by Newman-Keuls multiple
comparison test. P<0.05 was considered to indicate a
statistically significant difference.
Results
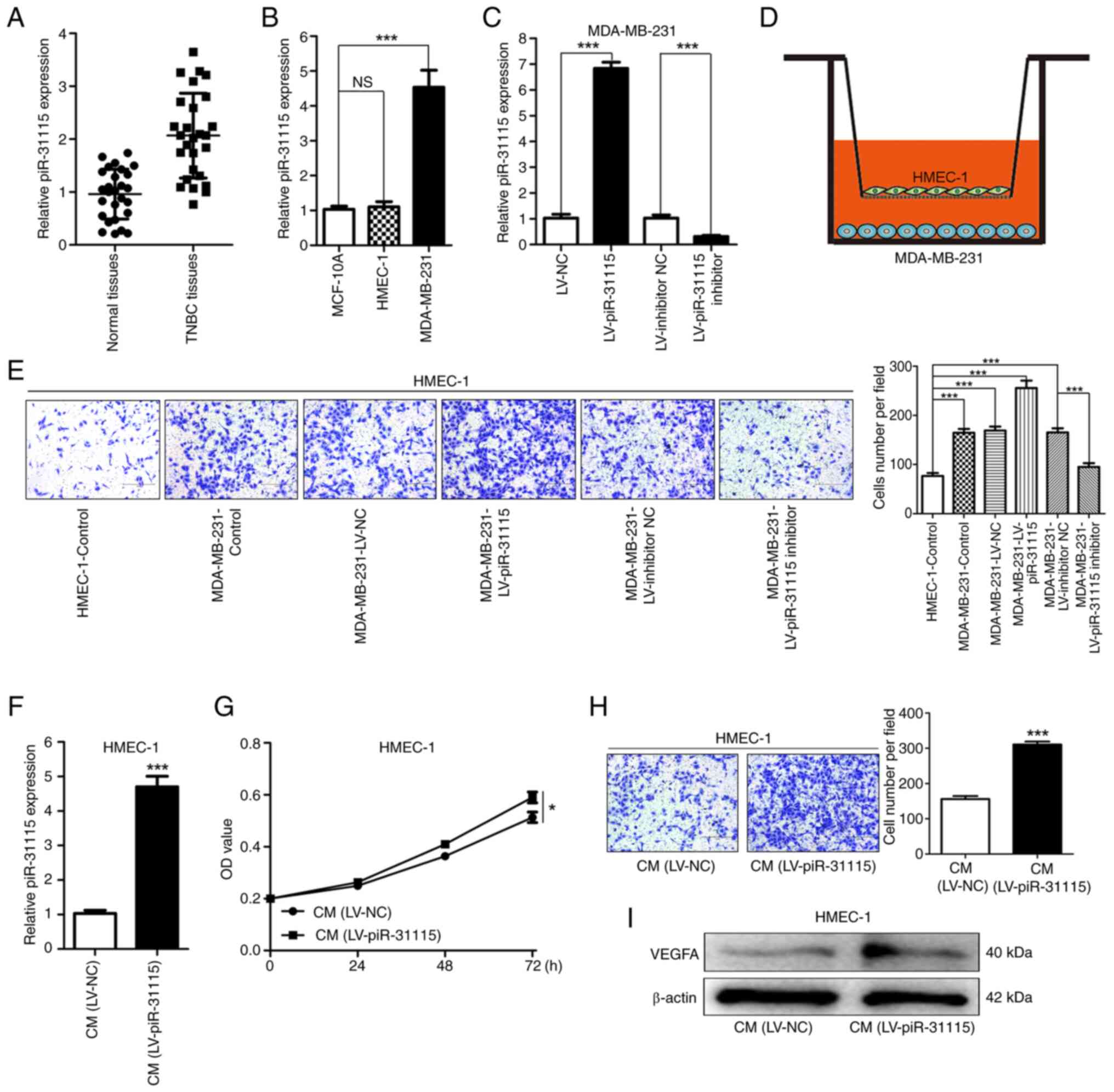
MDA-MB-231 cell-derived piR-31115
promotes the proliferation and migration of HMEC-1 cells
To investigate the potential role of piR-31115 in
TNBC, RT-qPCR was conducted on breast cancer and adjacent normal
tissues from 27 patients with TNBC to detect its expression. The
results evidently indicated that piR-31115 was significantly more
highly expressed in TNBC tissues compared with adjacent normal ones
(Fig. 1A), preliminarily suggesting
its potential involvement in TNBC development. Using MCF-10A cells
as a control, the expression levels of piR-31115 were evaluated in
MDA-MB-231 and HMEC-1 cells. The results demonstrated that the
expression level of piR-31115 in MDA-MB-231 cells was higher than
that in MCF-10A cells. Additionally, no significant difference was
detected in piR-31115 expression between MCF-10A cells and HMEC-1
cells (Fig. 1B). Lentiviral vector
transduction was carried out to obtain MDA-MB-231 cells in which
piR-31115 was stably overexpressed or knocked down (Fig. 1C). Through cell co-culture
experiments, it was discovered that MDA-MB-231 cells exhibited
chemotactic effects on HMEC-1 cells. After the expression of
piR-31115 in MDA-MB-231 cells was upregulated, its chemotactic
effect on HEMC-1 cells was markedly enhanced. When the expression
of piR-31115 in MDA-MB-231 cells was interfered with, its
chemotactic effect on HMEC-1 cells was reduced (Fig. 1D and E). These findings suggested
that MDA-MB-231 cell-derived piR-31115 can regulate the biological
behaviour of HMEC-1 cells. HMEC-1 cells were treated with medium
from MDA-MB-231 cells with overexpression of piR-31115. The RT-qPCR
results showed that the piR-31115 expression level was increased in
HMEC-1 cells (Fig. 1F). Further
examination of HMEC-1 cells after CM treatment revealed that their
proliferation and migration abilities were enhanced (Fig. 1G and H). WB analysis demonstrated
that the VEGFA protein expression level increased in HMEC-1 cells
after CM treatment (Fig. 1I). These
data suggest that MDA-MB-231 cell-derived piR-31115 plays a crucial
role in promoting angiogenesis.
MDA-MB-231 cell-derived piR-31115
increases the m6A level in HMEC-1 cells via METTL3
In recent years, m6A methylation has been
established to play a significant role in the metastasis of cancer
(23,24). A substantial increase was detected
in total methylation in HMEC-1 cells treated with CM (Fig. 2A). These findings indicate that
piR-31115 derived from MDA-MB-231 cells can regulate m6A
methylation levels in HMEC-1 cells. m6A methylation is a dynamic
and reversible modification regulated by methyltransferases
(METTL3, METTL14 and WTAP) and demethylases (ALKBH5 and FTO). The
mRNA expression levels of METTL3, METTL14, WTAP, ALKBH5 and FTO
were determined, and the results showed that only the expression of
METTL3 increased in HMEC-1 cells under CM treatment (Fig. 2B). Further detection of METTL3
protein expression levels by WB demonstrated that METTL3 protein
expression levels rose under CM treatment (Fig. 2C). piRNAs need to bind to the PIWI
family of proteins to exert a variety of biological effects
(11). Wang et al (25) have experimentally confirmed that the
expression of PIWIL4 is increased in MDA-MB-231 and promotes its
metastasis. Meanwhile, they also proposed the possibility that
PIWIL4 cooperates with piRNA to achieve its functions (25). In the present study, co-IP
experiments were utilized and it was found that PIWIL4 and METTL3
were bound to each other in HMEC-1 cells treated with CM. When
HMEC-1 cells were treated with CM from MDA-MB-231 cells with
piR-31115 knockdown, the binding effect of PIWIL4 and METTL3 was
diminished (Fig. 2D). These results
suggest that METTL3 is a target gene regulated by the
piR-31115-PIWIL4 complex. The upregulation of METTL3 expression in
HMEC-1 cells not only augmented the overall level of m6A
modification but also significantly enhanced their proliferative
and migratory capacities (Fig.
2E-H). After interfering with the expression of METTL3 in
HMEC-1 cells, the stimulatory effect of CM on cell proliferation
and migration was attenuated, concurrently leading to a
downregulation of VEGFA expression (Fig. 2I-M). These results comprehensively
indicate that METTL3 is an important regulatory gene of piR-31115
and that CM elevates its m6A modification level through the
upregulation of METTL3 expression in HMEC-1 cells, thereby
resulting in increased cell proliferation and migration, which is
beneficial for the formation of microvessels.
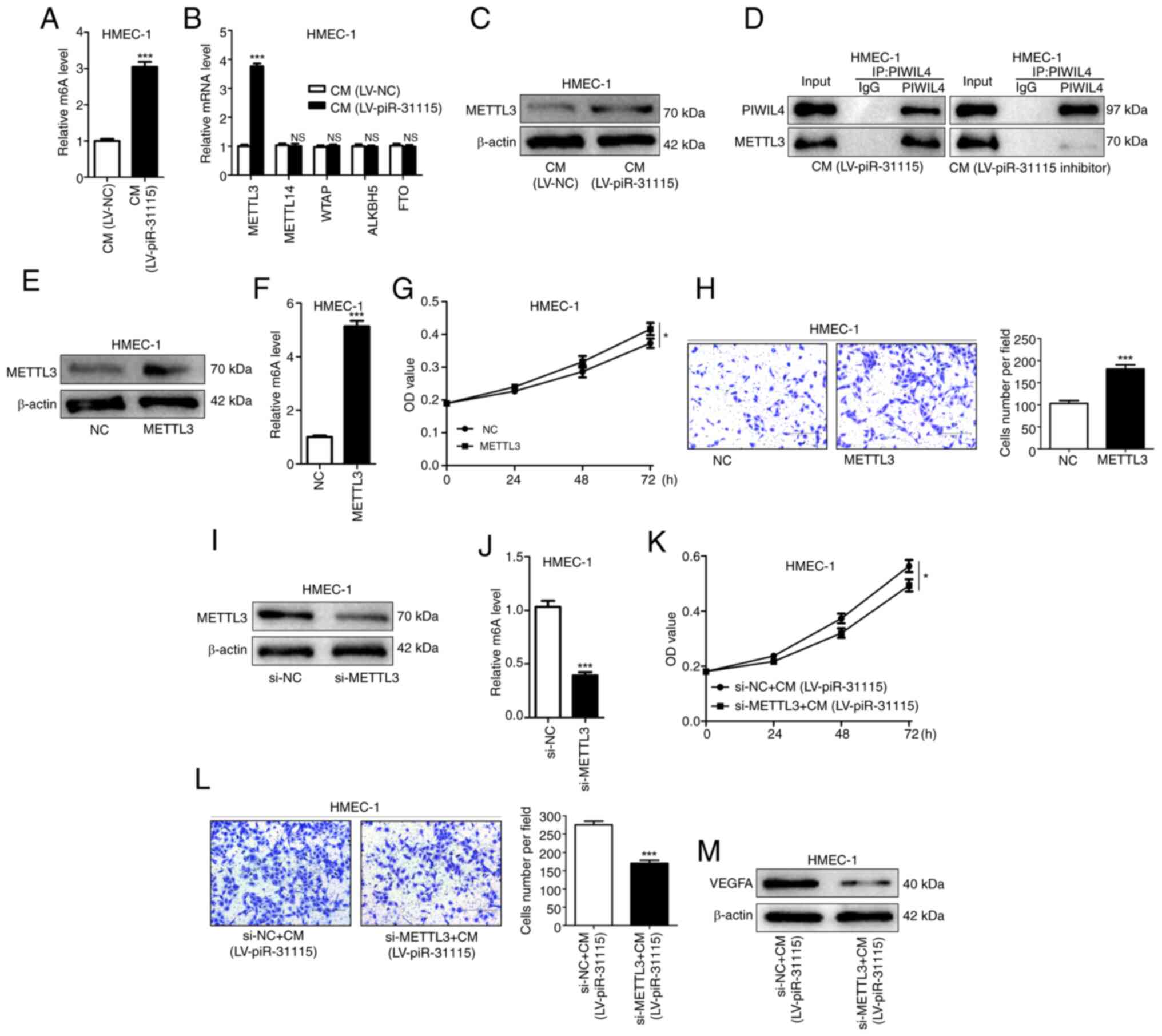 | Figure 2.piR-31115 mediates m6A methylation
via METTL3 regulation. (A) m6A RNA methylation quantification kit
was used to detect m6A levels in CM-cultured HMEC-1 cells. (B)
Reverse transcription-quantitative PCR ascertained METTL3, METTL14,
WTAP, ALKBH5 and FTO mRNA levels. (C) METTL3 protein levels after
CM treatment were analysed by WB. (D) Co-immunoprecipitation of
endogenous PIWIL4 and METTL3 from CM-treated cell lysates, then
analysed by WB. (E and F) Kit-detected m6A levels in
METTL3-overexpressing cells. (G) CCK-8 assay was used to assess
proliferation of METTL3-overexpressing cells. (H) Transwell assays
evaluating migration of METTL3-overexpressing cells (magnification,
×100). (I and J) Kit-detected m6A levels in METTL3-knockdown cells.
(K) CCK-8 assay gauged proliferation of METTL3-knockdown,
CM-treated cells. (L) Transwell assays measured their migration.
(M) WB was utilized to analyse VEGFA levels in METTL3-knockdown,
CM-treated cells (magnification, ×100). Data are derived from three
independent experiments. *P<0.05 and ***P<0.001. piR,
Piwi-interacting RNA; m6A, N6-methyladenosine; CM, conditioned
medium; WB, western blotting; CCK-8, Cell Counting Kit-8; NC,
negative control; LV, lentivirus; NS, not significant; si-, small
interfering. |
The influence of METTL3 on signaling
pathways in HMEC-1 Cells treated with CM
To clarify the influence of METTL3 on signaling
pathways in HMEC-1 cells after CM treatment, an RNA-sequencing
analysis was carried out. Remarkably, the KEGG analysis showed that
the Hippo pathway was significantly related to METTL3 (Fig. 3A). The Hippo pathway is an important
signalling pathway that regulates cell proliferation (26). To identify changes within this
pathway, RT-qPCR was used to detect the expression levels of key
regulators and downstream targets. Our research results suggest
that when HMEC-1 cells were treated with CM, interference with
METTL3 significantly inhibited YAP1 and its downstream targets
(Fig. 3B). When METTL3 was
overexpressed, it significantly increased the expression level of
YAP1 and its downstream targets (Fig.
3C). The results of MeRIP-qPCR demonstrated that the presence
of METTL3 was directly associated with an increase in the m6A level
of YAP1 in HMEC-1 cells (Fig. 3D).
Moreover, it was also found that the m6A level of YAP1 was
increased in CM-treated HMEC-1 cells (Fig. 3E). After interfering with METTL3
expression in HMEC-1 cells, the m6A level of YAP1 in HMEC-1 cells
decreased (Fig. 3F). Additionally,
interfering with METTL3 expression reduced the increase in the m6A
level in HMEC-1 cells induced by CM treatment (Fig. 3G). These results indicate that YAP1
is the target gene of METTL3 in HMEC-1 cells with CM treatment.
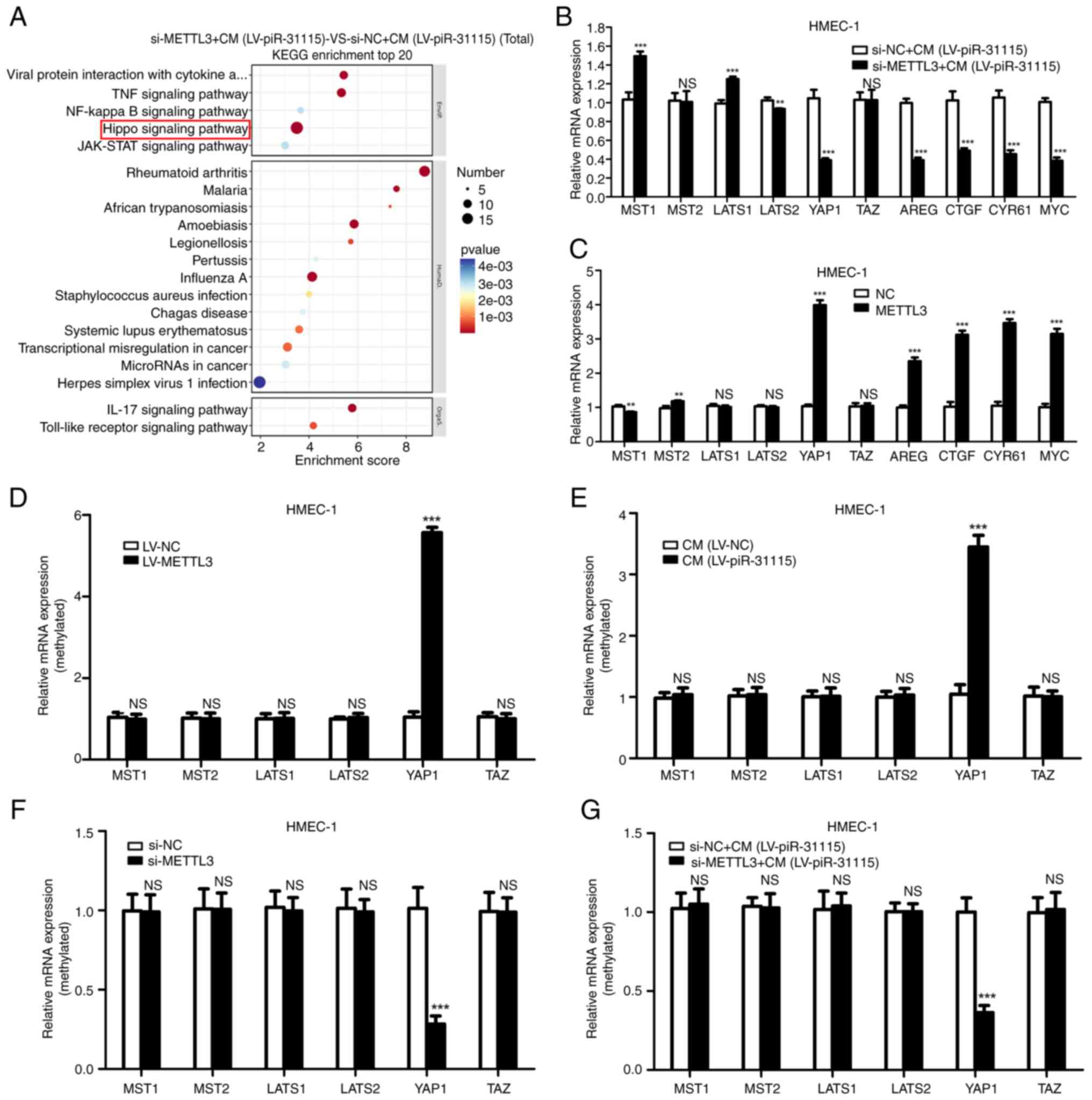 | Figure 3.Identification of METTL3 target genes
via RNA-sequencing. (A) KEGG pathway enrichment analysis on
METTL3-knockdown, CM-treated HMEC-1 cells. (B and C) Reverse
transcription-quantitative PCR was used to examine the Hippo
signalling pathway-related genes (MST1, MST2, LATS1, LATS2, YAP1,
TAZ, AREG, CTGF, CYR61 and MYC) expression in METTL3-knockdown,
CM-treated cells. (D-G) Detected effects of METTL3 and CM on
N6-methyladenosine levels of Hippo signalling pathway-related genes
in HMEC-1 cells. **P<0.01 and ***P<0.001. KEGG, Kyoto
Encyclopedia of Genes and Genomes; CM, conditioned medium; YAP1,
Yes-associated protein 1; NC, negative control; LV, lentivirus; NS,
not significant; si-, small interfering. |
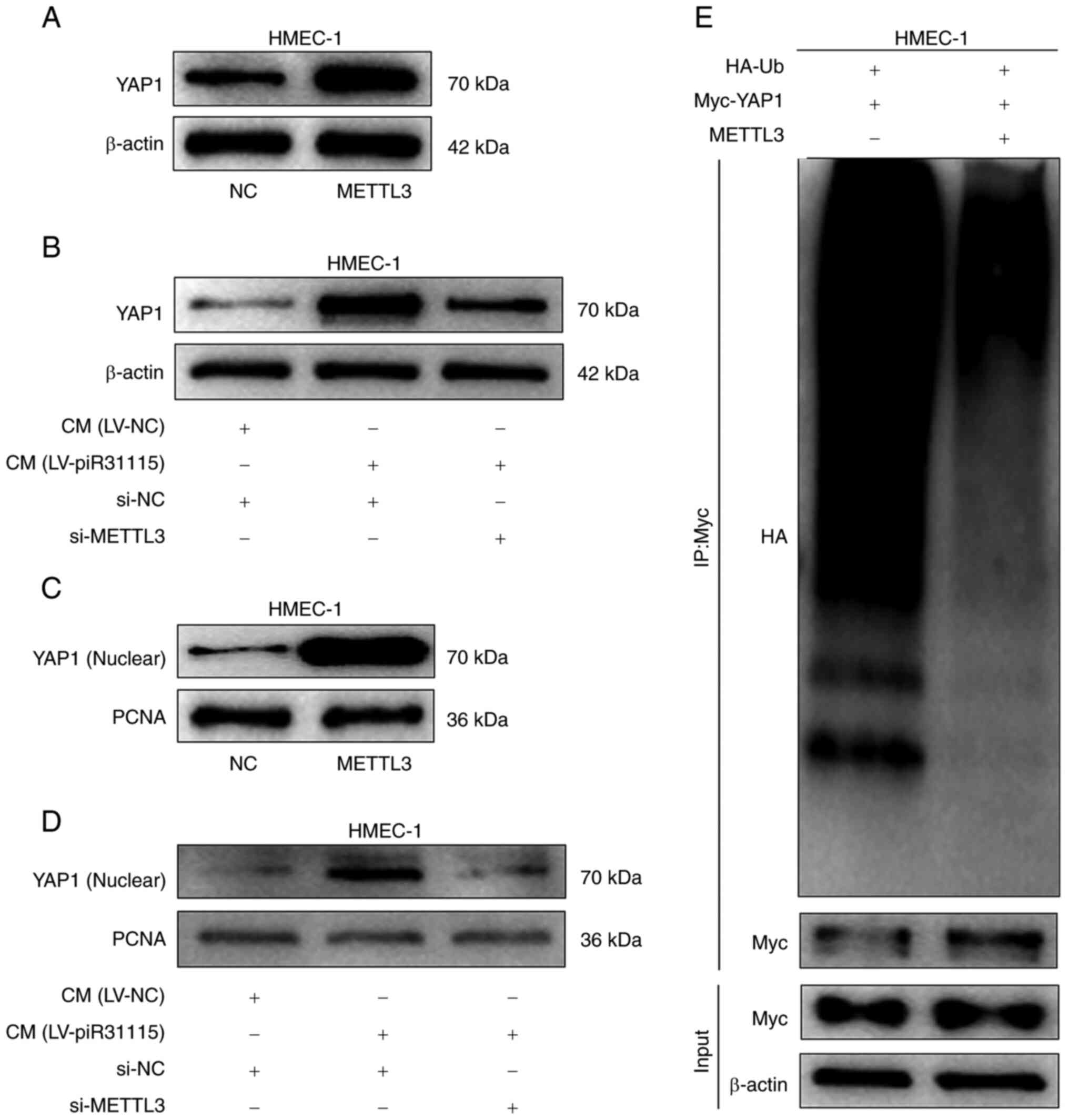
METTL3 promotes the nuclear
translocation of the YAP1 protein
The regulatory effect of METTL3 on the YAP1 protein
level was examined. WB results demonstrated that overexpression of
METTL3 significantly increased the YAP1 expression level in HMEC-1
cells (Fig. 4A). Meanwhile, CM also
enhanced the YAP1 protein expression level. However, interference
with METTL3 suppressed YAP1 expression level when the cells were
treated with CM (Fig. 4B). Nuclear
protein was extracted from HMEC-1 cells with overexpression of
METTL3 or treated with CM and it was found that YAP1 expression
increased. Nevertheless, interfering with METTL3 could inhibit the
promoting effect of CM on the expression of YAP1 nuclear protein
(Fig. 4C and D). The detection of
YAP1 ubiquitination in HMEC-1 cells revealed that the level of
ubiquitinated YAP1 protein decreased after overexpression of METTL3
(Fig. 4E). The aforementioned
results indicated that the upregulated expression of METTL3 in
HMEC-1 cells promotes nuclear translocation of the YAP1 protein
under CM treatment.
METTL3 affects the proliferation and
migration of HEMC-1 cells by regulating YAP1
When the expression of YAP1 in HMEC-1 cells was
interfered with, the stimulatory effect of CM on cell proliferation
and migration was significantly diminished (Fig. 5A-C). Moreover, it was also observed
that when the expression of YAP1 in HMEC-1 cells was interfered
with, the promoting influence of METTL3 on cell proliferation and
migration was significantly weakened (Fig. 5D and E). Meanwhile, the expression
level of VEGFA in HMEC-1 cells decreased (Fig. 5F). These results indicate that
METTL3 affects the proliferation and migration of HEMC-1 cells by
regulating YAP1.
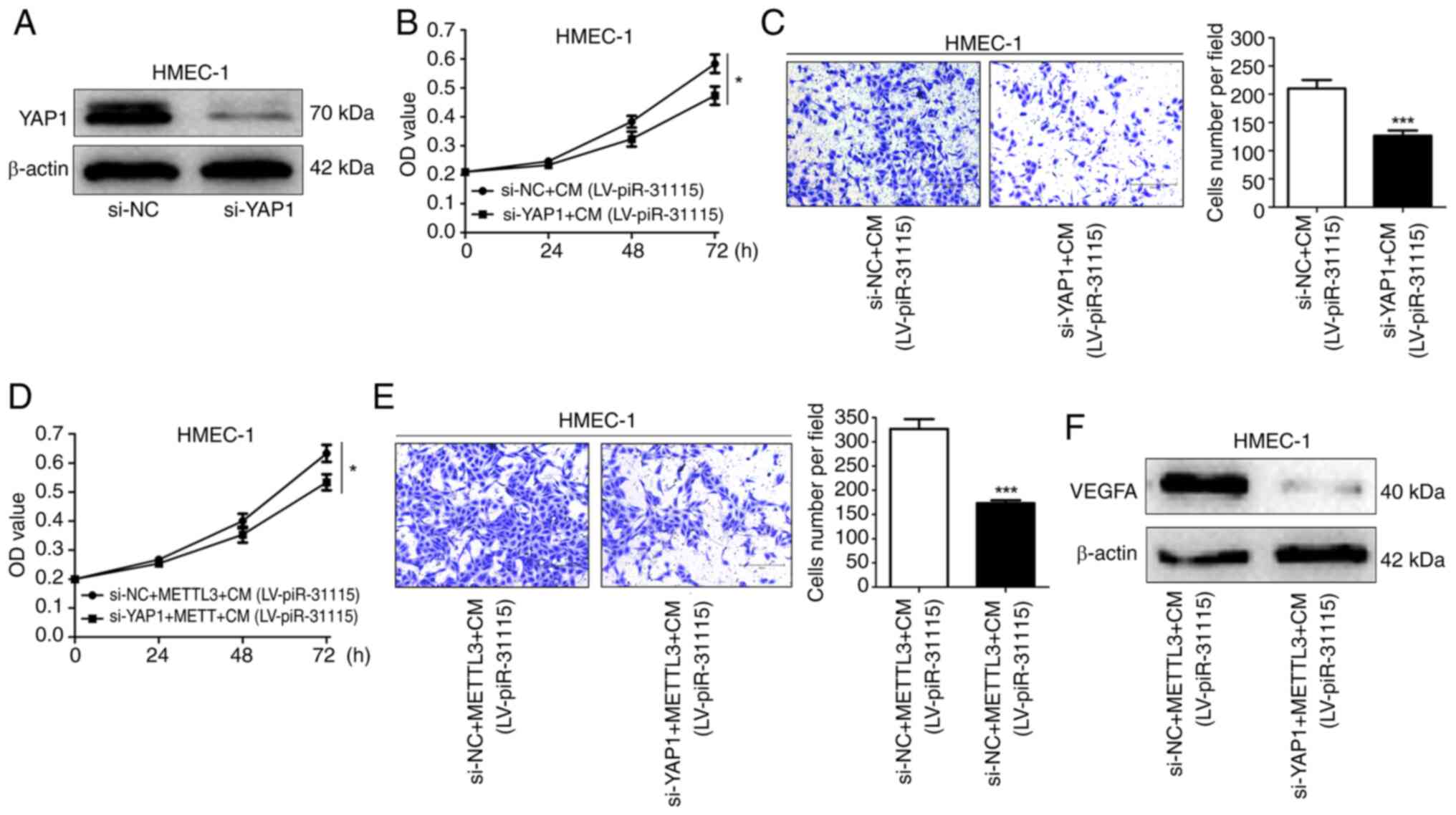 | Figure 5.YAP1 promotes HMEC-1 cell
proliferation and migration. (A) WB was used to determine YAP1
expression in YAP1-knockdown cells. (B) CCK-8 assay was used to
assess proliferation after CM treatment and YAP1 knockdown. (C)
Migration after CM treatment and YAP1 knockdown was evaluated by
Transwell assays (magnification, ×100). (D) Cell proliferation
after CM treatment, METTL3 overexpression or YAP1 knockdown was
detected using CCK-8 assay. (E) Transwell assays were utilized to
measure migration after CM treatment, METTL3 overexpression or YAP1
knockdown (magnification, ×100). (F) VEGFA expression after CM
treatment, METTL3 overexpression or YAP1 knockdown was analysed by
WB. Data are derived from three independent experiments. *P<0.05
and ***P<0.001. YAP1, Yes-associated protein 1; WB, western
blotting; CCK-8, Cell Counting Kit-8; CM, conditioned medium; NC,
negative control; LV, lentivirus; si-, small interfering. |
M6A-mediated regulation of YAP1
expression is identified by IGF2BP2 in HMEC-1 cells
Previous investigations have identified the IGF2BP
family as m6A readers. This family comprises IGF2BP1, IGF2BP2 and
IGF2BP3 (27). In order to
elucidate the role of the IGF2BP family within the present study,
siRNAs were employed to interfere with the expression of IGF2BP1,
IGF2BP2 and IGF2BP3 in HMEC-1 cells (Fig. 6A-C). It is evident that only in the
HMEC-1 cells with IGF2BP2 knockdown, the YAP1 protein expression
level was markedly diminished (6D-F). Interestingly, when IGF2BP2
expression was simultaneously interfered with and METTL3 was
overexpressed, the elevation in YAP1 expression level was
suppressed, indicating that IGF2BP2 plays a crucial role in the
process of METTL3 promoting YAP1 expression (Fig. 6G). Additionally, through co-IP
experiments, it was discovered that the ubiquitination level of
YAP1 in the si-IGF2BP2 group was increased compared with that in
the si-NC group, suggesting that IGF2BP2 may maintain the stability
of YAP1 by reducing its ubiquitination (Fig. 6H). Further investigations into the
effects of IGF2BP2 on the proliferation and migration of HMEC-1
cells revealed that interfering with IGF2BP2 expression eliminated
the promoting effect of METTL3 on the proliferation and migration
of HMEC-1 cells, further confirming the pivotal role of IGF2BP2 in
regulating YAP1-related cellular processes (Fig. 6I and J).
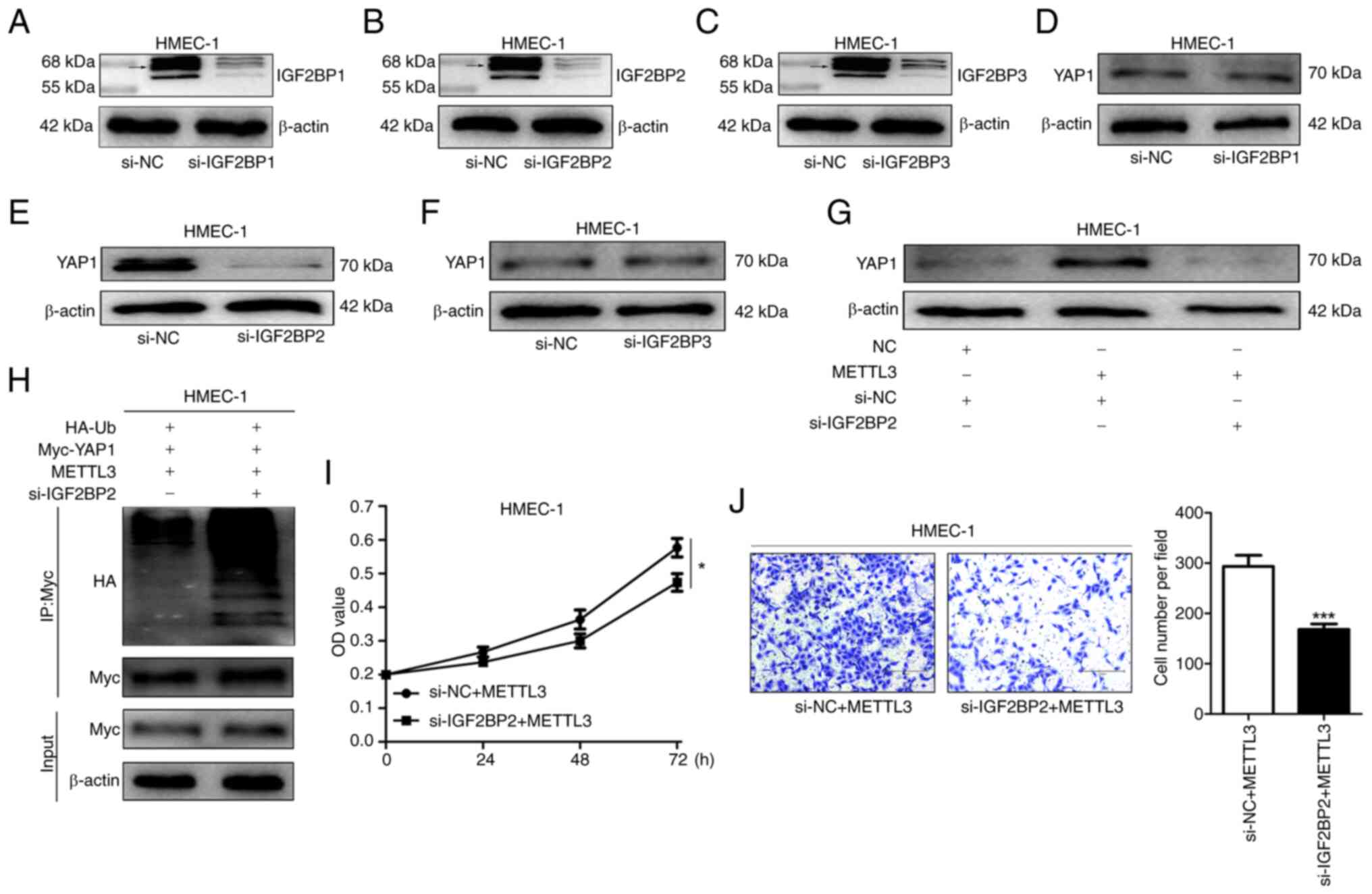 | Figure 6.IGF2BP2 promotes YAP1 expression in
HMEC-1 cells. (A-C) WB was used to detect IGF2BP1/2/3 expression in
IGF2BP1/2/3-knockdown cells Due to the existence of multiple
isoforms of IGF2BP1, IGF2BP2 and IGF2BP3 (possibly resulting from
alternative splicing or post-translational modifications), multiple
bands are presented in the WB. Among them, the bands around 65 kDa
(as indicated by arrows) are the ones that were mainly considered
to be the primary candidate bands representing the IGF2BP1, IGF2BP2
and IGF2BP3 proteins, and the subsequent analysis is mainly based
on these bands. (D-F) WB was utilized to analyse IGF2BP1/2/3
expression in YAP1-expressing cells. (G) YAP1 expression in
METTL3-overexpressing or IGF2BP2-knockdown cells was examined by
WB. (H) Ubiquitinated YAP1 level in METTL3-overexpressing or
IGF2BP2-knockdown cells was determined using WB. (I) Cell Counting
Kit-8 assay was used to assess proliferation of
METTL3-overexpressing or IGF2BP2-knockdown cells. (J) Transwell
assays were used to evaluate migration of METTL3-overexpressing or
IGF2BP2-knockdown cells (magnification, ×100). Data are derived
from three independent experiments. *P<0.05 and ***P<0.001.
YAP1, Yes-associated protein 1; WB, western blotting; si-, small
interfering; NC, negative control. |
Discussion
Cancer metastasis represents a formidable hurdle in
the realm of cancer treatment, with angiogenesis, the formation of
new blood vessels, assuming a pivotal role. In the context of TNBC,
an especially aggressive subtype that lacks specific therapeutic
targets, comprehending the mechanisms that underpin angiogenesis is
of paramount significance. TNBC cells possess the propensity to
detach from the primary tumour and metastasize to distant organs,
and this process of theirs is highly dependent on the establishment
of new blood vessels to sustain their growth and dissemination, as
recently described (28,29). Angiogenesis not only furnishes the
requisite nutrients and oxygen for the rapidly proliferating cancer
cells but also functions as a conduit for their spread (30,31).
By delving into the regulatory mechanisms of angiogenesis in TNBC,
there is a chance of identifying novel therapeutic targets and
devising more efficacious strategies to combat this lethal disease.
Recent studies have found that ncRNAs, as tissue-specific
molecules, play both oncogenic and tumour-suppressive roles in
cancer progression, including cancer cell proliferation,
metastasis, chemoresistance and stemness (14,32).
In the present study, our focus lies on the role of piR-31115
within the TNBC cell-induced angiogenesis process and its
associated molecular mechanisms.
piR-31115, being a constituent of the ncRNA family,
is a member of the piRNA family (21). Du et al (20) have established that the expression
of piRNA-31115 is abnormally upregulated in renal clear cell
carcinoma tissues and have illustrated that it functions as an
oncogene, thereby facilitating the progression of renal clear cell
carcinoma. In the present research centered around TNBC, it was
ascertained that the expression of piR-31115 was augmented in TNBC
samples and TNBC cell lines. Concurrently, the outcomes of the cell
co-culture experiment have disclosed an intriguing phenomenon: TNBC
cells inherently possess the capacity to recruit HMEC-1 cells.
Remarkably, when piR-31115 is overexpressed, this recruitment
effect on HMEC-1 cells is conspicuously intensified. On the
contrary, when the expression of piR-31115 is perturbed, the
recruitment of HMEC-1 cells is diminished. Zhao et al
(33) found through the research on
the in vitro co-culture of MDA-MB-231 cells and human
umbilical vein endothelial cells as well as the CD31 staining of
tumour endothelial cells in vivo that piR-2158 has an
inhibitory effect on the angiogenesis of breast cancer (33). Taken together, these results suggest
that some piRNAs play a role in promoting angiogenesis in the
context of TNBC.
Information exchange serves as the bedrock of
interactions between cells. Cells are capable of secreting
‘signalling molecules’, such as exosomes, in a variety of ways
(34). Upon other cells ingesting
these ‘signalling molecules’, they proceed to regulate the relevant
signalling pathways within the cell, prompting the cell to respond
(35). In the present study, the
expression of piR-31115 in HMEC-1 cells was significantly increased
after being treated with CM. This finding suggests that HMEC-1
cells take up piR-31115, which is secreted by MDA-MB-231 cells.
Currently, research on piRNAs in tumours is primarily focused on
the regulatory mechanism of intracellular signaling pathways in
cancer cells (17,18). However, there are scarce studies on
the regulatory effects of cancer cell-derived piRNAs on other
cells. In the present study, it was found that CM can increase the
total m6A modification level and the expression level of METTL3 in
HMEC-1 cells. These results suggest that piR-31115 derived from
MDA-MB-231 cells may regulate HMEC-1 cells through m6A
modification.
With the deepening of research in RNA epigenetics,
RNA methylation has emerged as a pivotal factor in the intricate
processes of tumorigenesis (36).
Among them, m6A methylation, which refers to the methylation of the
nitrogen atom N6 on the sixth carbon of adenosine (A) in the RNA
molecule, is the most common form of mRNA modification (37,38).
Research has demonstrated that it plays an important role in the
progression of cancer (39–41). METTL3 is a key protein for m6A
methylation in various types of cancers (42). However, there are few studies on
METTL3 regulating tumour angiogenesis. Previous studies have shown
that piRNAs need to bind to PIWI family proteins to exert their
biological effects (43). The
current results showed that piR-31115 derived from MDA-MB-231 cells
could strengthen the binding of PIWIL4 to METTL3, thus increasing
METTL3 expression level. Moreover, it was found that overexpression
of METTL3 promotes the proliferation and migration of HMEC-1 cells.
These results suggest that METTL3 plays an important role in
promoting the angiogenesis.
The Hippo signalling pathway plays a significant
role in organ development. Stimulated by extracellular growth
inhibition signals, a cascade of kinases leads to the
phosphorylation of the effector YAP1 and transcriptional
coactivator TAZ, which remain in the cytoplasm and are degraded by
ubiquitination to control cell proliferation and organ size
(26). In the present study, it was
found that interference with METTL3 expression in CM-treated HMEC-1
cells significantly affected the Hippo signalling pathway.
Interestingly, only the m6A methylation level of the YAP1 gene was
found to be regulated by METTL3. These findings confirmed that YAP1
was the target gene of METTL3 methylation. This is consistent with
the study by Ni et al (44).
The detection of YAP1 protein expression showed that METTL3 could
prevent the ubiquitination-mediated degradation of YAP1 by altering
its nuclear translocation, thereby enhancing the transcription of
downstream target genes and leading to the proliferation and
migration of HMEC-1 cells. m6A methylation-binding proteins often
determine the fate of modified target genes. The IGF2BP protein
family is a group of m6A reading proteins whose members enhance the
stability of target genes (45).
The present results showed that interference with IGF2BP2
expression results in an elevation of YAP1 ubiquitination levels,
significantly inhibiting the proliferation and migration of HMEC-1
cells. This indicates that IGF2BP2 plays an important role in
modulating the stability of YAP1 that undergoes m6A
modification.
The findings of the present study strongly suggest
that piR-31115 holds great potential for development as a drug
target. From the perspective of drug research and development,
multiple strategies can be explored to target piR-31115. For
instance, small molecule inhibitors or antisense oligonucleotides
that specifically bind to piR-31115 can be designed. By binding to
piR-31115, they can prevent its interaction with PIWI family
proteins or promote its degradation, thereby blocking the
signalling pathway that promotes angiogenesis. Gene editing
technologies such as the CRISPR-Cas system can also be used to
directly regulate the expression of the piR-31115 gene, suppressing
its abnormal function at the source. However, there are inevitably
some challenges in the development process. For example, ensuring
the high specificity of drug molecules for piR-31115 and avoiding
affecting the functions of other normal RNAs. Meanwhile, it is
necessary to construct an effective drug delivery system to
increase the concentration and efficacy of drug molecules in tumour
tissues and overcome problems such as difficulties in cell uptake
and poor in vivo stability.
Our findings suggest that targeting the
piR-31115/METTL3/YAP1/IGF2BP2 signalling pathway may offer a
promising strategy for inhibiting tumour angiogenesis in TNBC. By
interfering with this pathway, it is conceivable to suppress the
proliferation and migration of vascular endothelial cells, thus
impeding tumour growth and metastasis. This might potentially
result in an augmentation of the PFS of patients with TNBC, which
is of preponderant clinical importance. Future enquiries should
focus on validating these findings in vivo and exploring the
potential of devising novel therapies based on this mechanism.
Moreover, the improvement in PFS could potentially translate into
an enhanced quality of life for patients with TNBC, as it may defer
the recurrence of the disease and the exigency for further
aggressive treatments. Comprehending the role of this signalling
pathway in angiogenesis not only affords insights into the
pathophysiology of TNBC but also offers a glimmer of hope for more
efficacious therapeutic interventions in the future.
However, it is of utmost importance to note that the
current study is subject to certain limitations. Firstly, in our
experiments, the MDA-MB-231 cell line was solely utilized. Whilst
this cell line is widely employed in TNBC research, it is incapable
of fully representing the heterogeneity of TNBC or other breast
cancer subtypes. Different cell lines may display distinct genetic
profiles, signalling pathways and responses to diverse factors.
Consequently, the findings pertaining to the role of the
piR-31115/METTL3/YAP1/IGF2BP2 signalling pathway in tumour
angiogenesis may not be directly applicable to all TNBC cases or
other breast cancer types. Future studies ought to incorporate a
more extensive range of cell lines to augment the generalisability
of the results. Secondly, the present study was conducted entirely
in vitro. The in vitro environment lacks the
complexity and dynamic interactions that take place in vivo. In
vivo, tumours interact with surrounding stromal cells, immune
cells and the extracellular matrix, which can exert a significant
influence on angiogenesis and tumour progression. The absence of
these factors in our in vitro model may have given rise to
an incomplete comprehension of the physiological processes. As a
result, in vivo studies are indispensable for validating and
supplementing our in vitro findings and for providing a more
comprehensive understanding of the role of this signalling pathway
in TNBC angiogenesis.
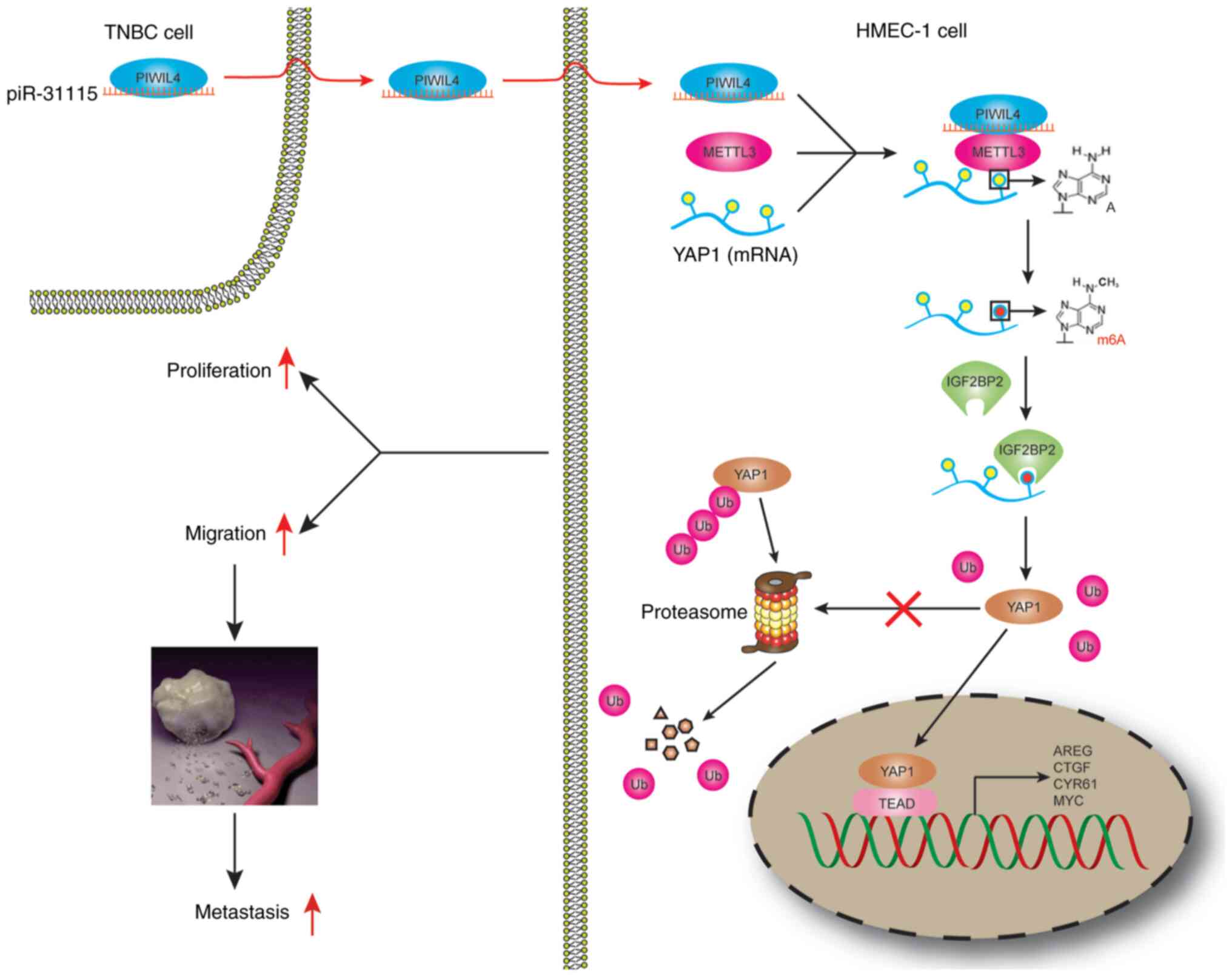
In conclusion, it was revealed that piR-31115
derived from MDA-MB-231 cells promotes HMEC-1 cells proliferation
and migration. The internalization of piR-31115 into HMEC-1 cells
augmented the m6A modification level of the METTL3-regulated gene
YAP1, which is recognized by IGF2BP2. The increased expression of
YAP1 ultimately led to alterations in the biological behaviour of
HMEC-1 cells (Fig. 7).
Acknowledgements
The authors are grateful to Dr Z. Wang (Zhongda
Hospital, Affiliated Hospital of Southeast University, Naning,
China), Dr XC. Sun (Jiangsu University, Zhenjing, China) and Dr H.
Yang (Tai'an City Central Hospital, Affiliated Hospital of Qingdao
University, Tai'an, China) for providing the cells.
Funding
The present study was supported by the Shandong Natural Science
Foundation of China (grant nos. ZR2021MH024 and ZR2023MC181), the
National Natural Science Foundation of China (grant no. 81602330),
the Zibo Municipal Medical and Health Technology Project (grant no.
20240309032) and the General Project of Nanjing Health Commission
(grant no. YKK20233).
Availability of data and materials
The data generated in the present study can be
requested from the corresponding author.
Authors' contributions
SMD carried out the investigation and developed
methodology. NL performed the formal analysis and wrote the initial
draft. WJX conceptualized, curated data and edited the manuscript.
KL provided resources, curated data, supervised the study,
developed methodology, obtained funding and reviewed the
manuscript. All authors read and approved the final version of the
manuscript. WJX and KL confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The current study was approved (approval no.
202102005) by the Ethics Committee of Zibo Central Hospital (Zibo,
China). Written informed consent for the utilization of tissues in
scientific research was duly obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lu B, Natarajan E, Balaji Raghavendran HR
and Markandan UD: Molecular classification, treatment, and genetic
biomarkers in triple-negative breast cancer: A review. Technol
Cancer Res Treat. 22:153303382211452462023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Geurts V and Kok M: Immunotherapy for
metastatic triple negative breast cancer: Current paradigm and
future approaches. Curr Treat Options Oncol. 24:628–643. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee J: Current treatment landscape for
early triple-negative breast cancer (TNBC). J Clin Med.
12:15242023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ribatti D, Nico B, Ruggieri S, Tamma R,
Simone G and Mangia A: Angiogenesis and antiangiogenesis in
triple-negative breast cancer. Transl Oncol. 9:453–457. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goggins E, Mironchik Y, Kakkad S, Jacob D,
Wildes F, Bhujwalla ZM and Krishnamachary B: Reprogramming of
VEGF-mediated extracellular matrix changes through autocrine
signaling. Cancer Biol Ther. 24:21841452023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Medina MA, Oza G, Sharma A, Arriaga LG,
Hernández Hernández JM, Rotello VM and Ramirez JT: Triple-negative
breast cancer: A review of conventional and advanced therapeutic
strategies. Int J Environ Res Public Health. 17:20782020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Y, Zhan Z, Yin X, Fu S and Deng X:
Targeted therapeutic strategies for triple-negative breast cancer.
Front Oncol. 11:7315352021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Alahdal M and Elkord E: Non-coding RNAs in
cancer immunotherapy: Predictive biomarkers and targets. Clin
Transl Med. 13:e14252023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Beňačka R, Szabóová D, Guľašová Z,
Hertelyová Z and Radoňak J: Non-coding RNAs in human cancer and
other diseases: Overview of the diagnostic potential. Int J Mol
Sci. 24:162132023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Isachesku E, Braicu C, Pirlog R,
Kocijancic A, Busuioc C, Pruteanu LL, Pandey DP and Berindan-Neagoe
I: The role of non-coding RNAs in epigenetic dysregulation in
glioblastoma development. Int J Mol Sci. 24:163202023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Girard A, Sachidanandam R, Hannon GJ and
Carmell MA: A germline-specific class of small RNAs binds mammalian
Piwi proteins. Nature. 442:199–202. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuramochi-Miyagawa S, Watanabe T, Gotoh K,
Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri
TW, et al: DNA methylation of retrotransposon genes is regulated by
Piwi family members MILI and MIWI2 in murine fetal testes. Genes
Dev. 22:908–917. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu Z, Yu X, Zhang S, He Y and Guo W: Novel
roles of PIWI proteins and PIWI-interacting RNAs in human health
and diseases. Cell Commun Signal. 21:3432023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Deng X, Liao T, Xie J, Kang D, He Y, Sun
Y, Wang Z, Jiang Y, Miao X, Yan Y, et al: The burgeoning importance
of PIWI-interacting RNAs in cancer progression. Sci China Life Sci.
67:653–662. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Garcia-Borja E, Siegl F, Mateu R, Slaby O,
Sedo A, Busek P and Sana J: Critical appraisal of the piRNA-PIWI
axis in cancer and cancer stem cells. Biomark Res. 12:152024.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng J, Deng H, Xiao B, Zhou H, Zhou F,
Shen Z and Guo J: piR-823, a novel non-coding small RNA,
demonstrates in vitro and in vivo tumor suppressive activity in
human gastric cancer cells. Cancer Lett. 315:12–17. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan H, Wu QL, Sun CY, Ai LS, Deng J, Zhang
L, Chen L, Chu ZB, Tang B, Wang K, et al: piRNA-823 contributes to
tumorigenesis by regulating de novo DNA methylation and
angiogenesis in multiple myeloma. Leukemia. 29:196–206. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yao J, Wang YW, Fang BB, Zhang SJ and
Cheng BL: piR-651 and its function in 95-D lung cancer cells.
Biomed Rep. 4:546–550. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Weng W, Liu N, Toiyama Y, Kusunoki M,
Nagasaka T, Fujiwara T, Wei Q, Qin H, Lin H, Ma Y and Goel A: Novel
evidence for a PIWI-interacting RNA (piRNA) as an oncogenic
mediator of disease progression, and a potential prognostic
biomarker in colorectal cancer. Mol Cancer. 17:162018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Du X, Li H, Xie X, Shi L, Wu F, Li G, Lai
C and Heng B: piRNA-31115 promotes cell proliferation and invasion
via PI3K/AKT pathway in clear cell renal carcinoma. Dis Markers.
2021:69153292021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Koduru SV, Tiwari AK, Leberfinger A,
Hazard SW, Kawasawa YI, Mahajan M and Ravnic DJ: A comprehensive
NGS data analysis of differentially regulated miRNAs, piRNAs,
lncRNAs and sn/snoRNAs in triple negative breast cancer. J Cancer.
8:578–596. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mao Z, Wang B, Zhang T and Cui B: The
roles of m6A methylation in cervical cancer: Functions, molecular
mechanisms, and clinical applications. Cell Death Dis. 14:7342023.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu ZM, Huo FC, Zhang J, Shan HJ and Pei
DS: Crosstalk between m6A modification and alternative splicing
during cancer progression. Clin Transl Med. 13:e14602023.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Z, Liu N, Shi S, Liu S and Lin H: The
role of PIWIL4, an argonaute family protein, in breast cancer. J
Biol Chem. 291:10646–10658. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhong Z, Jiao Z and Yu FX: The Hippo
signaling pathway in development and regeneration. Cell Rep.
43:1139262024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Duan M, Liu H, Xu S, Yang Z, Zhang F, Wang
G, Wang Y, Zhao S and Jiang X: IGF2BPs as novel m6A
readers: Diverse roles in regulating cancer cell biological
functions, hypoxia adaptation, metabolism, and immunosuppressive
tumor microenvironment. Genes Dis. 11:890–920. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cambria E, Coughlin MF, Floryan MA,
Offeddu GS, Shelton SE and Kamm RD: Linking cell mechanical memory
and cancer metastasis. Nat Rev Cancer. 24:216–228. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carrera-Aguado I, Marcos-Zazo L,
Carrancio-Salán P, Guerra-Paes E, Sánchez-Juanes F and Muñoz-Félix
JM: The inhibition of vessel co-option as an emerging strategy for
cancer therapy. Int J Mol Sci. 25:9212024. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Feng Y, Luo S, Fan D, Guo X and Ma S: The
role of vascular endothelial cells in tumor metastasis. Acta
Histochem. 125:1520702023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peng C, Xu Y, Wu J, Wu D, Zhou L and Xia
X: TME-related biomimetic strategies against cancer. Int J
Nanomedicine. 19:109–135. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zou Y, Yang A, Chen B, Deng X, Xie J, Dai
D, Zhang J, Tang H, Wu T, Zhou Z, et al: crVDAC3 alleviates
ferroptosis by impeding HSPB1 ubiquitination and confers
trastuzumab deruxtecan resistance in HER2-low breast cancer. Drug
Resist Updat. 77:1011262024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao Q, Qian L, Guo Y, Lü J, Li D, Xie H,
Wang Q, Ma W, Liu P, Liu Y, et al: IL11 signaling mediates piR-2158
suppression of cell stemness and angiogenesis in breast cancer.
Theranostics. 13:2337–2349. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cunha ERK, Ying W and Olefsky JM:
Exosome-mediated impact on systemic metabolism. Annu Rev Physiol.
86:225–253. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yao J, Chen Y and Lin Z: Exosomes:
Mediators in microenvironment of colorectal cancer. Int J Cancer.
153:904–917. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tang Q, Li L, Wang Y, Wu P, Hou X, Ouyang
J, Fan C, Li Z, Wang F, Guo C, et al: RNA modifications in cancer.
Br J Cancer. 129:204–221. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang L, Tian S, Zheng X, Zhang M, Zhou X,
Shang Y and Han Y: N6-methyladenosine RNA methylation in liver
diseases: From mechanism to treatment. J Gastroenterol. 58:718–733.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhu L, Zhang H, Zhang X and Xia L: RNA m6A
methylation regulators in sepsis. Mol Cell Biochem. 479:2165–2180.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Deng X, Qing Y, Horne D, Huang H and Chen
J: The roles and implications of RNA m6A modification in
cancer. Nat Rev Clin Oncol. 20:507–526. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ding SQ, Zhang XP, Pei JP, Bai X, Ma JJ,
Zhang CD and Dai DQ: Role of N6-methyladenosine RNA modification in
gastric cancer. Cell Death Discov. 9:2412023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu X, Xie X, Sui C, Liu X, Song M, Luo Q,
Zhan P, Feng J and Liu J: Unraveling the cross-talk between
N6-methyladenosine modification and non-coding RNAs in breast
cancer: Mechanisms and clinical implications. Int J Cancer.
154:1877–1889. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jin Q, Qu H and Quan C: New insights into
the regulation of METTL3 and its role in tumors. Cell Commun
Signal. 21:3342023. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li Y, Wang K, Liu W and Zhang Y: The
potential emerging role of piRNA/PIWI complex in virus infection.
Virus Genes. 60:333–346. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ni XF, Xie QQ, Zhao JM, Xu YJ, Ji M, Hu
WW, Wu J and Wu CP: The hepatic microenvironment promotes lung
adenocarcinoma cell proliferation, metastasis, and
epithelial-mesenchymal transition via METTL3-mediated
N6-methyladenosine modification of YAP1. Aging (Albany NY).
13:4357–4369. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ying Y, Ma X, Fang J, Chen S, Wang W, Li
J, Xie H, Wu J, Xie B, Liu B, et al: EGR2-mediated regulation of
m6A reader IGF2BP proteins drive RCC tumorigenesis and
metastasis via enhancing S1PR3 mRNA stabilization. Cell Death Dis.
12:7502021. View Article : Google Scholar : PubMed/NCBI
|