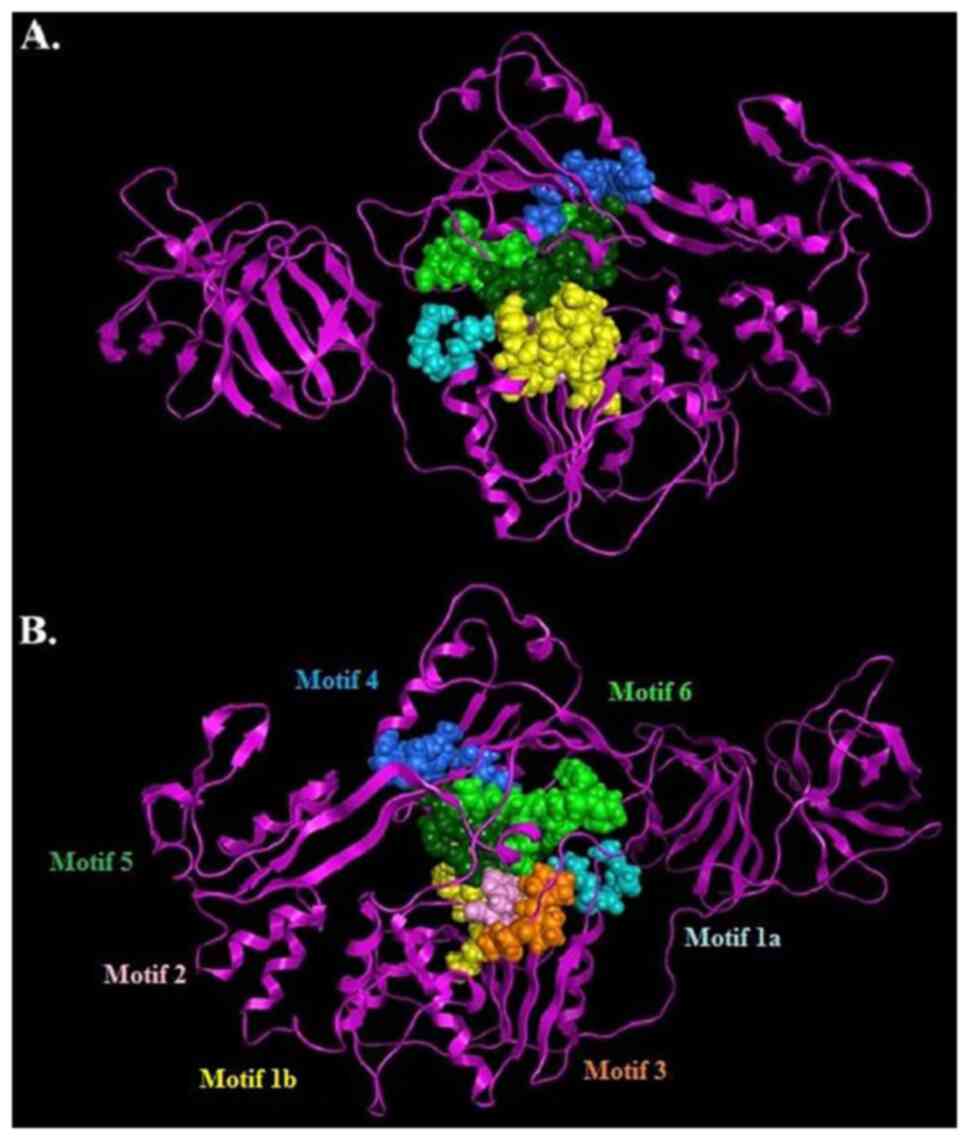
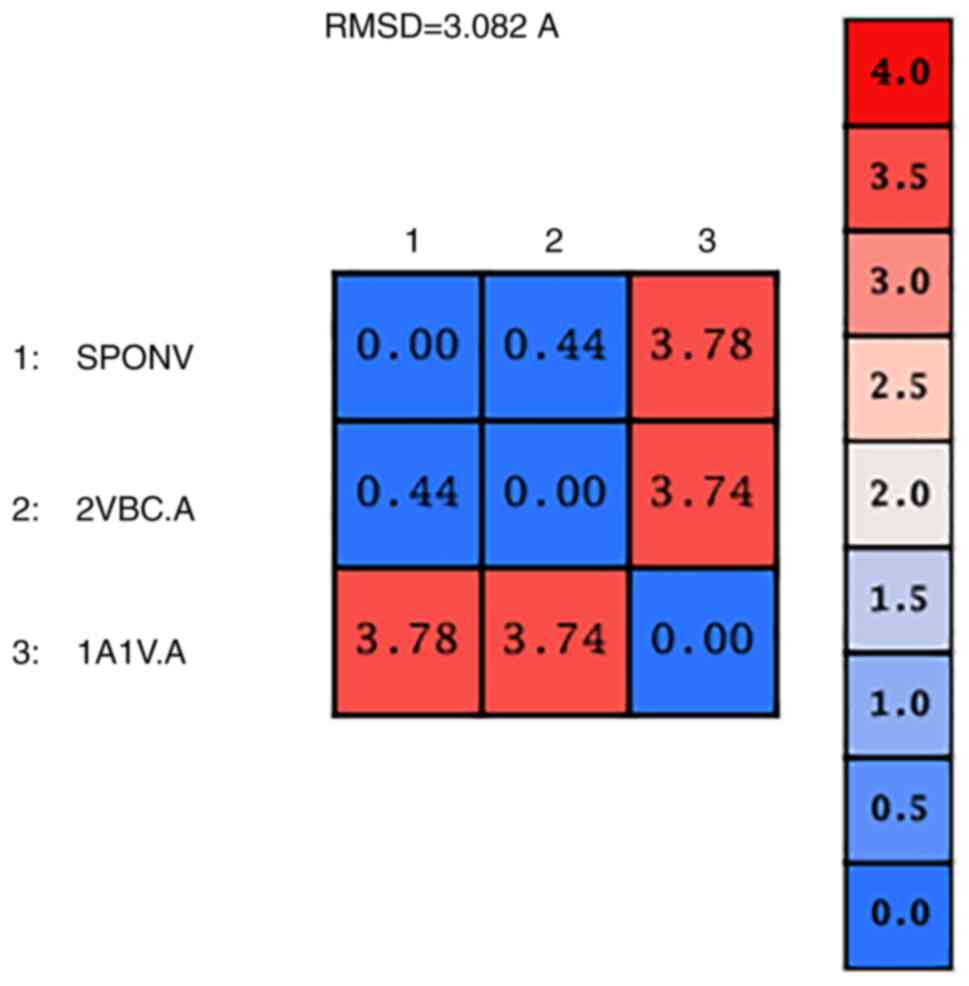
|
1
|
Papageorgiou L, Loukatou S, Sofia K,
Maroulis D and Vlachakis D: An updated evolutionary study of
Flaviviridae NS3 helicase and NS5 RNA-dependent RNA polymerase
reveals novel invariable motifs as potential pharmacological
targets. Mol BioSyst. 12:2080–2093. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Papageorgiou L, Loukatou S, Koumandou VL,
Makałowski W, Megalooikonomou V, Vlachakis D and Kossida S:
Structural models for the design of novel antiviral agents against
greek goat encephalitis. PeerJ. 2(e664)2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Vlachakis D and Kossida S: Molecular
modeling and pharmacophore elucidation study of the classical swine
fever virus helicase as a promising pharmacological target. PeerJ.
1(e85)2013.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
White SK, Lednicky JA, Okech BA, Morris JG
and Dunford JC: Spondweni virus in field-caught culex
quinquefasciatus mosquitoes, Haiti, 2016. Emerg Infect Dis.
24:1765–1767. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Pierson TC and Diamond MS: The continued
threat of emerging flaviviruses. Nat Microbiol. 5:796–812.
2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Luo D, Xu T, Hunke C, Grüber G, Vasudevan
SG and Lescar J: Crystal structure of the NS3 protease-helicase
from dengue virus. J Virol. 82:173–183. 2008.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kim JL, Morgenstern KA, Griffith JP, Dwyer
MD, Thomson JA, Murcko MA, Lin C and Caron PR: Hepatitis C virus
NS3 RNA helicase domain with a bound oligonucleotide: The crystal
structure provides insights into the mode of unwinding. Structure.
6:89–100. 1998.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Pickett BE, Sadat EL, Zhang Y, Noronha JM,
Squires RB, Hunt V, Liu M, Kumar S, Zaremba S, Gu Z, et al: ViPR:
An open bioinformatics database and analysis resource for virology
research. Nucleic Acids Res. 40:D593–D598. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
NCBI Resource Coordinators. Database
resources of the national center for biotechnology information.
Nucleic Acids Res. 44:D7–D19. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Vlachakis D, Papageorgiou L, Papadaki A,
Georga M, Kossida S and Eliopoulos E: An updated evolutionary study
of the notch family reveals a new ancient origin and novel
invariable motifs as potential pharmacological targets. PeerJ.
8(e10334)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Papageorgiou L, Shalzi L, Pierouli K,
Papakonstantinou E, Manias S, Dragoumani K, Nicolaides NC,
Giannakakis A, Bacopoulou F, Chrousos GP, et al: An updated
evolutionary study of the nuclear receptor protein family. World
Acad Sci. J 3:2021.
|
|
12
|
Korostensky C and Gonnet G: Near optimal
multiple sequence alignments using a traveling salesman problem
approach. 6th International Symposium on String Processing and
Information Retrieval. 5th International Workshop on Groupware
(Cat. No. PR00268): 105-114, 1999.
|
|
13
|
Mailund T, Brodal GS, Fagerberg R,
Pedersen CN and Phillips D: Recrafting the neighbor-joining method.
BMC Bioinformatics. 7(29)2006.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Waterhouse AM, Procter JB, Martin DM,
Clamp M and Barton GJ: Jalview version 2-a multiple sequence
alignment editor and analysis workbench. Bioinformatics.
25:1189–1191. 2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Mitsis T, Papageorgiou L, Efthimiadou A,
Bacopoulou F, Vlachakis D, Chrousos GP and Eliopoulos E: A
comprehensive structural and functional analysis of the ligand
binding domain of the nuclear receptor superfamily reveals highly
conserved signaling motifs and two distinct canonical forms through
evolution. World Acad Sci J. 1:264–274. 2019.
|
|
16
|
Henikoff S and Henikoff JG: . Amino acid
substitution matrices from protein blocks. Proc Natl Acad Sci USA.
89:10915–10919. 1992.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Altschul SF, Gish W, Miller W, Myers EW
and Lipman DJ: Basic local alignment search tool. J Mol Biol.
215:403–410. 1990.PubMed/NCBI View Article : Google Scholar
|
|
18
|
wwPDB consortium: Protein data bank: The
single global archive for 3D macromolecular structure data. Nucleic
Acids Res. 47:D520–D528. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Papakonstantinou E, Vlachakis D, Thireou
T, Vlachoyiannopoulos PG and Eliopoulos E: A holistic evolutionary
and 3d pharmacophore modelling study provides insights into the
metabolism, function, and substrate selectivity of the human
monocarboxylate transporter 4 (Hmct4). Int J Mol Sci.
22(2918)2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Molecular Operating Environment (MOE),
2016.0801 Chemical Computing Group ULC, 1010 Sherbooke St. West,
Suite #910, Montreal, QC, Canada, H3A 2R7, 2022.
|
|
21
|
Vangelatos I, Vlachakis D, Sophianopoulou
V and Diallinas G: Modelling and mutational evidence identify the
substrate binding site and functional elements in APC amino acid
transporters. Mol Membr Biol. 26:356–370. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Diakou KI, Mitsis T, Pierouli K,
Papakonstantinou E, Megalooikonomou V, Efthimiadou A and Vlachakis
D: Study of the langat virus RNA-dependent RNA polymerase through
homology modeling. EMBnet J. 26(e944)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Coutsias EA and Wester MJ: RMSD and
symmetry. J Comput Chem. 40:1496–1508. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Gooch JW: Ramachandran Plot. In: Gooch
J.W. (eds) Encyclopedic Dictionary of Polymers. Springer, New York,
NY, 2011.
|
|
25
|
Kandil S, Biondaro S, Vlachakis D, Cummins
AC, Coluccia A, Berry C, Leyssen P, Neyts J and Brancale A:
Discovery of a novel HCV helicase inhibitor by a de novo drug
design approach. Bioorg Med Chem Lett. 19:2935–2937.
2009.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Vlachakis D, Koumandou VL and Kossida S: A
holistic evolutionary and structural study of flaviviridae provides
insights into the function and inhibition of HCV helicase. PeerJ.
1(e74)2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wadood A, Riaz M, Uddin R and Ul-Haq Z: In
silico identification and evaluation of leads for the simultaneous
inhibition of protease and helicase activities of HCV NS3/4A
protease using complex based pharmacophore mapping and virtual
screening. PLoS One. 9(2)(e89109)2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Saraste M, Sibbald PR and Wittinghofer A:
The P-loop-a common motif in ATP- and GTP-binding proteins. Trends
Biochem Sci. 15:430–434. 1990.PubMed/NCBI View Article : Google Scholar
|
|
29
|
delToro D, Ortiz D, Ordyan M, Sippy J, Oh
CS, Keller N, Feiss M, Catalano CE and Smith DE: Walker-A motif
acts to coordinate ATP hydrolysis with motor output in viral DNA
packaging. J Mol Biol. 428:2709–2729. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ruff M, Krishnaswamy S, Boeglin M,
Poterszman A, Mitschler A, Podjarny A, Rees B, Thierry JC and Moras
D: Class II aminoacyl transfer Rna synthetases: Crystal structure
of yeast aspartyl-trna synthetase complexed with trna(Asp).
Science. 252:1682–1689. 1991.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Utama A, Shimizu H, Hasebe F, Morita K,
Igarashi A, Shoji I, Matsuura Y, Hatsu M, Takamizawa K, Hagiwara A
and Miyamura T: Role of the DExH Motif of the Japanese encephalitis
virus and hepatitis C virus NS3 proteins in the ATPase and RNA
helicase activities. Virology. 273:316–324. 2000.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Lee T and Pelletier J: The biology of DHX9
and its potential as a therapeutic target. Oncotarget.
7:42716–42739. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Luo D, Xu T, Watson RP, Scherer-Becker D,
Sampath A, Jahnke W, Yeong SS, Wang CH, Lim SP, Strongin A, et al:
Insights into RNA unwinding and ATP hydrolysis by the flavivirus
NS3 protein. EMBO J. 27:3209–3219. 2008.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hollingsworth SA and Karplus PA: A fresh
look at the Ramachandran plot and the occurrence of standard
structures in proteins. BioMolecular Concepts. 1:271–283.
2010.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Rathi PC, Ludlow RF and Verdonk ML:
Practical high-quality electrostatic potential surfaces for drug
discovery using a graph-convolutional deep neural network. J Med
Chem. 63:8778–8790. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Papageorgiou L, Vlachakis C, Dragoumani K,
Raftopoulou S, Brouzas D, Nicolaides NC, Chrousos GP, Charmandari
E, Megalooikonomou V and Vlachakis D: HCV genetics and genotypes
dictate future antiviral strategies. J Mol Biochem. 6:33–40.
2017.PubMed/NCBI
|
|
37
|
Li K, Frankowski KJ, Belon CA,
Neuenswander B, Ndjomou J, Hanson AM, Shanahan MA, Schoenen FJ,
Blagg BS, Aubé J and Frick DN: Optimization of potent hepatitis C
virus NS3 helicase inhibitors isolated from the yellow dyes
thioflavine S and primuline. J Med Chem. 55:3319–3330.
2012.PubMed/NCBI View Article : Google Scholar
|