Introduction
Liver cancer is recognized as the sixth most
prevalent malignancy worldwide, and its incidence rate has been
rising in recent decades (1).
Statistical studies indicated that 905,677 new cases of liver
cancer and 830,180 fatal cases occurred in 2020 worldwide (2). Despite advancements in therapeutic
regimens, including antiangiogenic drugs, radiotherapy,
chemotherapy, immunotherapy and targeted therapy (3,4),
almost 80% of patients with advanced-stage liver cancer are
characterized by low response and high recurrence rates, limited
overall survival (OS) improvement, and a relative 5-year survival
rate of merely 18% (5,6). Hence, exploring the molecular
mechanisms underlying liver tumorigenesis is crucial for developing
effective treatment strategies.
Liver cancer occurs predominantly in individuals
infected with hepatitis B (HBV) and/or C viruses (7). The HBV DNA consists of four regions:
C, P, S and X, encoding HBcAg, hepatitis B virus DNA polymerase,
HBsAg and hepatitis B virus X protein (HBx), respectively (8,9). HBx,
an important small regulatory protein, has attracted researchers'
attention. It mainly mediates the transcriptional process by
binding to the HBV covalently-closed circular DNA and promotes HBV
replication (10). HBx also induces
the abnormal expression of proto-oncogenes or oncogenes, regulates
autophagy, facilitates cell proliferation, and accelerates the
migration and invasion of liver cancer, thereby promoting liver
cancer development (11–13). HBx modulates a series of oncogenes
that promote the initiation and development of liver cancer and are
closely associated with autophagy (14). Chronic HBV infection is one of the
main causative factors for liver cancer; however, the mechanism of
HBV-associated liver tumorigenesis is not fully understood and
deserves further exploration.
Protein regulator of cytokinesis 1 (PRC1) is
localized to the nucleus, cytoplasm and plasma membrane and is an
important factor for cytokinesis and cell cleavage (15). Studies have shown that PRC1 is
associated with the malignant occurrence and progression of various
tumors, including lung adenocarcinoma, ovarian cancer, liver
hepatocellular carcinoma (LIHC) and prostate cancer (16–19).
However, the role and functional mechanisms of PRC1 in
HBV-associated liver cancer remain unknown. An improved
understanding could contribute to the development of effective and
specific drugs. The present study aimed to explore the effect of
the PRC1 gene on autophagy function in human liver cancer cells and
elucidate whether it participates in the occurrence and development
of HBV-associated liver cancer in an autophagy-dependent
manner.
Materials and methods
Patients and samples
Tumor tissues were obtained from 86 patients with
liver cancer undergoing primary tumor resection at the Hongqi
Hospital Affiliated to Mudanjiang Medical University from June 2018
to April 2022. Among the 86 patients, 43 patients were
HBV-positive, and 43 patients were HBV-negative. The present study
was approved (approval no. 202052) by the Medical Ethics Committee
of Hongqi Hospital Affiliated to Mudanjiang Medical University
(Mudanjiang, China), and written informed consent was obtained from
each participant. Clinical characteristics (including sex and age
distribution) are included in Table
I.
 | Table I.Relationship between PRC1 protein
expression and clinicopathological characteristics of liver
cancer. |
Table I.
Relationship between PRC1 protein
expression and clinicopathological characteristics of liver
cancer.
|
| Expression level of
PRC1 |
|
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Low (n=44) (%) | High (n=42)
(%) | Chi-square | P-value |
|---|
| Age, years |
|
| 0.330 | 0.566 |
|
≤55 | 14 (31.8) | 11 (26.2) |
|
|
|
>55 | 30 (68.2) | 31 (73.8) |
|
|
| Sex |
|
| 0.361 | 0.548 |
|
Male | 31 (70.5) | 32 (76.2) |
|
|
|
Female | 13 (29.5) | 10 (23.8) |
|
|
| Tumor volume,
cm3 |
|
| 0.171 | 0.679 |
| ≤5 | 19 (43.2) | 20 (47.6) |
|
|
|
>5 | 25 (56.8) | 22 (52.4) |
|
|
| Pathological
stage |
|
| 4.872 | 0.027 |
|
I~II | 34 (77.3) | 23 (54.8) |
|
|
|
III~IV | 10 (22.7) | 19 (45.2) |
|
|
| Differentiation
grade |
|
| 4.769 | 0.029 |
|
High | 33 (75) | 22 (52.4) |
|
|
|
Low | 11 (25) | 20 (47.6) |
|
|
| Cirrhosis |
|
| 0.385 | 0.535 |
|
Yes | 27 (61.4) | 23 (54.8) |
|
|
| No | 17 (38.6) | 19 (45.2) |
|
|
| Lymph node
metastasis |
|
| 3.965 | 0.046 |
|
Yes | 10 (22.7) | 18 (42.9) |
|
|
| No | 34 (77.3) | 24 (57.1) |
|
|
| Alpha-fetoprotein,
µg/l |
|
| 5.18 | 0.023 |
|
≤400 | 36 (81.8) | 25 (59.5) |
|
|
|
>400 | 8 (18.2) | 17 (40.5) |
|
|
Bioinformatics analyses
PRC1 expression profiles in liver cancer were
compiled from The Cancer Genome Atlas (http://portal.gdc.cancer.gov) and the Gene Expression
Profiling Interactive Analysis (http://gepia.cancer-pku.cn/) databases. Based on the
Kaplan-Meier plotter (http://kmplot.com/analysis), the OS rate of patients
with liver cancer was determined.
Cell culture
The human liver cancer cell lines, Huh7
(HBV-negative) and HepG2 (HBV-negative), were purchased from
Chinese Academy of Sciences Cell Bank of Type Culture Collection.
Cells were cultured in Dulbecco's modified Eagle's medium (DMEM;
Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum
(FBS; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (100 IU/ml; Thermo Fisher Scientific, Inc.)
at 37°C in a humidified incubator under 5% CO2. The Huh7
and HepG2 cells underwent annual short tandem repeat (STR)
profiling analysis, population doubling time and morphology for
genetic confirmation. All experiments were performed with
early-passage cells (passages 3–10).
Transfection of small interfering RNA
(siRNA)
The siRNA oligonucleotides were obtained from
General Biology Co., Ltd. (www.generalbiol.com). The primer sequences of the
siRNAs were as follows: PRC1-Homo-1816 sense,
5′-GGAAAGCGCUGCAAUUAGATT-3′ and antisense,
5′-UCUAAUUGCAGCGCUUUCCTT-3′; negative siRNA sense,
5′-GUACCGCACGUCAUUCGUAUC-3′ and antisense,
5′-UACGAAUGACGUGCGGUACGU-3′. According to the manufacturer's
protocol, 50 nM of siRNAs were mixed with 125 µl of
Opti-MEMTM. Additionally, 5 µl of lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) was added to 125 µl of
Opti-MEMTM and incubated for 5 min at 25°C. The solution
containing siRNA was then mixed with solution containing
lipofectamine 2000 and incubated for 15 min at ambient temperature.
The siRNA/liposome 2000 complex was added to six-well plates at 250
µl per well and incubated for 48 h at 37°C. Cells were collected
for further analysis and the silencing efficiency of the PRC1 siRNA
vector have been confirmed (Fig.
S1).
Plasmids and recombinant
adenoviruses
PRC1 cDNA or HBx DNA fragments were subcloned into a
pcDNA3.1 mammalian expression vector (Invitrogen; Thermo Fisher
Scientific, Inc.). The primer sequences of the pcDNA3.1-PRC1
(pc-PRC1) plasmid were as follows: PRC1 sense,
5′-ACACTCTGTGCAGCGAGTTAC-3′ and antisense,
5′-TTCGCATCAATTCCACTTGGG-3′. The primers for the pcDNA3.1-HBx
(pc-HBx) plasmid were as follows: HBx sense,
5′-GGGACGTCCTTTGTCTACGT-3′ and antisense,
5′-GGGAGACCGCGTAAAGAGAG-3′. The reconstituted pc-HBx, pc-PRC1, and
empty [pc-negative control (NC)] plasmids were transfected into
Huh7 and HepG2 cells. Briefly, pc-HBx, pc-PRC1, or pc-NC plasmids
mixed with lipofectamine 2000 were incubated with cells for 6 h and
then cultured in DMEM containing 20% FBS for 48 h. Plasmids (2.5
µg/ml) were used for cellular viability, migration, invasion and
apoptosis assays. The overexpression efficiency of the pc-PRC1 and
pc-HBx plasmid was confirmed (Figs.
S2 and S3).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNAiso Plus (Takara Biotechnology Co., Ltd.) was
applied to extract total RNA from liver cancer tissues.
Subsequently, reverse transcription was performed using the
HiScript III qRT SuperMix kit (Vazyme Biotech Co. Ltd.) according
to the manufacturer's protocol. The relative level of PRC1 was
normalized to β-actin using the threshold cycle (2−ΔΔCT)
method (20). The specific primer
sequences were as follows: PRC1 sense, 5′-GCTGAGATTGTGCGGTTA-3′ and
antisense, 5′-GCCTTCAACTCTTCTTCCA-3′; β-actin sense,
5′-ACGTGGACATCCGCAAAGAC-3′ and antisense,
5′-CTGCTGTCACCTTCACCGTTC-3′. The following optimal thermal cycling
conditions were used: 95°C for 3 min, followed by 30 cycles of 95°C
for 1 min, 60°C for 1 min and 72°C for 1 min.
Western blot analysis
Huh7 and HepG2 cells were harvested and lysed in
RIPA buffer supplemented with 1 mM phenyl-methyl-sulfonyl fluoride
(Beyotime Institute of Biotechnology). Protein concentrations were
assessed using the bicinchoninic acid (BCA) method. An equal amount
of protein (20 µg) was separated using 12% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis, transferred to a
polyvinyl difluoride membrane (MilliporeSigma). After blocking with
5% skimmed milk for 2 h at room temperature, the membranes were
immunoblotted with primary antibodies specific to PRC1 (1:3,000;
cat. no. PA1-27244; Thermo Fisher Scientific, Inc.) and ATG5
(1:500; cat. no. NB110-53818; Novus Biologicals, LLC) overnight at
4°C. Afterward, the membranes were incubated with horseradish
peroxidase-conjugated secondary antibodies (1:1,000; cat. no.
A0216; Beyotime Institute of Biotechnology) for 1 h at room
temperature. Subsequently, an enhanced chemiluminescence western
detection system (Cell Signaling Technology, Inc.) was used to
detect band intensities, and the bands densities were analyzed with
ImageJ software 1.8.0 (National Institutes of Health).
Immunofluorescence analysis
Huh7 and HepG2 cells (5.0×106 cells/ml)
climbing slices were placed in a 6-well plate and fixed with 4%
paraformaldehyde for 30 min at room temperature, permeabilized, and
blocked with 0.1% Triton X-100 in 5% bovine serum albumin (Beijing
Solarbio Science & Technology Co., Ltd.) solution for 30 min.
The samples were treated with primary antibody against PRC1 (1:500;
cat. no. 15617-1-AP; ProteinTech Group, Inc.) or LC3B (1:500; cat.
no. 14600-1-AP; ProteinTech Group, Inc.) overnight at 4°C, followed
by incubation with FITC-conjugated secondary antibody (1:200; cat.
no. SA00003-2; ProteinTech Group, Inc.) for 1 h at ambient
temperature. Nuclei were counterstained with 100 µl
4′-6-diamidino-2-phenylindole (DAPI; 1 µg/ml; Beyotime Institute of
Biotechnology). Cells were visualized with a confocal laser
scanning microscope (Olympus Corporation).
Autophagy flux assay
An mCherry-GFP-LC3 reporter was used to observe
autophagosome formation and autophagic flux. Autophagosome
formation leads to an increase in yellow spots, while autolysosome
formation leads to an increase in red spots. Briefly, cells were
infected with adenovirus encoding mCherry-GFP-LC3 for 2 h and then
visualized with a confocal laser scanning microscope. The number of
LC3-positive puncta per cell in GFP-positive or
mCherry-GFP-positive cells was determined.
Electron microscopy
Huh7 and HepG2 cells were harvested, centrifuged in
1 mm3 blocks, and soaked in 2.5% glutaraldehyde solution
for 2 h. The pellets were post-fixed in 1% osmium tetroxide (Ted
Pella; www.tedpella.com) for 1 h, dehydrated in
graded alcohol solutions (50, 70, 90, 96 and 100%), and embedded in
Eponate 12 resin (Ted Pella). Ultrathin (70~80 nm) sections were
prepared and stained with 5% uranyl acetate (Ted Pella) and lead
citrate (Ted Pella) for 1 h at ambient temperature. The
ultrastructure of Huh7 and HepG2 cells was visualized using a
transmission electron microscope (JEM 1011; JEOL, Ltd.).
Cell viability assay
The Cell Counting Kit-8 (CCK-8; Dojindo
Laboratories, Inc.) assay was used to assess cell viability.
Briefly, Huh7 and HepG2 cells were transfected with 2.5 µg/ml
pc-PRC1 or pc-NC. After 48 h, the cells were plated into a 96-well
plate at 5,000 cells per well. After 48-h incubation, 100 µl of
culture medium supplemented with 10 µl CCK-8 solution was added
into each well and then incubated for 2 h at 37°C. The optical
density at 450 nm was detected using a BioTek Synergy H1
Microplate Reader (BioTek Instrument Inc.).
Cell apoptosis assay
Huh7 and HepG2 cells were transfected with 50 nM
PRC1 siRNA or negative control siRNA or exposed to 5 mM 3-MA for 48
h, and then transfected with 2.5 µg/ml pc-HBx for another 48 h.
Cells (5×105 cells/ml) were harvested and incubated with
5 µl of Annexin V and 5 µl of propidium iodide (PI) (Vazyme Biotech
Co., Ltd.) for 25 min in the dark. Then, apoptotic cells were
detected using a FacsCalibur flow cytometer (BD Biosciences), and
the results were analyzed using the software CellQuest version 3.3
(BD Biosciences).
Cell invasion assay
The invasion of cells was detected using Transwell
chambers (6-well plate, 8-µm pore size). The cell invasion chambers
were precoated by placing 100 µl Matrigel (1:5 dilution) onto the
filter, and incubating the filter for 30 min at 37°C to allow
Matrigel polymerization. Huh7 and HepG2 cells were transfected with
2.5 µg/ml pc-PRC1 or pc-NC. After 48 h, starved cells
(5×105 cells/ml) were seeded into the top chamber, while
500 µl of DMEM containing 20% FBS was filled into the lower
chamber. After 48 h, the invasive cells were stained with 0.1%
crystal violet for 20 min at ambient temperature. The invasive
ability of Huh7 and HepG2 cells was then determined under a light
microscope (magnification, ×200; Olympus Corporation).
Cell migration assay
The wound-healing assays was used to assess the
migratory ability of Huh7 and HepG2 cells. Cells were transfected
with 2.5 µg/ml pc-PRC1 or pc-NC for 48 h and then reseeded into
6-well plates. Once cells reached ~80-90% confluence, the monolayer
of cells was scratched in a straight line on the surface (at time
point 0). Subsequently, the cells were cultured in serum-free
medium in a 5% CO2 incubator at 37°C (21). The healing width of the wound was
observed at 48 h under a light microscope (magnification, ×200;
Olympus Corporation).
In vivo subcutaneous xenograft
assay
HepG2 cells were transfected with PRC1 siRNA (50 nM)
or negative control siRNA (50 nM) or exposed to the autophagosome
inhibitor 3-methyladenine (3-MA, 5 mM) for 48 h, and then
transfected with pc-HBx (2.5 µg/ml) for another 48 h. A total of 20
5-week-old male BALB/c-nu mice weighing 18–23 g (four mice per
group, Liaoning Changsheng biotechnology Co., Ltd.) were used. All
animals were housed in a specific pathogen-free animal facility and
maintained in a temperature and humidity-controlled environment
(~20–22°C, 40~60% humidity) with free access to food and water in a
lighting cycle of 12 h darkness/12 h light. A total of 100 µl
sterile PBS containing 1×107 HepG2 cells were processed
and then subcutaneously injected into the right flank of the mice.
Xenograft tumor growth was recorded every 2 days; when the tumor
volume reached 100~150 mm3, the mice were humanely
sacrificed by intraperitoneal injection of pentobarbital (100
mg/kg). Tumor tissues were collected and weighed.
Paraformaldehyde-fixed and paraffin-embedded tumor tissues were cut
into sections for immunohistochemistry (IHC). The animal care
committee at Mudanjiang Medical University (Mudanjiang, China)
approved the present study (approval no. 2022-500-177). All animals
received human care, and the study protocols complied with the
institution's guidelines.
IHC
The liver cancer tissues were fixed in 4%
paraformaldehyde and embedded in paraffin, and then processed and
sectioned. IHC was conducted by Wuhan Servicebio Technology Co.,
Ltd. Primary monoclonal antibodies against PRC1 (1:500) were added
to the paraffinized slides, probed with HRP-labeled antibodies
(1:50; cat. no. A0216; Beyotime Institute of Biotechnology), and
visualized with 3, 3′-diaminobenzidine. Subsequently, the cell
nuclei were stained with blue hematoxylin for 5 min at ambient
temperature for histochemical assessment. The expression level of
PRC1 was calculated based on the staining intensity score (strong
staining: 3; moderate staining: 2; weak staining: 1) multiplied by
a score of the percentage of positive cells (4, >75%; 3, 51~75%;
2, 25~50%; 1, <25%), resulting in an IHC score of between 1 and
12. As outlined in Table I, an IHC
score ≥6 indicated a high expression of PRC1, whereas a score <6
was considered a low expression of PRC1.
Paraffin-embedded slides of subcutaneous tumors from
xenograft mice were treated with ATG5 (1:400; cat. no. NB110-53818;
Novus Biologicals, LLC), Bax (1:500; cat. no. 3351; ProSci;
www.prosci-inc.com), and Bcl-2 (1:1,000;
cat. no. NB100-56098; Novus Biologicals, LLC) antibodies overnight
at 4°C. A light microscope (Carl Zeiss AG) was used to observe and
acquire images of the stained tissues. The H-score in the
measurement area was calculated as follows: H-Score=(percentage of
strong-intensity area ×3) + (percentage of moderate-intensity area
×2) + (percentage of weak-intensity area ×1).
Terminal deoxynucleotidyl transferase
dUTP nick end labeling (TUNEL) analysis
The apoptotic rate in the xenograft tumor tissues
was evaluated using a TUNEL assay kit (cat. no. 11684795910; Roche
Diagnostics GmbH) following the manufacturer's protocols.
Paraffin-embedded slides of subcutaneous tumors were treated with
4% H2O2 for 25 min and then incubated with 50
µl of TUNEL reaction solution and 50 µl of peroxidase-conjugated
antibodies for another 25 min. The slides were reacted with 50 µl
of 0.05% diaminobenzidine solution, and the apoptotic rate was
observed using inverted fluorescence microscope (Olympus
Corporation).
Enzyme-linked immunosorbent assay
(ELISA)
The levels of Th1 cytokines tumor necrosis factor-α
(TNF-α; cat. no. MTA00B), interleukin (IL)-2 (cat. no. QK202),
interferon-γ (IFN-γ; cat. no. QK285) and Th2 cytokines IL-6 (cat.
no. D6050B), IL-10 (cat. no. M1000B) and IL-4 (cat. no. M4000B) in
mice serum were detected using ELISA kits (R&D Systems, Inc.)
according to the manufacturer's protocols.
Statistical analysis
Statistical analysis was performed using SPSS
software version 13.0 (SPSS Inc.). Quantitative results are
presented as the mean ± standard deviation. Unpaired Student's
t-test was used to analyze the differences between two groups, and
one-way analysis of variance was used to compare more than two
groups, and Tukey's post-hoc test was employed for further
statistical assessment. In Table I,
the Chi-square test was performed to analyze clinical data.
P<0.05 was considered to indicate a statistically significant
difference.
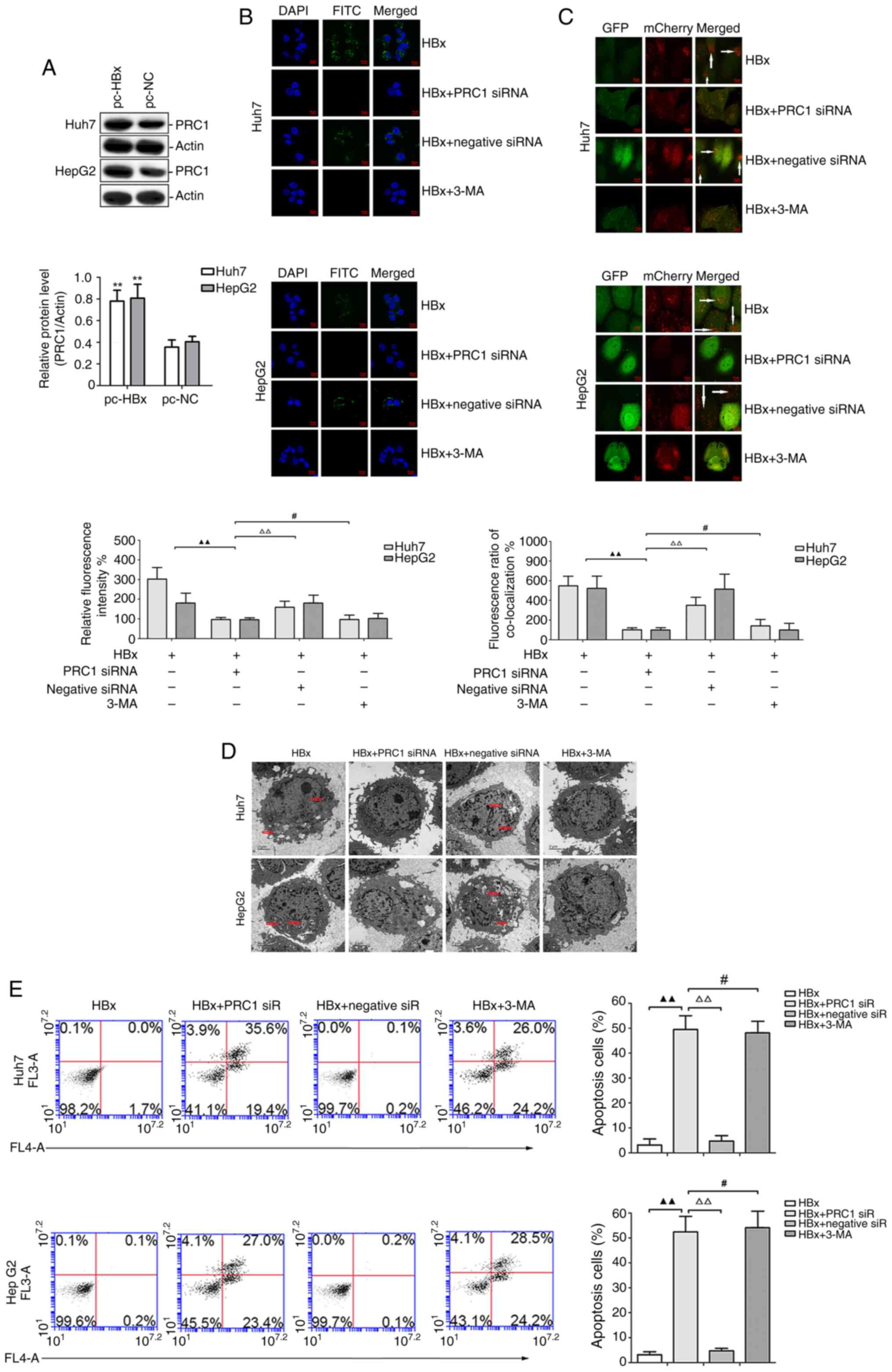
Results
Characteristics of the PRC1 gene in
liver cancer tissues
The expression of the PRC1 gene was evaluated
in human pan-cancer using dot plot analysis (Fig. 1A). The results indicated that
compared with adjacent normal tissues, the PRC1 gene was highly
expressed in most tumor tissues, including bladder urothelial
carcinoma, breast invasive carcinoma, head and neck squamous cell
carcinoma, cervical squamous cell carcinoma, LIHC and lung squamous
cell carcinoma. Box plots revealed that PRC1 gene expression in
liver cancer tissues (369 cases) was significantly higher than that
in normal liver tissue (160 cases, Fig.
1B). Pathological stage plot indicated an association between
PRC1 mRNA expression and the tumor pathological stage (Pr
(>F)=0.000142; Fig. 1C). The
association between PRC1 gene expression and OS in patients with
liver cancer (364 cases) was analyzed using the Kaplan-Meier
plotter. High PRC1 gene expression was associated with shorter OS
in patients with liver cancer (Log rank P=0.00038; Fig. 1D). Moreover, Kaplan-Meier plots
demonstrated a PRC1 gene hazard ratio (HR) of 1.9 (where factors
with an HR >1 are considered a risk factor for the disease),
supporting its importance as a hazard factor for liver cancer. IHC
and RT-qPCR indicated that PRC1 expression in HBV-positive tumor
tissues was higher than in HBV-negative tumor tissues (Fig. 1E and F). An association between PRC1
expression level and baseline demographics of patients with liver
cancer is presented in Table I. The
chi-square test indicated that high PRC1 expression levels were
significantly associated with pathological stage III–IV, low
differentiation, lymph node metastasis and alpha-fetoprotein (AFP)
>400 µg/l.
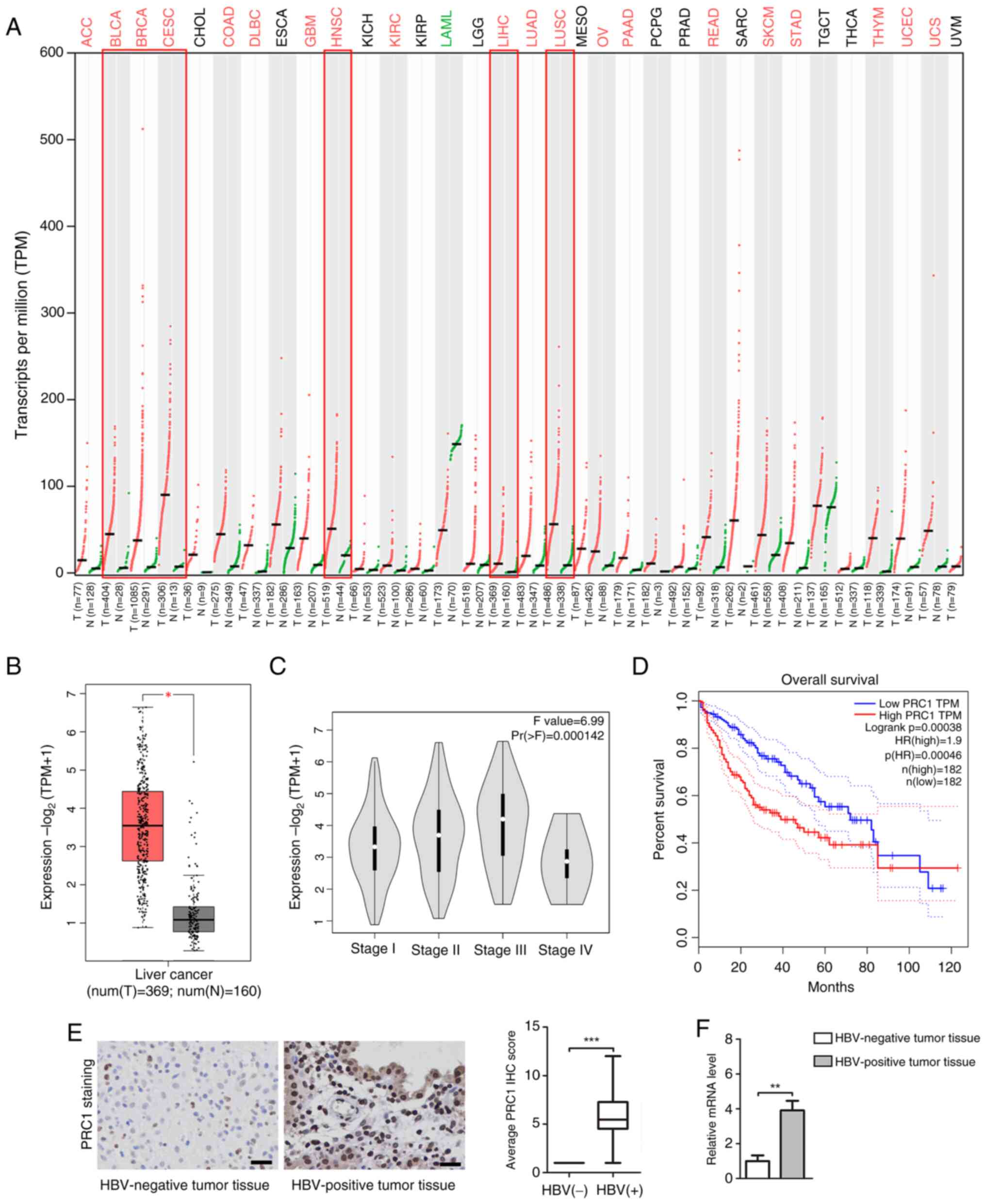 | Figure 1.Characteristics of the PRC1 gene in
liver cancer tissues. (A) The expression of the PRC1 gene was
evaluated in human pan-cancer using dot plot analysis. (B) PRC1
gene expression in box plot, including 369 cases of liver cancer
tissues and 160 cases of normal tissues. Data were acquired from
the Genotype-Tissue Expression project and are shown as the mean ±
quartile values. *P<0.05 by the unpaired Student's t-test. (C)
Relationship between PRC1 gene expression and tumor pathological
stage was presented in pathological stage plot. (D) The association
between PRC1 gene expression and overall survival in patients with
liver cancer (364 cases) was analyzed using the Kaplan-Meier
plotter. (E) The expression of PRC1 protein in HBV-positive tumor
tissues and HBV-negative tumor tissues was measured using
immunohistochemical staining (magnification, ×400). Brown indicates
positive staining (Scale bars, 50 µm). (F) The mRNA level of PRC1
was detected using the reverse transcription-quantitative PCR
assay. **P<0.01 and ***P<0.001 vs. the HBV-negative tumor
tissues. PRC1, protein regulator of cytokinesis 1; HBV, hepatitis B
virus; ACC, adenoid cystic carcinoma; BLCA, bladder urothelial
carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous
cell carcinoma and endocervical adenocarcinoma; CHOL,
cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, diffuse large
B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma
multiforme; HNSC, head and neck squamous cell carcinoma; KICH,
kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP,
kidney renal papillary cell carcinoma; LGG, low-grade glioma; LIHC,
liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC,
Lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian
serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG,
pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma;
READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous
melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell
tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine
corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM,
uveal melanoma. |
PRC1 gene enhances cell autophagy and
induces malignant phenotypes in vitro
To identify the role of the PRC1 gene in
liver cancer development, the effect of its upregulation on
autophagy was assessed in Huh7 and HepG2 cells. The
autophagy-associated ATG5 protein level was determined using
western blot analysis (Fig. 2A).
The present findings demonstrated that the pc-PRC1 treatment
significantly increased the level of ATG5 relative to the mock
treatment (P<0.01). Further examination using a plasmid encoding
mCherry-GFP-LC3B identified higher autophagic flux (the rate of red
puncta formation in each cell) in the cytoplasm of Huh7 and HepG2
cells treated with pc-PRC1 but low (basal) autophagic flux in
mock-treated cells or cells treated with pc-NC (Fig. 2B). The viability of Huh7 and HepG2
cells transfected with pc-PRC1 was significantly higher than in the
mock treatment (P<0.01; Fig.
2C). Moreover, the Transwell and scratch tests showed that
compared with mock treatment, the pc-PRC1 treatment significantly
increased the number of invasive cells and enhanced the migration
distance (P<0.01; Fig. 2D and
E).
Effect of the PRC1 gene on autophagy
and apoptosis in HBx-treated Huh7 and HepG2 cells
To evaluate whether the PRC1 gene plays an important
role in HBV-positive liver cancer, the effect of pc-HBx treatment
was first assessed on PRC1 protein expression in Huh7 and HepG2
cells. As shown in Fig. 3A, PRC1
protein expression in the pc-HBx treatment group was significantly
higher than in the pc-NC group (P<0.01). Furthermore, the
FITC-labelled PRC1 protein was primarily located in the cytoplasm
of Huh7 and HepG2 cells in the HBx and HBx + negative siRNA
treatments, while it rarely appeared in cells treated with HBx +
PRC1 siRNA or HBx + 3-MA (Fig. 3B).
To further assess the role of the PRC1 gene in HBx-treated Huh7 and
HepG2 cells, the autophagic flux was detected using the
mCherry-GFP-LC3B plasmid. The data revealed that the HBx and HBx +
negative siRNA treatments potently enhanced the autophagic flux,
while autophagic flux was rarely observed in the HBx + PRC1 siRNA
and HBx + 3-MA treatments (Fig.
3C). Consistent with these results, electron microscopy
demonstrated autophagosomes in the HBx and HBx + negative siRNA
treatments. By contrast, the HBx + PRC1 siRNA and HBx + 3-MA
treatments eliminated these adverse effects and restored normal
mitochondrial morphology (Fig. 3D).
Finally, cell apoptotic level was measured by annexin V-fluorescein
isothiocyanate/PI staining. As shown in Fig. 3E, the cell apoptotic rate in the HBx
+ PRC1 siRNA group was significantly higher than in the HBx and HBx
+ negative siRNA groups (P<0.01). However, the HBx + PRC1 siRNA
and HBx + 3-MA groups had similar cell apoptotic rates.
Effect of the PRC1 gene on autophagy
and apoptosis in an HBx-treated murine xenograft model
HepG2 cells overexpressing the HBx gene or with
silenced PRC1 gene expression were subcutaneously injected into
nude mice to assess the role of PRC1 in liver cancer in
vivo. TUNEL assays indicated that the apoptotic rate in the HBx
+ PRC1 siRNA group was significantly higher than in the HBx and HBx
+ negative siRNA groups (P<0.01). However, similar apoptotic
rates were observed in the HBx + PRC1 siRNA or HBx + 3-MA groups
(P>0.05; Fig. 4A). IHC assays
demonstrated that the expression of ATG5 and Bcl-2 after the HBx +
PRC1 siRNA treatment was significantly lower than compared with HBx
and HBx + negative siRNA treatments (P<0.01). However, the HBx +
PRC1 siRNA and HBx + 3-MA treatment groups had similar ATG5 and
Bcl-2 expression levels (P>0.05). Bax protein levels in the HBx
+ PRC1 siRNA treatment group were significantly higher than those
in the HBx and HBx + negative siRNA treatment groups (P<0.01).
However, the HBx + PRC1 siRNA and HBx + 3-MA groups had similar Bax
expression levels (P>0.05; Fig.
4B).
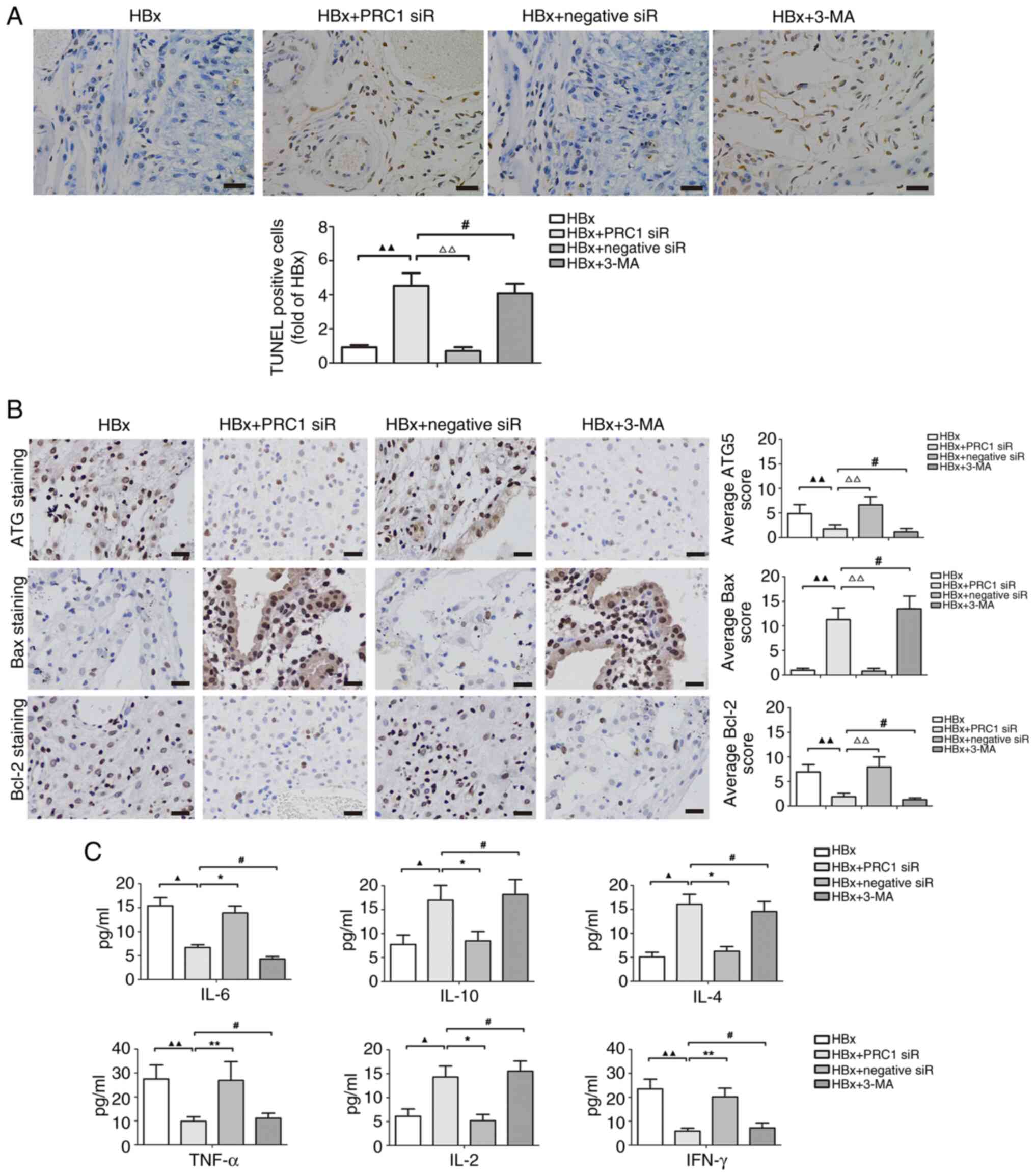 | Figure 4.Effect of the PRC1 gene on autophagy
and apoptosis in an HBx-treated murine xenograft model. (A)
Apoptosis of tumor tissues was measured by TUNEL assay. The
apoptotic cells are stained brown. Scale bars, 50 µm. (B) The
expression of autophagy-related protein ATG5 and apoptosis-related
proteins Bax, Bcl-2 in tumor tissues was observed using
immunohistochemical staining. Brown indicates positive staining.
Scale bars, 50 µm. ▲▲P<0.01 vs. HBx treatment;
#P>0.05 and ΔΔP<0.01 vs. HBx and PRC1
siRNA co-treatment. (C) The levels of Th1 cytokines (TNF-α, IL-2
and IFN-γ) and Th2 cytokines (IL-6, IL-10 and IL-4) in the mice
serum were detected by ELISA. ▲P<0.05 and
▲▲P<0.01 vs. HBx treatment; #P>0.05,
*P<0.05 and **P<0.01 vs. HBx and PRC1 siRNA co-treatment.
PRC1, protein regulator of cytokinesis 1; HBx, hepatitis B protein
x; siRNA, small interfering RNA; TNF-α, tumour necrosis factor-α;
IL, interleukin; IFN-γ, interferon-γ; 3-MA, 3-methyladenine. |
Cytokines are an important factor in inflammatory
responses during liver tumorigenesis. Treatment with HBx + PRC1
siRNA significantly induced the levels of IL-10, IL-4 and IL-2 and
suppressed the levels of IL-6, TNF-α and IFN-γ relative to
treatment with HBx or HBx + negative siRNA (all P<0.05).
However, the HBx + PRC1 siRNA and HBx + 3-MA treatment groups had
similar levels of the Th1 and Th2 cytokines (P>0.05; Fig. 4C). These results suggest that the
PRC1 gene regulates the release of cytokines in HBx-associated
liver cancer.
Discussion
There is a significant association between the
sustained replication of HBV and the malignant phenotypes of liver
cancer in humans (14). The present
study explored the role of the PRC1 gene in the occurrence and
development of HBV-associated liver cancer. Exploring target
molecules that simultaneously regulate HBV and liver cancer could
be a feasible strategy to overcome these two closely related
diseases (22). In summary, the
findings of the present study indicate that PRC1 gene expression
sensitively responds to HBV infection and hence serves as an ideal
prospective target for liver cancer treatment.
HBV is a major risk factor for liver tumorigenesis
and is closely related to the survival and prognosis of patients
with liver cancer. HBV-associated liver cancer accounts for ~50~60%
of liver cancer cases (23,24). Numerous HBV genotypes have exhibited
varying degrees of autophagic activity augmentation (25). It has been reported that HBV
activates autophagy mainly in an HBx protein-dependent manner
(26). HBx is a multifunctional
regulatory protein involved in regulating viral replication and
cellular functions, such as cell proliferation, apoptosis,
differentiation, transformation, autophagy and DNA repair (10,27).
Autophagy is a crucial homeostatic pathway that promotes the
degradation and recycling of cellular substances (28). However, autophagy has dual functions
in cancer, in the tumor cells (intrinsic) and in the surrounding
matrix (extrinsic), depending on the tumor staging, specific
oncogenic mutations and the surrounding microenvironment (29). It was previously indicated that
autophagy plays a suppressive role in liver cancer development. The
deletion of the ATG5 or liver-specific ATG7 gene in mice resulted
in the development of liver tumors (30). Furthermore, expression of the
autophagy-related Beclin 1 protein in liver tumors was lower than
in adjacent normal tissues (31).
However, Qian et al (32)
pointed out that autophagy shifted from a suppressive role to a
promoter role in the late stage of liver cancer by suppressing the
expression of p53 and p21. The current study demonstrated the
mechanism by which autophagy affects HBV-related tumorigenesis and
its significance for potential treatments.
Autophagy maintains cell metabolism and enhances the
resistance to hypoxia and nutrient deprivation. By alleviating
cellular stress, autophagy allows cancer cells to survive through
therapy or, occasionally, enter a dormant state (33). Activation of the
epithelial-mesenchymal transition-related transcription factors
Slug and Snail causes the acquisition of a cancer-initiating cells
phenotype and activates autophagy (34). Subsequent studies indicated that
autophagy activation in the liver causes tissue destruction and
regeneration cycles, resulting in the emergence of progenitor cells
from hepatocytes, thereby driving the early stages of liver
tumorigenesis (35). Consistent
with these studies, western blot and immunofluorescence analysis in
present study revealed that HBx induced cell autophagy in liver
cancer cells (Figs. 3 and S4). These results signified that
autophagy activation might be essential to the occurrence of
HBV-associated liver cancer. The present findings also demonstrated
that HBx enhanced the expression of PRC1 protein. PRC1 is a
substrate of cyclin-dependent kinases and an exhibitor of E3
ubiquitin ligase activity, which participates in cell cycle
regulation (36). It was found that
deubiquitinating enzymes could reverse the process of protein
ubiquitin degradation, affecting the occurrence and prognosis of
liver cancer through tumor signaling pathways, apoptosis, autophagy
and cell cycle regulation (37).
However, the effect of PRC1 on autophagy in HBV-associated liver
cancer formation remained unclear. It was concluded that the PRC1
gene participates in HBV-associated liver cancer development by
regulating autophagy. The present study found that PRC1 gene
overexpression significantly upregulated the expression of the
autophagy-related protein ATG5 and increased the autophagic flux in
the cytoplasm of Huh7 and HepG2 cells. In vitro functional
and in vivo animal model studies revealed that silencing of
the PRC1 gene or using the autophagosome inhibitor 3-MA reversed
HBx-induced autophagy, resulting in rarely observed autophagic flux
and autophagosomes, and promotion of cell apoptosis. The present
findings indicate that the PRC1 gene plays an oncogenic role in
HBx-associated HCC, and HBX affects autophagy in HCC through PRC1
(Figs. 3 and S5). The PRC1 gene could be a prognostic
marker for HBV-associated HCC.
It was reported that PRC1 expression in lung cancer
tissues was higher than in normal tissues (38). Furthermore, silencing the PRC1 gene
in lung cancer cells suppressed their proliferating, migratory and
invasive capabilities. Silencing the PRC1 gene in liver cancer
significantly induced apoptosis of Hep3B and Huh7 cells (39). The current data revealed that PRC1
was overexpressed in HBV-positive liver cancer tissues and
HBx-treated cell lines but was downregulated in HBV-negative liver
cancer tissues because HBx was absent. High expression levels of
PRC1 were associated with pathological stage III~IV, low
differentiation, lymph node metastasis and AFP >400 µg/l. In
vivo animal model experiments indicated that the HBx treatment
significantly decreased IL-10, IL-4 and IL-2 production and
increased IL-6, TNF-α and IFN-γ generation; silencing the PRC1 gene
reversed this effect in an autophagy-dependent manner. These
findings indicate that the PRC1 gene plays an oncogenic role in
HBx-associated liver cancer. It might be possible to treat
HBx-associated liver cancer by inhibiting the expression of the
PRC1 gene. The PRC1 gene could be a prognostic marker for
HBV-associated liver cancer.
In conclusion, a previously unknown relationship
between autophagy and the PRC1 gene was demonstrated in
HBV-associated liver cancer. HBx induced the expression of the PRC1
gene, a crucial carcinogenic factor that facilitates cell
proliferation, migration and invasion in an autophagy-dependent
manner. Therefore, PRC1 could serve as an important prognostic
biomarker for patients with liver cancer. Manipulation of
PRC1-regulated autophagy could be a potential clinical therapeutic
approach for HBV-associated liver cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the basic scientific research
projects of provincial higher education institutions in
Heilongjiang (grant no. 2020-KYYWFMY-0011).
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article.
Authors' contributions
JJH, XZC, CW and FYG made substantial contributions
to the conception. FYG made substantial contributions to the study
design. JJH and XZC participated in the laboratory work and data
analyses. JJH and FYG confirm the authenticity of all the raw data.
All authors participated in drafting or revising the content for
important intellectual content, and read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Human studies were approved (approval no. 202052) by
the Medical Ethics Committee of Hongqi Hospital Affiliated to
Mudanjiang Medical University (Mudanjiang, China), and written
informed consent was obtained from each participant. Animal
experiments were approved (approval no. 2022-500-177) by the animal
care committee at Mudanjiang Medical University (Mudanjiang,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HBV
|
hepatitis B virus
|
|
HBx
|
hepatitis B protein x
|
|
OS
|
overall survival
|
|
PRC1
|
protein regulator of cytokinesis 1
|
|
RT-qPCR
|
reverse transcription quantitative
polymerase chain reaction
|
|
DAPI
|
4′-6-diamidino-2-phenylindole
|
|
CCK-8
|
Cell Counting Kit-8
|
|
PI
|
propidium iodide
|
|
3-MA
|
3-methyladenine
|
|
IHC
|
immunohistochemistry
|
|
TUNEL
|
terminal deoxynucleotidyl
transferase-mediated nick-end tagging of UTP
|
|
ELISA
|
enzyme-linked immunosorbent assay
|
|
TNF-α
|
tumour necrosis factor-α
|
|
IL
|
interleukin
|
|
IFN-γ
|
interferon-γ
|
References
|
1
|
Brown ZJ, Tsilimigras DI, Ruff SM, Mohseni
A, Kamel IR, Cloyd JM and Pawlik TM: Management of hepatocellular
carcinoma: A review. JAMA Surg. 158:410–420. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Peng Z, Fan W, Zhu B, Wang G, Sun J, Xiao
C, Huang F, Tang R, Cheng Y, Huang Z, et al: Lenvatinib combined
with transarterial chemoembolization as First-line treatment for
advanced hepatocellular carcinoma: A phase III, randomized clinical
trial (LAUNCH). J Clin Oncol. 41:117–127. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang C, Zhang H, Zhang L, Zhu AX, Bernards
R, Qin W and Wang C: Evolving therapeutic landscape of advanced
hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol.
20:203–222. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ajoolabady A, Tang D, Kroemer G and Ren J:
Ferroptosis in hepatocellular carcinoma: Mechanisms and targeted
therapy. Br J Cancer. 128:190–205. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhan X, Wu R, Kong XH, You Y, He K, Sun
XY, Huang Y, Chen WX and Duan L: Elevated neutrophil extracellular
traps by HBV-mediated S100A9-TLR4/RAGE-ROS cascade facilitate the
growth and metastasis of hepatocellular carcinoma. Cancer Commun
(Lond). 43:225–245. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shen C, Jiang X, Li M and Luo Y: Hepatitis
virus and hepatocellular carcinoma: Recent advances. Cancers
(Basel). 15:5332023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Costante F, Stella L, Santopaolo F,
Gasbarrini A, Pompili M, Asselah T and Ponziani FR: molecular and
clinical features of hepatocellular carcinoma in patients with
HBV-HDV Infection. J Hepatocell Carcinoma. 10:713–724. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou Q, Li L, Sha F, Lei Y, Tian X, Chen
L, Chen Y, Liu H and Guo Y: PTTG1 reprograms asparagine metabolism
to promote hepatocellular carcinoma progression. Cancer Res.
83:2372–2386. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Z, Li N, Cai P, Zhang C, Cao G and
Yin J: Mechanism of HBx carcinogenesis interaction with non-coding
RNA in hepatocellular carcinoma. Front Oncol. 13:12491982023.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li Y, Gan L, Lu M, Zhang X, Tong X, Qi D,
Zhao Y and Ye X: HBx downregulated decorin and decorin-derived
peptides inhibit the proliferation and tumorigenicity of
hepatocellular carcinoma cells. FASEB J. 37:e228712023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu X, Ni Z, Song T, Lv W, Chen Y, Huang D,
Xie Y, Huang W and Niu Y: C-Terminal truncated HBx facilitates
oncogenesis by modulating cell cycle and glucose metabolism in
FXR-deficient hepatocellular carcinoma. Int J Mol Sci. 24:51742023.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
You H, Zhang N, Yu T, Ma L, Li Q, Wang X,
Yuan D, Kong D, Liu X, Hu W, et al: Hepatitis B virus X protein
promotes MAN1B1 expression by enhancing stability of GRP78 via
TRIM25 to facilitate hepatocarcinogenesis. Br J Cancer.
128:992–1004. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dobrinić P, Szczurek AT and Klose RJ: PRC1
drives Polycomb-mediated gene repression by controlling
transcription initiation and burst frequency. Nat Struct Mol Biol.
28:811–824. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu P, Cui N, Song ZY, Yong WX, Luo XX,
Wang GC, Wang X, Wu YN, Xu Q, Zhang LM, et al: PRC1 plays an
important role in lung adenocarcinoma and is potentially targeted
by fostamatinib. Eur Rev Med Pharmacol Sci. 26:8924–8934.
2022.PubMed/NCBI
|
|
17
|
Bu H, Li Y, Jin C, Yu H, Wang X, Chen J,
Wang Y, Ma Y, Zhang Y and Kong B: Overexpression of PRC1 indicates
a poor prognosis in ovarian cancer. Int J Oncol. 56:685–696.
2020.PubMed/NCBI
|
|
18
|
Huang R, Liu J, Li H, Zheng L, Jin H,
Zhang Y, Ma W, Su J, Wang M and Yang K: Identification of hub genes
and their correlation with immune infiltration cells in
hepatocellular carcinoma based on GEO and TCGA databases. Front
Genet. 12:6473532021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Luo HW, Chen QB, Wan YP, Chen GX, Zhuo YJ,
Cai ZD, Luo Z, Han ZD, Liang YX and Zhong WD: Protein regulator of
cytokinesis 1 overexpression predicts biochemical recurrence in men
with prostate cancer. Biomed Pharmacother. 78:116–120. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li M, Xiao Y, Liu P, Wei L, Zhang T, Xiang
Z, Liu X, Zhang K, Zhong Q and Chen F: 4–Methoxydalbergione
inhibits esophageal carcinoma cell proliferation and migration by
inactivating NF–κB. Oncol Rep. 49:422023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Y, Liu X, Luo M, Li Y and Li H: HBx
modulates drug resistance of Sorafenib-resistant hepatocellular
carcinoma cells. Discov Med. 35:1035–1042. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu J, Li W, Hou GJ, Sun DP, Yang Y, Yuan
SX, Dai ZH, Yin HZ, Sun SH, Huang G, et al: Circular RNA cFAM210A,
degradable by HBx, inhibits HCC tumorigenesis by suppressing YBX1
transactivation. Exp Mol Med. 55:2390–2401. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He L, Shen H, Deng H, Zhang X, Xu Y, Shi C
and Ouyang Z: Identification of critical residues in the regulatory
protein HBx for Smc5/6 interaction and hepatitis B virus
production. Antiviral Res. 211:1055192023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lei Y, Xu X, Liu H, Chen L, Zhou H, Jiang
J, Yang Y and Wu B: HBx induces hepatocellular carcinogenesis
through ARRB1-mediated autophagy to drive the G1/S cycle.
Autophagy. 17:4423–4441. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Peantum J, Kunanopparat A, Hirankarn N,
Tangkijvanich P and Kimkong I: Autophagy Related-protein 16-1
Up-regulated in hepatitis B Virus-related hepatocellular carcinoma
and impaired apoptosis. Gastroenterology Res. 11:404–410. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin X, Li AM, Li YH, Luo RC, Zou YJ, Liu
YY, Liu C, Xie YY, Zuo S, Liu Z, et al: Silencing MYH9 blocks
HBx-induced GSK3β ubiquitination and degradation to inhibit tumor
stemness in hepatocellular carcinoma. Signal Transduct Target Ther.
5:132020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kimkong I and Kunanopparat A: Autophagy
related protein 9A increase in hepatitis B virus-associated
hepatocellular carcinoma and the role in apoptosis. World J
Hepatol. 12:1367–1371. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Debnath J, Gammoh N and Ryan KM: Autophagy
and autophagy-related pathways in cancer. Nat Rev Mol Cell Bio.
24:560–575l. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gong K, Chen C, Zhan Y, Chen Y, Huang Z
and Li W: Autophagy-related gene 7 (ATG7) and reactive oxygen
species/extracellular signal-regulated kinase regulate
tetrandrine-induced autophagy in human hepatocellular carcinoma. J
Biol Chem. 287:35576–35588. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun H, Yu J, Wen Z, Wang M and Chen W:
Decreased expression of Beclin-1 in patients with hepatocellular
carcinoma. J BUON. 24:634–641. 2019.PubMed/NCBI
|
|
32
|
Qian ZM, Chen YJ and Bao YX:
Pharmacological mechanisms of norcantharidin against hepatocellular
carcinoma. Am J Cancer Res. 13:5024–5038. 2023.PubMed/NCBI
|
|
33
|
Vargas JNS, Hamasaki M, Kawabata T, Youle
RJ and Yoshimori T: The mechanisms and roles of selective autophagy
in mammals. Nat Rev Mol Cell Biol. 24:167–185. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yu JL and Gao X: MicroRNA 1301 inhibits
cisplatin resistance in human ovarian cancer cells by regulating
EMT and autophagy. Eur Rev Med Pharmacol Sci. 24:1688–1696.
2020.PubMed/NCBI
|
|
35
|
Kudo Y, Sugimoto M, Arias E, Kasashima H,
Cordes T, Linares JF, Duran A, Nakanishi Y, Nakanishi N, L'Hermitte
A, et al: PKCλ/ι loss induces autophagy, oxidative phosphorylation,
and NRF2 to promote liver cancer progression. Cancer Cell.
38:247–262. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Perchey RT, Serres MP, Nowosad A, Creff J,
Callot C, Gay A, Manenti S, Margolis RL, Hatzoglou A and Besson A:
p27Kip1 regulates the microtubule bundling activity of PRC1.
Biochim Biophys Acta Mol Cell Res. 1865:1630–1639. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang J, Zhan Y, Jiang L, Gao Y, Zhao B,
Zhang Y, Zhang W, Zheng J and Yu J: Identification of the potential
prognosis biomarkers in hepatocellular carcinoma: An analysis based
on WGCNA and PPI. Int J Gen Med. 14:9555–9565. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shi J, Hao S, Liu X, Li Y and Zheng X:
Feiyiliu Mixture sensitizes EGFRDel19/T790M/C797S mutant non-small
cell lung cancer to osimertinib by attenuating the PRC1/Wnt/EGFR
pathway. Front Pharmacol. 14:10930172023. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen W, Chen M, Zhao Z, Weng Q, Song J,
Fang S, Wu X, Wang H, Zhang D, Yang W, et al: ZFP36 binds with PRC1
to inhibit tumor growth and increase 5-Fu chemosensitivity of
hepatocellular carcinoma. Front Mol Biosci. 7:1262020. View Article : Google Scholar : PubMed/NCBI
|