Introduction
Type 2 diabetes mellitus (T2DM) poses a significant
public health concern that has an impact on the quality of life of
affected individuals, thereby creating a financial burden on
society (1). Based on the World
Health Organization (WHO) data (2019), non-communicable diseases
are responsible for 74% of global deaths. Specifically, diabetes
was ranked as the tenth leading cause of mortality worldwide in
2019, with fatalities accounting for 1.6 million. It is estimated
that by 2035, ~592 million individuals could lose their lives due
to diabetes-related complications (2). T2DM, which comprises 90% of all
diabetes cases, used to be considered as a disease common in
wealthy, developed countries. However, it has currently spread
worldwide and is the leading cause of mortality and disability,
particularly in younger individuals (3). The susceptibility to type 2 diabetes
differs significantly worldwide, notably with Asian Indians, Native
Americans and Pacific Islanders facing a notably elevated
susceptibility to the condition. Since the 1990s, there has been a
consistent increase in the worldwide occurrence of type 2 diabetes.
In particular, there has been a substantial increase in the global
diabetic population since the beginning of 2000(4). According to the International
Diabetes Federation (IDF), 8.8% of adults globally have diabetes,
with males being slightly more affected (9.6%) than females (9.0%).
Presently, 463 million individuals have diabetes, and 374 million
have impaired glucose tolerance (IGT), which is a precursor of the
condition. Projections indicate these numbers could reach 700
million for diabetes and 548 million for IGT by the year 2045, a
51% increase from 2019(5). The
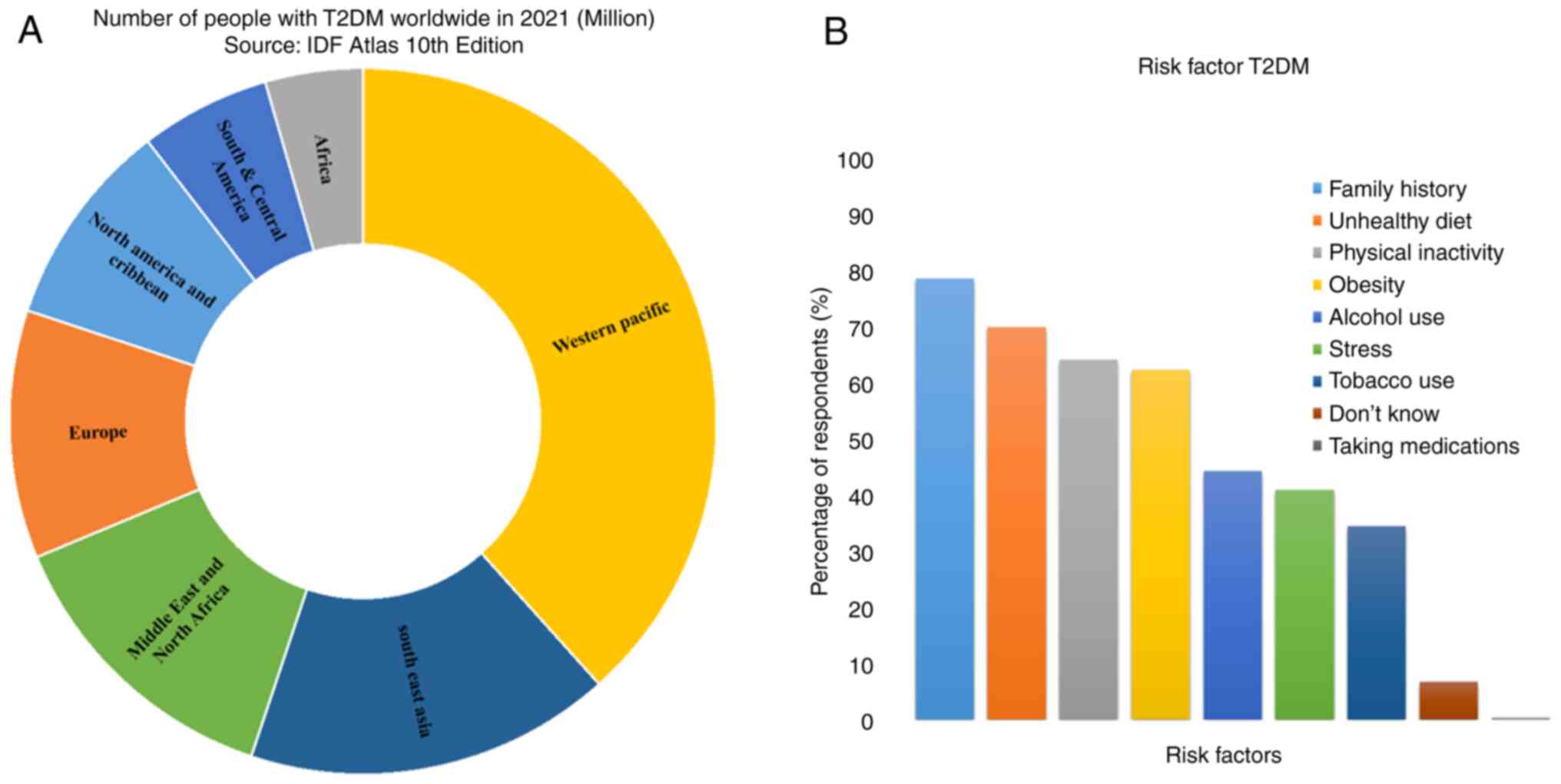
global prevalence and risk factors for T2DM are presented in
Fig. 1 (6,7).
T2DM is a long-term endocrine disorder marked by
‘hyperglycemia’, or the failure of the body to control blood sugar
levels due to insulin resistance and a decreased insulin output. It
is the most prevalent type of diabetes, affecting millions of
individuals globally (8). T2DM is
defined as a reduction in the responsiveness of the cells in the
body to insulin, a hormone produced by the pancreas that manages
blood sugar levels. To combat the resistance, the body generates
more insulin; however, over time, the pancreas becomes less able to
produce adequate insulin, which raises blood sugar levels (9). A variety of factors contribute to the
onset of T2DM, including family background, age, socioeconomic
status, ethnicity, weight issues, metabolic syndrome and specific
unhealthy habits. The interaction among these risk factors in
causing T2DM is a complex physiological process involving intricate
genetic and environmental influences, which appear to differ across
various population groups (10).
Several indicators of T2DM include an increased thirst, reduced
appetite, frequent urination and unexplained weight reduction. The
failure to address T2DM could lead to complications, such as
ulcers, heart-related issues, and harm to multiple organs such as
the kidneys and nerves, as well as vision and hearing impairments
(11). In addition to frequently
occurring alongside other conditions, such as dyslipidemia and
hypertension, T2DM poses a substantial standalone risk for
developing coronary artery disease (CAD). Patients with T2DM
experience elevated rates of cardiovascular disease, particularly
CAD, which is associated with increased morbidity and mortality
rates. While individuals without diabetes have a 25% chance of
developing CAD, those with T2DM are at a 2-4-fold greater risk
(12). Several research studies
have established a connection between mutations in specific genes
and the development of diabetes mellitus, particularly T2DM. These
genes, in combination with environmental factors, contribute to the
onset of T2DM through diverse pathways. The hereditary component of
risk factors typically involves multiple genes interacting with
each other or with environmental elements. Consequently, addressing
genetic risk factors for T2DM could enhance the comprehension of
these conditions and lead to improved clinical care.
Insulin-like growth factor (IGG) mRNA-2 binding
protein 2 (IGF2BP2), a protein found on chromosome 3q27,
plays roles in both embryonic development and the formation of the
pancreas. Additionally, it governs the transcription of IGF2, a
pivotal factor in the development of insulin action (13). IGF2BP2 has the potential to
reduce the expression of IGF2 in both adipose tissue and the
dysfunction of β-cells. This action affects a growth factor
essential for regulating pancreatic development and fat cell
formation. Moreover, IGF2BP2 is involved in T2DM, a
condition linked to reduced insulin secretion (14). Research has demonstrated a
significant association between IGF2BP2 and overweight or
obesity, which are known risk factors for T2DM. Given the link
between obesity and T2DM, it is hypothesized that the association
between IGF2BP2 and T2DM may be influenced by obesity
(15). This phenomenon is referred
to as the interaction between IGF2BP2 and obesity in
relation to T2DM. Moreover, IGF2BP2 is associated with
decreased insulin secretion and significantly contributes to the
development of T2DM across various ethnic populations.
Conversely, the sirtuin1 (SIRT1) gene,
situated on chromosome 10, operates the nicotinamide adenine
dinucleotide (NAD+)-dependent histone deacetylase
(16). It falls under the category
of enzymes known as silent information regulators, contributing to
ageing, longevity and the development of age-related metabolic
conditions, such as T2DM (17).
The protein regulates pancreatic growth by controlling pancreatic
and duodenal homeobox 1 transcription, modulates hepatic metabolism
and gluconeogenesis by reducing G6Pase and phosphoenolpyruvate
carboxykinase transcription, and inhibits adipogenesis by binding
to genes regulated by peroxisome proliferator-activated receptor-γ.
Notably, SIRT1 enhances insulin secretion by downregulating
uncoupling protein (Ucp2) expression in pancreatic β-cell
mitochondria (18). Numerous
studies have demonstrated that SIRT1 plays a crucial role in
glucose-dependent insulin secretion, gluconeogenesis, regulating
inflammation, lipolysis, and β-cell survival. Its association with
various histones and nonhistone substrates has a significant impact
on the development and advancement of diabetes, particularly T2DM
(19). However, additional
validation is necessary to establish definitive findings. Hence, in
the present study, a meta-analysis and statistical power assessment
were conducted to explore the association between gene
polymorphisms of IGF2BP2 and SIRT1 and their impact
on susceptibility to T2DM.
Materials and methods
The Preferred Reporting Item for Systematic Review
and Meta-Analysis (PRISMA) checklist was followed throughout the
investigation, using the widely accepted guidelines for systematic
reviews and meta-analyses. Furthermore, the International
Prospective Register of Systematic Reviews (PROSPERO) validated the
credibility of the study by confirming the registration of its
prospective review protocol (ID: CRD42024545234).
Inclusion criteria
In the present meta-analysis, the following criteria
were consistently applied to evaluate all the research articles.
These criteria include: i) The study design must be a case-control
with allelic and genotype frequencies; ii) the research must
involve exploring genetic variations in the IGF2BP2 and
SIRT1 genes that are linked to T2DM; and iii) only studies
involving human subjects were considered.
Exclusion criteria
The studies that did not meet the following criteria
were excluded: i) Studies are not related to IGF2BP2 and
SIRT1 genetic variants associated with susceptibility to
T2DM; ii) studies involving animal models; iii) studies lacking
information on allelic and genotype frequency; and iv) non-clinical
studies, abstracts and reviews were excluded.
Data extraction
The authors employed standardized criteria to gather
relevant articles. Upon encountering any discrepancies, they
discussed these with co-authors. Following this, the authors
rigorously examined the retrieved articles to establish the
genotype and allelic frequencies of both the case and control
subjects. In instances where certain studies lacked complete
genotypic data, missing values were estimated using available
allelic frequencies. Studies where crucial data could not be
extracted from the control and case groups were excluded. Each
study provided the following information: Study design, sample
size, first author name, ethnicity, publication year and
Hardy-Weinberg equilibrium (HWE) score. Rigorous criteria were
followed to meet the eligibility criteria and screen the data
comprehensively.
Quality of studies using risk
bias
A comprehensive evaluation of risk bias is crucial
for accurately evaluating the methodological quality and potential
biases of studies incorporated into a meta-analysis. In the present
study, the Cochrane risk of bias tool (ROB2) software was utilized
to assess the risk of bias in the research. The studies were
classified into three groups based on their bias levels: ‘high
risk’, ‘moderate risk’, and ‘low risk’.
Statistical analysis
The present study used a range of statistical
methods to examine the genetic variations in the IGF2BP2 and
SIRT1 gene polymorphisms and their impact on the development
of T2DM. The odds ratios (ORs) with 95% confidence intervals (CIs)
were calculated to evaluate the associations. The statistical
significance was determined with a threshold set at P<0.05. The
consistency of results across different studies was evaluated using
the index of inconsistency, which measures the proportion of total
variability across studies that is due to heterogeneity (I²) rather
than random chance. A random-effects model was applied for all
meta-analyses, regardless of the I² value observed, to account for
potential variations among studies from different groups and
geographical locations. Summary ORs were assessed using a Z-test
(P<0.05), with heterogeneity among studies determined by the Q
statistic and I2 value. In addition, a sensitivity
analysis was performed to evaluate the effects of excluding
specific studies, particularly those in which the controls did not
conform to the HWE. Funnel plot and Egger's regression method were
employed to identify potential publication bias. All statistical
analyses were performed using MetaGenyo software (https://metagenyo.genyo.es/).
Power analysis
The metadata were assessed using power analysis,
conducted with a 95% CI and a 0.05 α error. The power of the sample
size of each study, including both case and control groups, was
assessed individually for the selected genes. Power calculations
were executed using G*Power 3.1 software (version 3.1; https://download.cnet.com/g-power/3001-2054_4-10647044.html?dt=internalDownload).
Protein-protein interactions
(PPIs)
The online search tool database STRING (v11.0) can
predict functional proteins and PPIs associated with identified
T2DM-linked polymorphisms, achieving a score of ≥0.4.
Results
Search results
The present meta-analysis included a total of 16
studies, which were selected based on specific inclusion and
exclusion criteria. The study strategy for IGF2BP2 and
SIRT1 gene polymorphisms is illustrated in Fig. S1. Through a thorough search across
various databases, 10 studies involving 4,058 cases and 23,723
controls for the IGF2BP2 rs4402960 gene variant and five
studies with 978 cases and 1,066 controls for the IGF2BP2
rs1470579 gene variant were identified (20-30).
Additionally, five studies comprising 607 cases and 1,219 controls
for the SIRT1 rs7895833 gene polymorphism were identified
(31-35).
The key characteristics extracted from the included case-control
studies are presented in Table I,
Table II and Table III. The present study examined a
subset of articles to note the variability found in all genetic
models, including dominant (CC + CA vs. AA), allelic (C vs. A),
recessive (CC vs. AA), and over-dominant (CA vs. CC + AA).
 | Table ICharacteristics of the selected
case-control studies for the association between the IGF2BP2
rs4402960 gene polymorphism and type 2 diabetes mellitus and the
HWE score. |
Table I
Characteristics of the selected
case-control studies for the association between the IGF2BP2
rs4402960 gene polymorphism and type 2 diabetes mellitus and the
HWE score.
| | Genotype frequency
of cases | Genotype frequency
of controls | |
|---|
| SNP | Authors, year of
publication | Ethnicity | GG | GT | TT | GG | GT | TT | Total cases | Total controls | HWE-P-value | HWE-adjusted
P-value | (Refs.) |
|---|
| IGF2BP2 | El Taweel et
al, 2017 | Asian | 11 | 16 | 3 | 18 | 11 | 1 | 30 | 30 | 0.6605 | 0.8845 | (20) |
| rs4402960 | Altalalgah et
al, 2018 | Asian | 10 | 67 | 23 | 49 | 40 | 11 | 100 | 100 | 0.5158 | 0.8597 | (21) |
| | Tarnowski et
al, 2018 | Caucasian | 89 | 93 | 22 | 105 | 76 | 26 | 204 | 207 | 0.0432 | 0.1062 | (22) |
| | Nfor et al,
2020 | Asian | 543 | 416 | 71 | 12,265 | 7,584 | 1,160 | 1,030 | 21,009 | 0.7819 | 0.8845 | (23) |
| | Sargazi et
al, 2020 | Asian | 60 | 205 | 31 | 29 | 176 | 87 | 296 | 292 | 0 | 0 | (24) |
| | Verma et al,
2021 | Asian | 162 | 107 | 100 | 62 | 29 | 9 | 369 | 100 | 0.0531 | 0.1062 | (25) |
| | Li et al,
2021 | Asian | 728 | 484 | 62 | 635 | 473 | 86 | 1,274 | 1,194 | 0.8709 | 0.8845 | (26) |
| | Falih et al,
2022 | Asian | 177 | 177 | 46 | 217 | 156 | 27 | 400 | 400 | 0.8845 | 0.8845 | (27) |
| | Ajenah et
al, 2023 | Asian | 30 | 21 | 9 | 32 | 18 | 10 | 60 | 60 | 0.0175 | 0.0617 | (28) |
| | Zhou et al,
2023 | Asian | 164 | 102 | 29 | 219 | 92 | 20 | 295 | 331 | 0.0185 | 0.0617 | (29) |
 | Table IICharacteristics of selected
case-control studies for the association between the IGF2BP2
rs1470579 polymorphism with type 2 diabetes mellitus and HWE
score. |
Table II
Characteristics of selected
case-control studies for the association between the IGF2BP2
rs1470579 polymorphism with type 2 diabetes mellitus and HWE
score.
| | Genotype frequency
of cases | Genotype frequency
of controls | |
|---|
| SNP | Authors, year of
publication | Ethnicity | AA | AC | CC | AA | AC | CC | Total cases | Total controls | HWE-P-value | HWE-adjusted
P-value | (Refs.) |
|---|
| IGF2BP2 | Vatankhah et
al, 2020 | Asian | 55 | 11 | 40 | 67 | 55 | 40 | 106 | 162 | 0.0001 | 0.0005 | (30) |
| rs1470579 | El Taweel et
al, 2017 | Asian | 8 | 9 | 13 | 19 | 8 | 3 | 30 | 30 | 0.1631 | 0.2718 | (20) |
| | Sargazi et
al, 2020 | Asian | 41 | 64 | 42 | 35 | 86 | 22 | 147 | 143 | 0.0109 | 0.0272 | (24) |
| | Falih et al,
2022 | Asian | 186 | 174 | 40 | 216 | 155 | 29 | 400 | 400 | 0.869 | 0.869 | (27) |
| | Zhou et al,
2023 | Asian | 140 | 126 | 29 | 197 | 115 | 19 | 295 | 331 | 0.6832 | 0.854 | (29) |
 | Table IIICharacteristics of selected
case-control studies for the association between the SIRT1
rs7895833 gene polymorphism with Type 2 diabetes mellitus and HWE
score. |
Table III
Characteristics of selected
case-control studies for the association between the SIRT1
rs7895833 gene polymorphism with Type 2 diabetes mellitus and HWE
score.
| | Genotype frequency
of cases | Genotype frequency
of controls | |
|---|
| SNP | Authors, year of
publication | Ethnicity | AA | AG | GG | AA | AG | GG | Total cases | Total controls | HWE-P-value | HWE-adjusted
P-value | (Refs.) |
|---|
| SIRT 1 | Mahmoud et
al, 2016 | Asian | 25 | 33 | 22 | 60 | 65 | 25 | 80 | 150 | 0.3069 | 0.7672 | (31) |
| rs7895833 | Meneguette et
al, 2016 | Asian | 50 | 47 | 3 | 52 | 40 | 8 | 100 | 100 | 0.9367 | 0.9367 | (32) |
| | Wozny et al,
2017 | Caucasian | 103 | 41 | 6 | 195 | 80 | 9 | 150 | 284 | 0.8205 | 0.9367 | (33) |
| | Tao et al,
2022 | Asian | 19 | 48 | 74 | 37 | 237 | 275 | 141 | 549 | 0.1386 | 0.693 | (35) |
| | Dmitrenko et
al, 2022 | Asian | 55 | 77 | 4 | 85 | 46 | 5 | 136 | 136 | 0.6884 | 0.9367 | (34) |
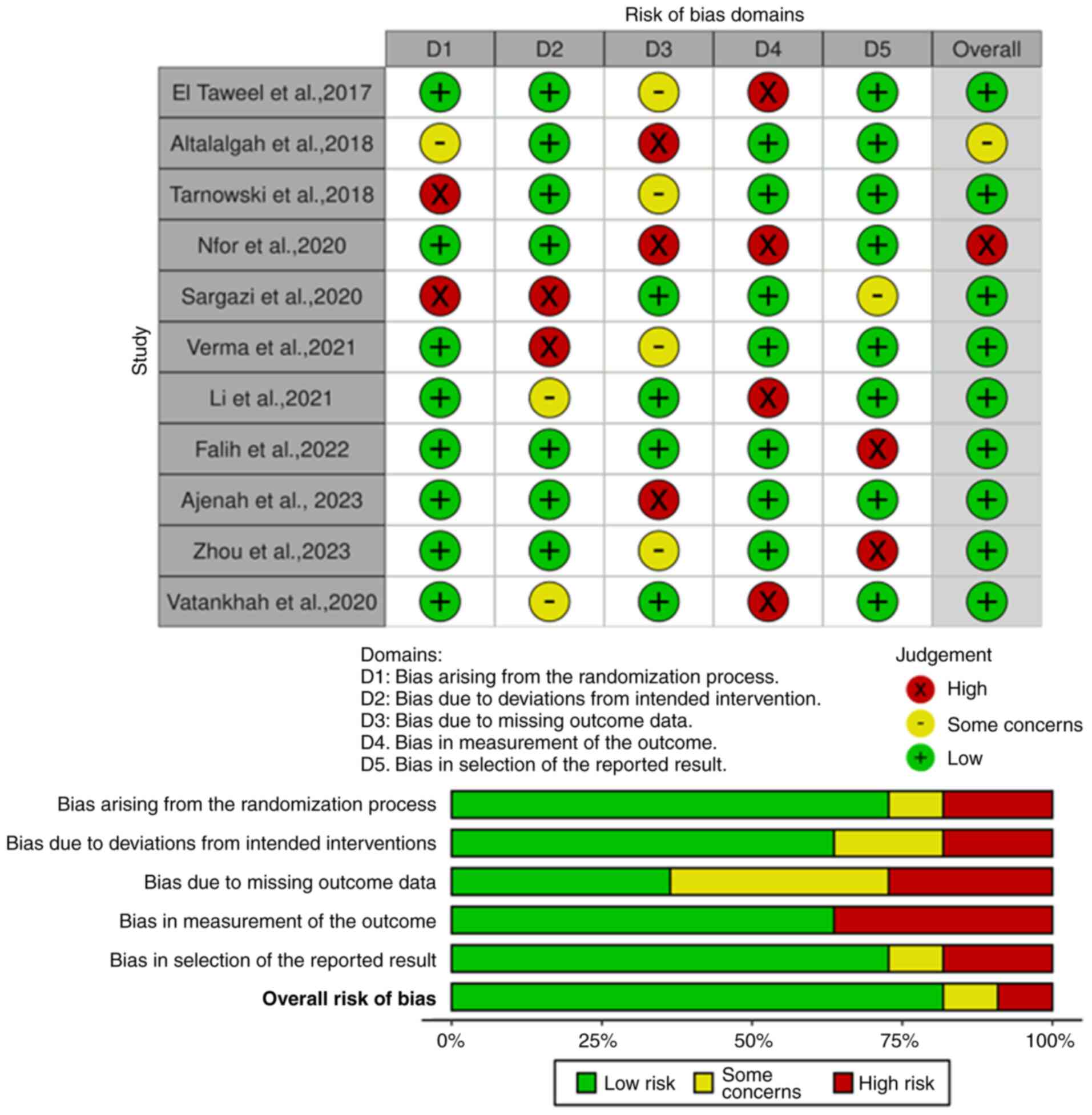
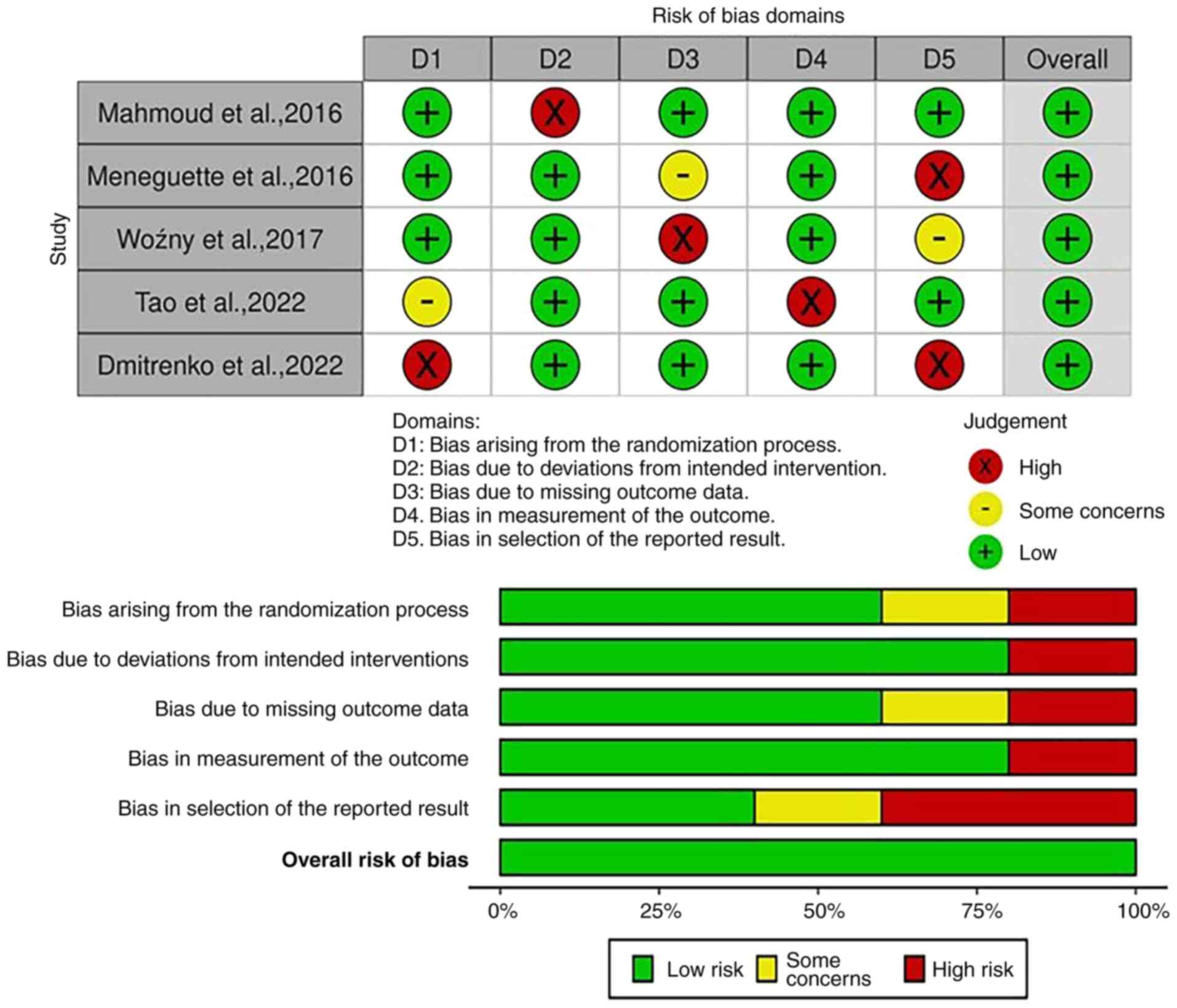
Risk of bias
The included studies underwent a comprehensive
assessment of their methodological quality using the Cochrane Risk
of Bias Tool 2. A row represents each study and each column
represents a different bias category. The color-coded scheme in the
figures indicates the level of risk associated with each type of
bias in each study; red represents high risk, yellow represents
moderate risk, and green represents low risk (Figs. 2 and 3). The analysis indicated that the
majority of the research had a low risk of bias, suggesting that it
was conducted and documented in a manner that effectively minimized
the potential for bias or systematic errors.
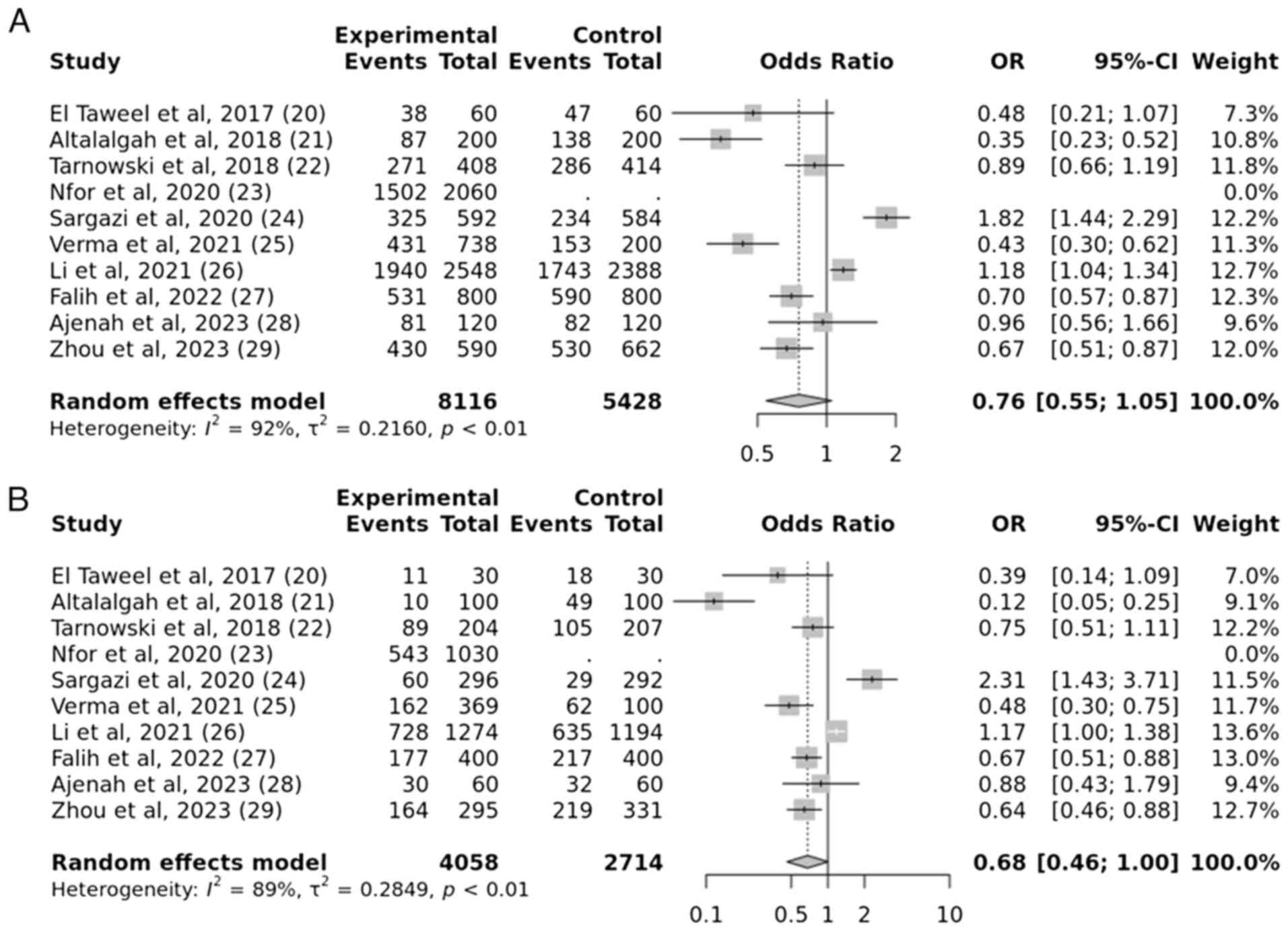
Quantitative data analysis
In total, 10 studies were evaluated to determine the
associations between the IGF2BP2 rs4402960 gene polymorphism
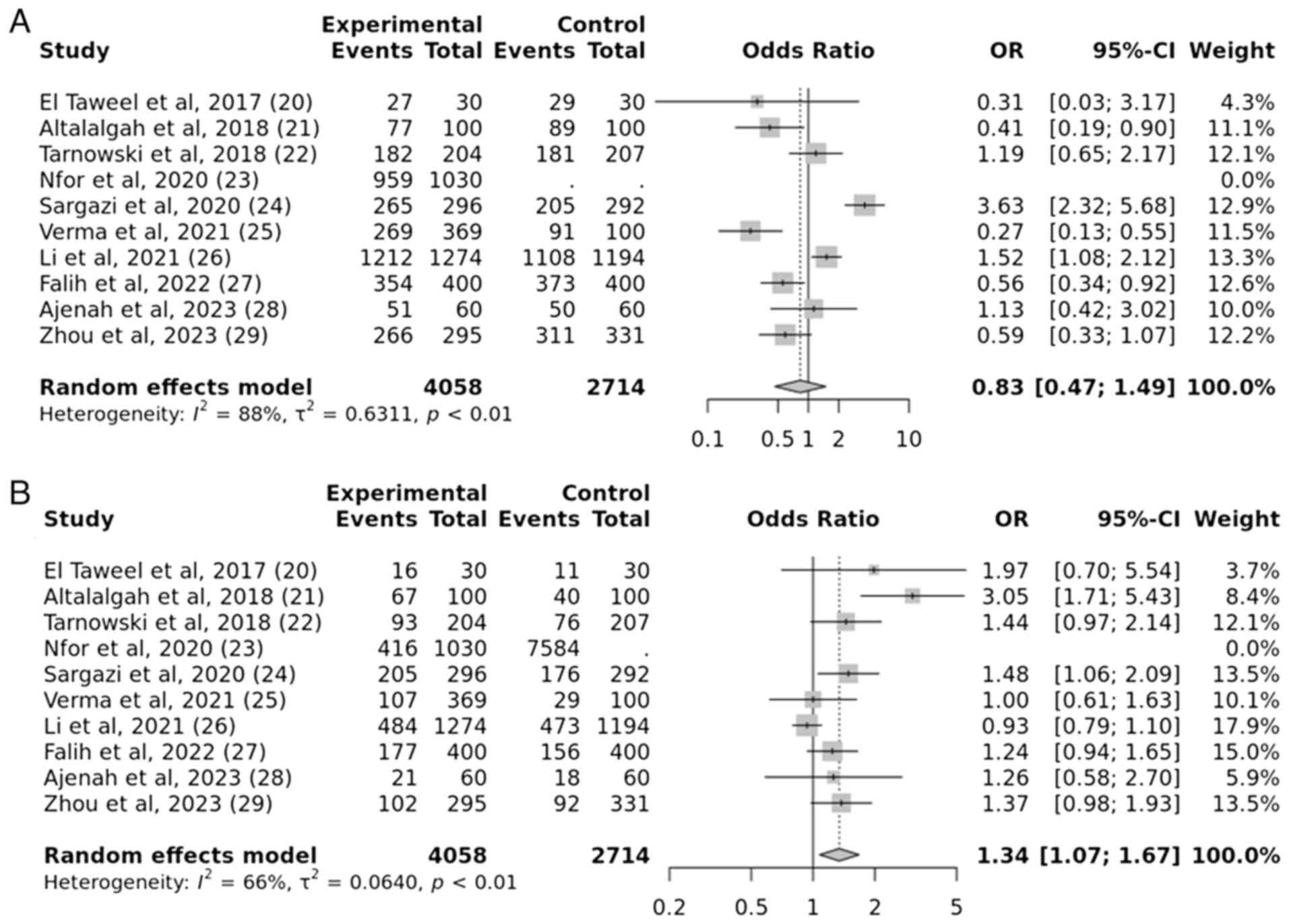
and the susceptibility to T2DM. The results revealed a strong
correlation between the IGF2BP2 rs4402960 gene mutation and
the onset of T2DM susceptibility, under the over-dominant model.
The obtained P-value was 0.009, indicating a statistically
significant association. The OR was calculated as 1.34, with a 95%
CI of 1.07-1.67. However, no significant association was observed
under other genetic models such as allelic, recessive and dominant
models. The obtained P-values are 0.09, 0.06 and 0.5, respectively,
which indicates no statistical significance (Figs. 4 and 5).
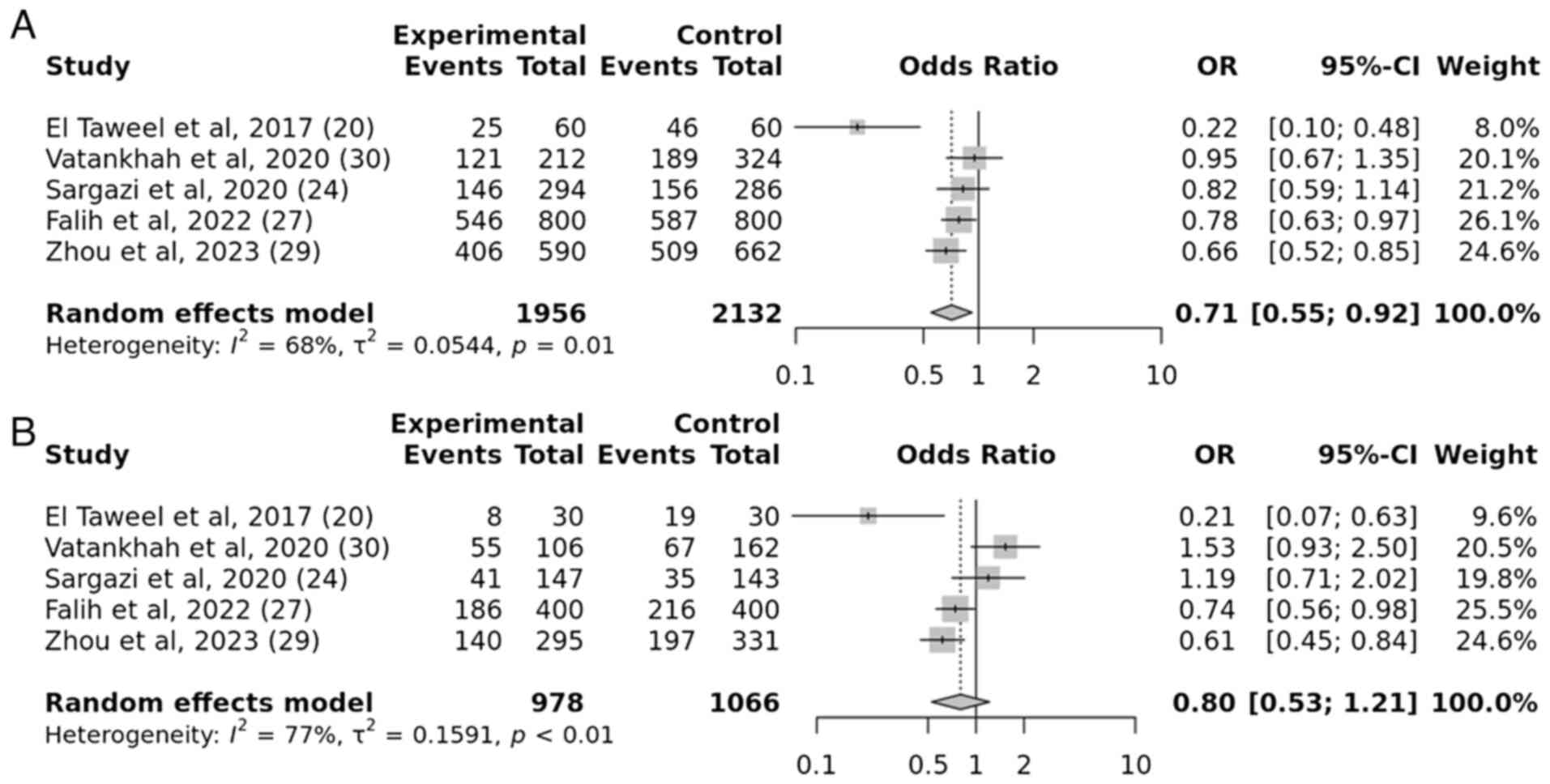
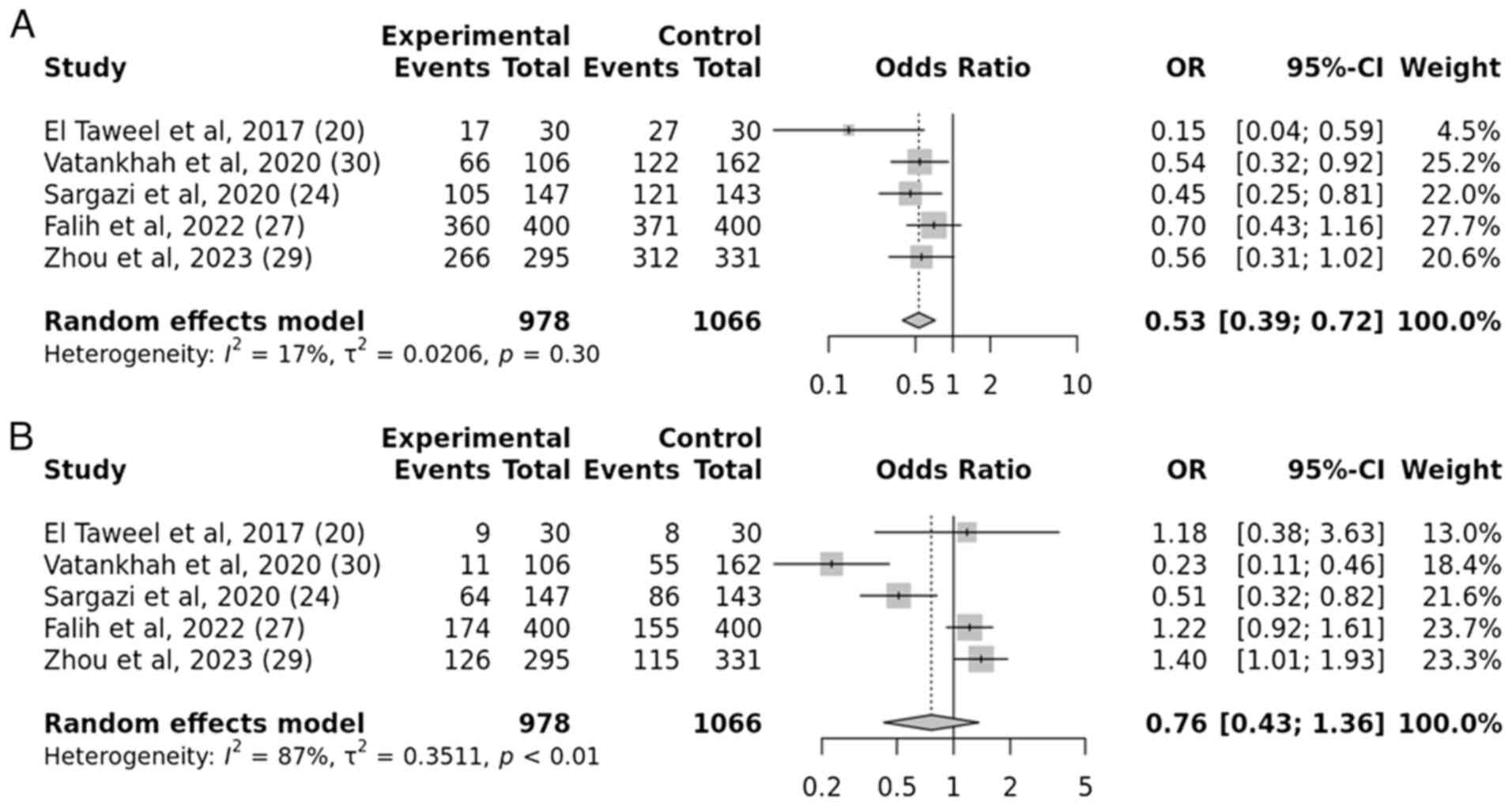
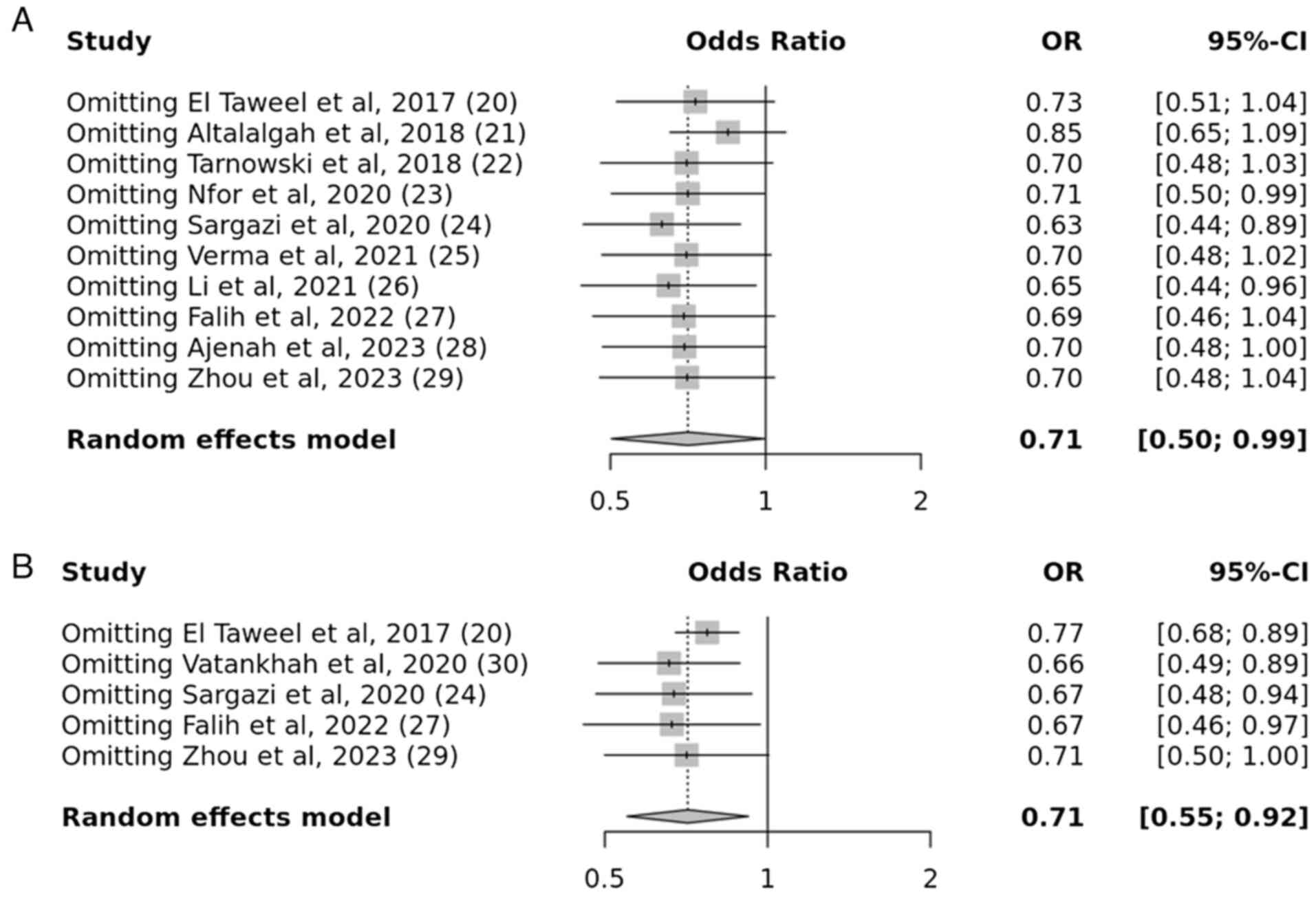
A total of five case-control studies were examined
to determine the association between the IGF2BP2 rs1470579 gene
variation and the susceptibility to T2DM. The results demonstrated
a substantial connection between the IGF2BP2 rs1470579 gene
variation and the development of T2DM, under the allelic and
recessive models. The obtained P-values, 0.009 and 0.007,
respectively, indicate a statistically significant. For the allelic
model, the OR was 0.71 with a 95% CI of 0.55-0.92. For the
recessive model, the OR was 0.80 with a 95% CI of 0.53-1.21.
However, no association was observed in the dominant and
over-dominant models. The resulting P-values were 0.3 and 0.6,
which indicated a statistically insignificant association (Figs. 6 and 7).
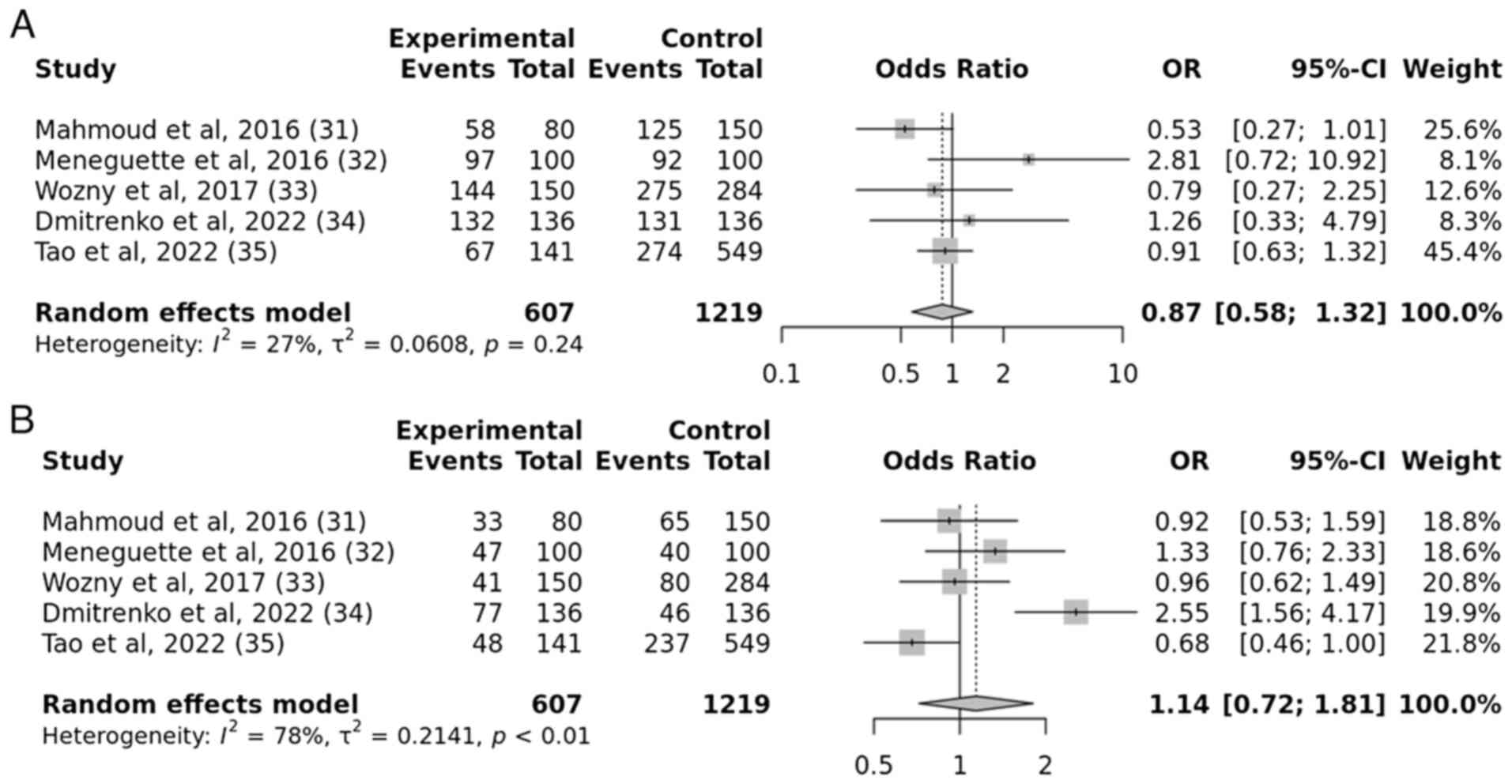
In order to evaluate the association between the
SIRT1 rs7895833 gene polymorphism and susceptibility to
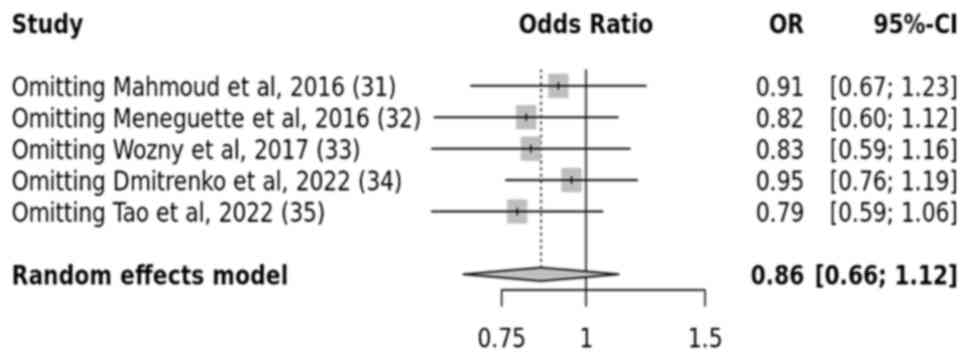
T2DM, five studies were analyzed. The results revealed no
significant link between the SIRT1 rs7895833 gene
polymorphism and T2DM across all genetic models, such as the
allelic, recessive, dominant and over-dominant models. The
resulting P-values, which were 0.2, 0.6, 0.3 and 0.5, respectively,
indicated a statistically insignificant association between the
SIRT1 rs7895833 gene polymorphism and the risk of developing
T2DM (Figs. 8 and 9).
Sensitivity analysis and publication
bias
A sensitivity analysis was carried out to explore
the inconsistent findings across multiple studies, particularly
those that deviated from the HWE. Studies that did not meet the HWE
or intervention criteria were excluded from the analysis. Notably,
the results revealed that removing these studies did not
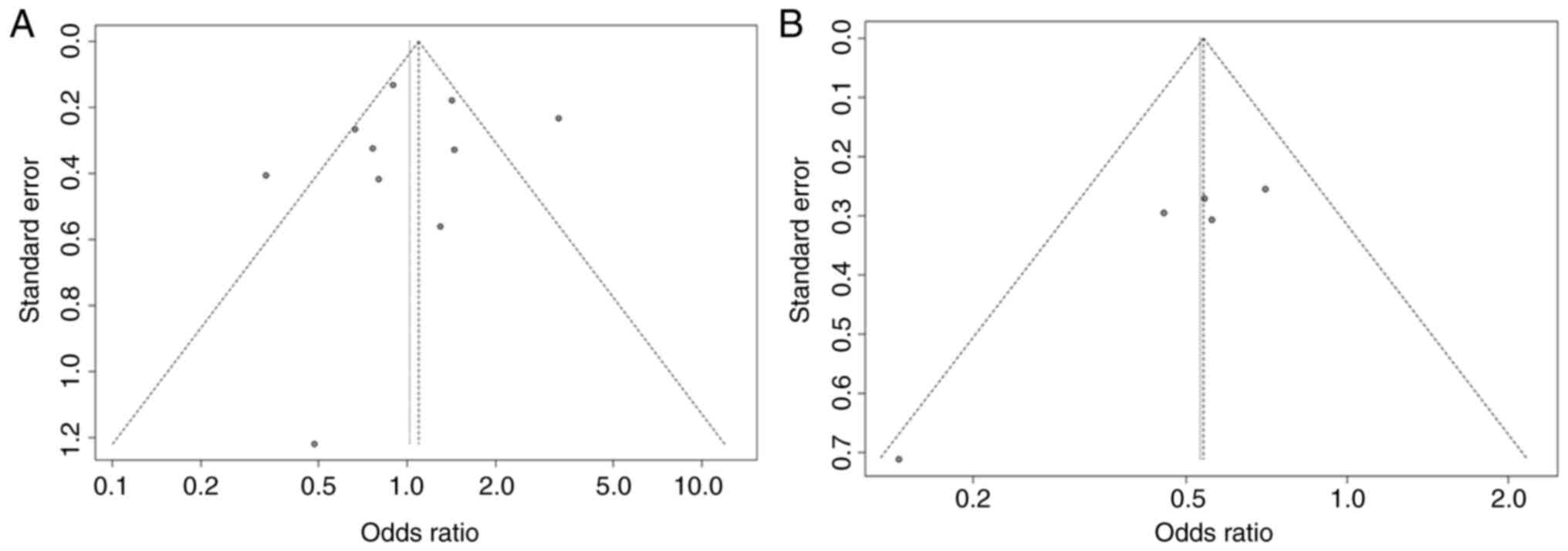
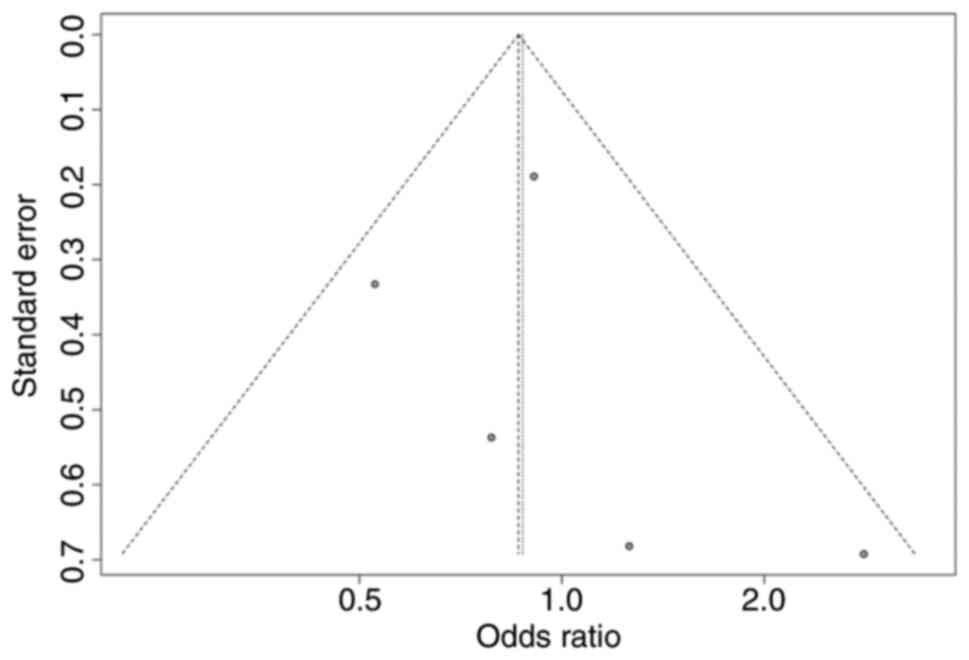
significantly affect the overall P-value (Figs. 10 and 11). To assess publication bias and
confirm the accuracy of the results, a funnel plot and Egger's test
were employed. Figs. 12 and
13 illustrate the funnel plot
generated for this assessment. The symmetrical distribution of the
data points around the average effect size indicates that there is
no significant publication bias among the included studies.
Power analysis and construction of the
PPI network
A power analysis was performed to establish the
significance level of each study for the selected single nucleotide
polymorphisms (SNPs). According to the findings, the sample size in
the selected literature met the significant level requirement,
encompassing an α error probability of 0.05. The findings of the
power analysis are presented in Table
IV. The power analysis plot depicts the power analysis results
for a two-tailed hypothesis test. Power analysis assesses the
likelihood of detecting an effect of a given size under specific
conditions, such as sample size, effect size and significance level
(Fig. S2).
 | Table IVResults of power analysis. |
Table IV
Results of power analysis.
| Gene | SNP | Number of
studies | Cases | Controls | α err prob | Power (1-β err
prob) |
|---|
| IGF2BP2 | rs4402960 | 10 | 4,058 | 23,723 | 0.05 | 1.0000000 |
| | rs11705701 | 5 | 978 | 1,066 | 0.05 | 0.9981868 |
| SIRT1 | rs7895833 | 5 | 607 | 1,219 | 0.05 | 0.9820296 |
The PPI of recognized polymorphic proteins related
to T2DM was mapped and examined using STRING to determine their hub
genes. The IGF2BP2 protein network has 136 edges and 21
nodes, with a PPI enrichment P-value of P<1.0e-16 and a
clustering coefficient of 0.92. The average node degree is 13. The
SIRT1 protein network has 11 nodes and 33 edges, with a PPI
enrichment p-value of 0.00166 and a clustering coefficient of 0.83.
The average node degree is 6 (Fig.
S3). The protein network exhibits a higher-than-expected level
of interactions among its constituents compared to a random
selection of proteins of similar size and degree of distribution
from the genome. This elevated enrichment implies a biological
interconnectedness among these proteins as a cohesive unit to some
degree.
Discussion
T2DM is a complex, polygenic disorder characterized
by numerous genes contributing to a higher risk of the disease. In
recent times, 16 new genetic sites associated with susceptibility
to T2DM have been identified by researchers (36). IGF2BP2 is a member of the
mRNA-binding protein group, which is crucial for RNA localization,
stability, and translation. It is prominently present in pancreatic
islets and interacts with IGF2, a significant molecule involved in
growth and insulin signaling (37). Based on prior research, it appears
that the IGF2BP2 gene mutation is linked to reduced initial
insulin secretion in response to glucose as determined by the
hyperglycemic clamps and affects pancreatic β-cell function in
individuals with T2DM, contributing to the development of T2DM
pathogenesis (38). In in
vivo research on fruit fly, Drosophila melanogaster,
IGF2BP2 serves as a signaling molecule essential for insulin
function, growth, and pancreas development. Elevated levels of both
fasting plasma glucose and serum insulin were observed.
Additionally, the gene diacylglycerol kinase gamma, similar to
IGF2BP2, has been shown to be associated with regulating
insulin secretion (39).
Therefore, further functional investigations into the
pathophysiological pathways of IGF2BP2 are required.
Ali et al (40) reported that in the Egyptian
population, the homozygous (TT) genotype of IGF2BP2
(rs4402960) and the homozygous (CC) genotype of rs1470579 variants
were identified as risk factors for the occurrence of T2DM.
Previous meta-analyses have explored the potential link between
mutations in the IGF2BP2 gene (rs4402960 and rs1470579) and
the susceptibility to T2DM (41,42).
The present meta-analysis encompasses articles from 2016 to 2024,
representing the most recent data. However, the present study did
not integrate the findings from earlier research. Conducting
updated meta-analyses is crucial for maintaining the relevance and
accuracy of scientific knowledge. By integrating the latest
research findings, these analyses ensure that conclusions are
founded on the most current and comprehensive evidence, thus
advancing our understanding of gene-trait associations and guiding
the effective utilization of biomarkers in diagnosis, prognosis and
treatment strategies.
The SIRT1 protein functions as an epigenetic
regulator in human physiology. Changes in the expression of SIRT1
are linked to various diseases, such as metabolic and
cardiovascular diseases (43).
SIRT1, a deacetylase-dependent NAD+, plays a
crucial role in regulating insulin signaling, metabolic rate and
glucose utilization. It is implicated in the pathogenesis of T2DM
by regulating insulin secretion and impacting glucose metabolism in
liver cells. By repressing Ucp2 and activating transcription
factors, SIRT1 maintains glucose homeostasis through the
modulation of downstream genes (44). The association between T2DM and
SIRT1 SNP variants has been examined across diverse ethnic
groups. Sadeghi et al (5 found that the SIRT1 gene
variants rs12778366 and rs3758391 may be linked to susceptibility
to T2DM in an Iranian population. It is essential to replicate
these findings across diverse ethnicities and with larger sample
sizes to improve result accuracy (45). Similarly, the study by Kaabi et
al (46) found the occurrence
rates of SIRT1 rs12778366 and rs3758391 SNPs within the
Saudi population. Their study indicated that there is no link
between these genetic variations and susceptibility to T2DM. This
discovery contributes to the expanding collection of studies
investigating the genetic factors associated with T2DM (46). Only a limited number of studies
have been carried out on the specific SIRT1 rs7895833 gene
variants. The findings across these studies are inconsistent.
Therefore, the present study compiled all the available data and
performed a meta-analysis. To the best of our knowledge, this is
the first meta-analysis on a SIRT 1 gene polymorphism in T2DM.
Based on the findings of the present meta-analyses,
a significant association was found between the IGF2BP2
rs4402960 gene and the development of T2DM in the over-dominant
model. However, no substantial association was found in the
allelic, recessive and dominant models. Similarly, the present
study indicates that the IGF2BP2 rs1470579 gene variant is
associated with an increased risk of developing T2DM in the allelic
and recessive models, while no significant association was observed
in the dominant and over-dominant models. Conversely, there was no
notable association between the SIRT1 rs7895833 gene variant
and T2DM susceptibility across all genetic models. The sensitivity
analysis indicates that no single study significantly affects the
overall results. The present meta-analysis confirms that both
IGF2BP2 and SIRT1 gene polymorphisms adhere to the
HWE principle. Egger's test and funnel plots were used to detect
publication bias, revealing no evidence of bias. The reliability of
the conclusions is supported by the methodological quality, which
was measured using the ROB2 tool. This demonstrates a decreased
risk level across different aspects of research design in each
included study. Thus, statistical data firmly supports our views.
Strict protocols were used to extract and analyze the data, and a
power analysis verified that the sample sizes of the selected
studies were sufficient.
Mohammed et al (47) concluded that the IGF2BP2
rs6777038 and rs6444082 variants may significantly contribute to
the onset of T2DM in the Iraqi population. However, Liu et
al (48) found that similar
variants were not associated with gestational diabetes. In the
study by Li et al (49), it
was found that IGF2BP2 gene mutations were not significantly
linked to gestational diabetes. They concluded that further
research was required to validate these findings in the future. Zhu
et al (50) discovered
connections between miRNA polymorphisms in the insulin signaling
pathways and susceptibility to T2DM, marking the first exploration
of such links. Their findings indicated that variants, such as
miR-133a-2 rs13040413, let-7a-1 rs13293512 and miR-27a rs895819
were associated with susceptibility to T2DM in either general or
subgroup analyses within the population in China (50). Similarly, Pang et al
(51) analyzed genetic variants in
the SIRT1 gene promoter region in patients with T2DM and
controls. The identified variants in patients with T2DM may
influence the development of T2DM by affecting SIRT1 levels.
Targeting these genetic variants through pharmacological aspects
could offer a novel therapy for patients with T2DM (51). In addition, the study by Letonja
et al (52) reported that
the SIRT1 rs7069102 polymorphism exhibited a significant
association with diabetic nephropathy in patients with T2DM,
suggesting its potential as a marker for susceptibility to diabetic
nephropathy in this population (52). Li et al (53) revealed that the SIRT1
variant rs10997866 exhibited a notable association with
susceptibility to type 1 diabetes, with the G minor allele
significantly increasing the risk of developing type 1 diabetes.
Additional research involving larger sample sizes, diverse ethnic
groups and investigations into various variants of the SIRT1
gene is essential for attaining precise conclusions.
Several proteins have been implicated in T2DM, as
strongly demonstrated in the study conducted by Chahar et al
(54), which revealed that the
measurement of adiponectin, IGF1 and IGF2 levels in serum, combined
with gene expression analysis, plays a crucial role in predicting
the progression from T2DM to diabetic nephropathy. Canto-Cetina
et al (55) demonstrated
that fibronectin type III domain-containing protein 5
(FNDC5/irisin) was significantly linked to susceptibility to T2DM
in Maya-Mestizo women under the dominant model. Their findings
indicate that the FNDC5 gene modulates the production of irisin, a
hormone released during exercise that aids in converting fat cells
to a type that burns energy. Variations in the FNDC5 gene may
affect irisin levels, thereby influencing how the body manages
blood sugar and increasing the chances of developing T2DM (55). The study by Atere et al
(56) revealed the association
between serum amyloid A (SAA), fasting blood sugar and lipid
profile markers in diabetes. Notably, SAA emerged as a superior
indicator, holding the potential to improve the diagnosis and
management of diabetic patients (56). Furthermore, Huang et al
(57) identified HMG20A and HNF1B
gene polymorphisms as being associated with an elevated risk of
developing T2DM in all genetic models. Variations in these genes
could affect glucose and insulin metabolism, potentially increasing
the risk of developing T2DM (57).
The present study sheds light on the genetic basis
of T2DM, potentially paving the way for improved diagnoses and
treatment strategies. Unveiling these genetic markers could hold
promise for risk assessment, early detection and personalized
treatment plans. These findings highlight the crucial role of
studies across diverse populations, which can enrich the
understanding of the IGF2BP2 genetic variant and its
contribution to the development of T2DM. Ultimately, the present
study provides valuable insight into these associations, enhancing
the current knowledge and emphasizing the need to unravel these
complexities.
In conclusion, the present meta-analysis reveals the
role of IGF2BP2 and SIRT1 gene polymorphisms in the
development of T2DM. The present study found that the
IGF2BP2 rs4402960 and rs1470579 variants were significant
risk factors for the onset of T2DM. However, there was no
significant association between the SIRT1 rs7895833 gene
polymorphism and susceptibility to T2DM. Understanding the
complexities of genetic predispositions can help improve risk
assessment, early detection and personalized treatment strategies.
The present study also emphasizes the need for ongoing
investigations to validate these findings and enhance the ability
to combat T2DM and mitigate its impact on public health. The
present meta-analysis has certain limitations, however. The present
study did not explore potential impacts from gene-environment
interactions and other demographic characteristics such as age, sex
and comorbidities, as the study mainly focused on gene
polymorphisms. Additionally, a subgroup analysis was not conducted
due to insufficient studies. The majority of the studies have
focused on Asian populations. Therefore, the study necessitates a
larger and more diverse sample size, encompassing various
ethnicities. Further research involving diverse populations is
recommended to enhance the relevance and applicability of the
results.
Supplementary Material
Flowchart of the literature screening
process for the selectoin of studies on gene polymorphisms. (A)
IGF2BP2 (rs4402960 and rs1470579) gene polymorphisms and (B)
SIRT1 rs7895833 gene polymorphism. IGF2BP2, insulin
growth factor-2 mRNA binding protein 2; SIRT1, sirtuin
1.
Power analysis plot illustrates the
relationship between sample size or effect size and statistical
power for a two-tailed hypothesis test. (A) IGF2BP2
rs4402960 gene polymorphism, (B) IGF2BP2 rs1470579 gene
polymorphism, and (C) SIRT1 rs7895833 gene polymorphism.
IGF2BP2, insulin growth factor-2 mRNA binding protein 2;
SIRT1, sirtuin 1.
The network of protein-protein
interactions among the differentially expressed genes within the
selected gene linked to type 2 diabetes mellitus. (A)
IGF2BP2, (B) SIRT1 gene. IGF2BP2, insulin
growth factor-2 mRNA binding protein 2; SIRT1, sirtuin
1.
Acknowledgements
The authors would like to thank the management of
Chettinad Academy of Research and Education (Deemed to be
University) for providing the facilities to perform this study.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SV conducted literature search, collected data,
developed the study methodology, contributed to the writing of the
manuscript, and created the tables and figures. VRD was involved in
the writing of the original draft of the manuscript, in data
validation, and in data curation. BRS was involved in the study
design, data analysis and interpretation of the results. CK was
involved in the preparation of the manuscript, editing assistance
and study supervision. GKS conducted the investigations, provided
editing assistance, supervised the study, and was also involved in
the conceptualization of the study. The authors confirm that all
raw data presented in this article authentic and accurately
represent the finding of study. SV and GKS confirm the authenticity
of all raw data. All authors have thoroughly reviewed and have read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Meng Y, Liu X, Ma K, Zhang L, Lu M, Zhao
M, Guan M-X and Qin G: Association of MTHFR C677T polymorphism and
type 2 diabetes mellitus (T2DM) susceptibility. Mol Genet Genomic
Med. 7(e1020)2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
World Health Organization The top 10
causes of death available from: http://www.who.int/en/news-room/fact-sheets/detail/the-top-10-causes-of-death
last accessed on 2021 Jun 04.
|
|
3
|
Pradeepa R and Mohan V: Epidemiology of
type 2 diabetes in India. Indian J Ophthalmol. 69:2932–2938.
2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Golden SH, Yajnik C, Phatak S, Hanson R
and Knowler WC: Racial/ethnic differences in the burden of type 2
diabetes over the life course: A focus on the USA and India.
Diabetologia. 62:1751–1760. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
International Diabetes Federation IDF
Diabetes Atlas. 9th edition. International Diabetes Federation,
Belgium, 2019.
|
|
6
|
Magliano DJ, Boyko EJ and Atlas ID: What
is diabetes? In IDF Diabetes Atlas. 10th edition, 2021.
International diabetes federation. Available from: https://diabetesatlas.org/idfawp/resourcefiles/2021/07/IDF_Atlas_10th_Edition_2021.pdf.
|
|
7
|
Sathish T, Thankappan KR, Panniyammakal J
and Oldenburg B: Knowledge of diabetes among adults at high risk
for type 2 diabetes in the Trivandrum district of Kerala, India.
Diabetology. 4:76–85. 2023.
|
|
8
|
Galicia-Garcia U, Benito-Vicente A, Jebari
S, Larrea-Sebal A, Siddiqi H, Uribe KB, Ostolaza H and Martín C:
Pathophysiology of type 2 diabetes mellitus. Int J Mol Sci.
21(6275)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Roden M and Shulman GI: The integrative
biology of type 2 diabetes. Nature. 576:51–60. 2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kyrou I, Tsigos C, Mavrogianni C, Cardon
G, Van Stappen V, Latomme J, Kivelä J, Wikström K, Tsochev K,
Nanasi A, et al: Sociodemographic and lifestyle-related risk
factors for identifying vulnerable groups for type 2 diabetes: A
narrative review with emphasis on data from Europe. BMC Endocr
Disord. 20:1–3. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yahaya TO and Salisu TF: A review of type
2 diabetes mellitus predisposing genes. Curr Diabetes Rev.
16:52–61. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wu L, Zhang Y, Zhao H, Rong G, Huang P,
Wang F and Xu T: Dissecting the association of apolipoprotein E
gene polymorphisms with type 2 diabetes mellitus and coronary
artery disease. Front Endocrinol (Lausanne).
13(838547)2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhao F, Wu L, Wang Q, Zhao X, Chen T, Yin
C, Yan L and Yang X: Insulin-like growth factor 2 mRNA-binding
protein 2-regulated alternative splicing of nuclear factor 1 C-type
causes excessive granulosa cell proliferation in polycystic ovary
syndrome. Cell Prolif. 55(e13216)2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Witka BZ, Oktaviani DJ, Marcellino M,
Barliana MI and Abdulah R: Type 2 diabetes-associated genetic
polymorphisms as potential disease predictors. Diabetes Metab Syndr
Obes. 12:2689–2706. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Berumen J, Orozco L, Betancourt-Cravioto
M, Gallardo H, Zulueta M, Mendizabal L, Simon L, Benuto RE,
Ramírez-Campos E, Marin M, et al: Influence of obesity, parental
history of diabetes, and genes in type 2 diabetes: A case-control
study. Sci Rep. 9(2748)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wu QJ, Zhang TN, Chen HH, Yu XF, Lv JL,
Liu YY, Liu YS, Zheng G, Zhao JQ, Wei YF, et al: The Sirtuin family
in health and disease. Signal Transduct Target Ther.
7(402)2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Khalil MM, Kasem HE and Genena SE:
Association of insulin receptor (INR) gene rs2252673 and Sirtuin1
rs7069102 polymorphisms with diabetic nephropathy in patients with
type 2 diabetes mellitus. Hum Gene. 33(201078)2022.
|
|
18
|
Ramirez A, Hernandez M, Suarez-Sanchez R,
Ortega C, Peralta J, Gomez J, Valladares A, Cruz M, Vázquez-Moreno
MA and Suárez-Sánchez F: Type 2 diabetes-associated polymorphisms
correlate with SIRT1 and TGF-β1 gene expression. Ann Hum
Genet. 84:185–194. 2020.
|
|
19
|
Li J, Yang Y, Xia Y, Luo S, Lin J, Xiao Y,
Li X, Huang G, Yang L, Xie Z and Zhou Z: Effect of SIRT1
gene single-nucleotide polymorphisms on susceptibility to type 1
diabetes in a Han Chinese population. J Endocrinol Invest.
47:819–826. 2024.PubMed/NCBI View Article : Google Scholar
|
|
20
|
El Taweel ME and Ali AA: Association of
the type 2 diabetes mellitus susceptibility gene (IGF2BP2)
with schizophrenia in an Egyptian sample. Middle East Curr
Psychiatry. 24:55–62. 2017.
|
|
21
|
Sharif FA, Shubair ME, Zaharna MM, Ashour
MJ, Altalalgah IO, Najjar M and Thalathini M: Genetic polymorphism
and risk of having type 2 diabetes in a Palestinian population: A
study of 16 gene polymorphisms. Insulin. 3:1–6. 2018.
|
|
22
|
Tarnowski M, Bujak J, Kopytko P, Majcher
S, Ustianowski P, Dziedziejko V, Safranow K and Pawlik A: Effect of
FTO and IGF2BP2 gene polymorphisms on duration of pregnancy
and Apgar scores in women with gestational diabetes. J Obstet
Gynaecol. 39:151–156. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Nfor ON, Ndzinisa NB, Tsai MH, Hsiao CH
and Liaw YP: Interactive effect of IGF2BP2 rs4402960
variant, smoking, and type 2 diabetes. Diabetes Metab Syndr Obes.
13:5097–5102. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sargazi S, Nia MH, Saravani R, Shahroudi
MJ, Jahantigh D and Shakiba M: IGF2BP2 polymorphisms as
genetic biomarkers for either schizophrenia or type 2 diabetes
mellitus: A case-control study. Gene Rep. 20(100680)2020.
|
|
25
|
Verma AK, Goyal Y, Bhatt D, Beg MM, Dev K,
Alsahli MA and Rahmani AH: Association between CDKAL1, HHEX,
CDKN2A/2B and IGF2BP2 gene polymorphisms and susceptibility
to type 2 diabetes in Uttarakhand, India. Diabetes Metab Syndr
Obes. 14:23–36. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li Y, He S, Li C, Shen K, Yang M, Tao W,
Yang Y, Shi L and Yao Y: Evidence of association between
single-nucleotide polymorphisms in lipid metabolism-related genes
and type 2 diabetes mellitus in a Chinese population. Int J Med
Sci. 18(356)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Falih Z, Khodair BA, Mohammed NI and
Mohammed TK: Insulin-like growth factor-2 binding protein-2 gene
polymorphisms in Iraqi patients with type 2 diabetes mellitus. Open
Access Maced J Med Sci. 10:1178–1183. 2022.
|
|
28
|
Ajenah T, Al-Gazally ME and Bayati AA:
Lack of association between IGF2BP2 genes (rs4402960 and
rs11705701) polymorphism and type two diabetes mellitus. J Kufa
Chem Sci. 3:11–22. 2023.
|
|
29
|
Zhou W, Gao Q, He C, Wang L, Wang Y, Feng
L, Li W, Liu W, Ma R and Liu L: Association between polymorphism in
diabetes susceptibility gene insulin-like growth factor
2mRNA–binding protein 2 and risk of diffuse large B-cell lymphoma.
Clin Med Insights Oncol. 17(11795549231201128)2023.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Vatankhah Yazdi K, Kalantar SM, Houshmand
M, Rahmanian M, Manaviat MR, Jahani MR, Kamalidehghan B and
Almasi-Hashiani A: SLC30A8, CDKAL1, TCF7L2, KCNQ1 and
IGF2BP2 are associated with type 2 diabetes mellitus in
Iranian patients. Diabetes Metab Syndr Obes. 13:897–906.
2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Mahmoud AA, Moghazy HM and Ezat MA:
Association of sirtuin 1 gene single nucleotide polymorphisms with
type 2 diabetes mellitus in essential hypertension patients. Meta
Gene. 10:8–12. 2016.
|
|
32
|
Meneguette MV, Oliveira CA, Lima MH, Pina
KN and Amaral ME: Polymorphism in the SIRT1 gene and
parameters of metabolic syndrome in a sample of the adult Brazilian
population. Rev Nutr. 29:1–10. 2016.
|
|
33
|
Wozny L, Stefanowicz M, Danikiewicz M and
Grzeszczak W: The influence of rs2273773 and rs7895833 SIRT1
gene polymorphisms on life expectancy in context of metabolic
factors in Silesian population. Ann Acad Med Siles. 71:162–172.
2017.
|
|
34
|
Dmitrenko OP, Karpova NS and Nurbekov MK:
Association of polymorphisms rs1801282 of the PPARG gene, rs8192678
of the PPARGC1A gene and rs7895833 of the SIRT1 gene with
the risk of preeclampsia in pregnant women with gestational
diabetes in the Russian population. J Genet Genomic Sci.
7(2)2022.
|
|
35
|
Tao TT, Lin XH, Tang SJ, Gui WW, Zhu WF
and Li H: Association of genetic variants in the Sirt1 and
Nrf2 genes with the risk of metabolic syndrome in a Chinese
Han population. BMC Endocr Disord. 22(84)2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Himanshu D, Ali W and Wamique M: Type 2
diabetes mellitus: pathogenesis and genetic diagnosis. J Diabetes
Metab Disord. 19:1959–1966. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Xu X, Shen HR, Zhang JR and Li XL: The
role of insulin-like growth factor 2 mRNA binding proteins in
female reproductive pathophysiology. Reprod Biol Endocrinol.
20(89)2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Cao J, Yan W, Ma X, Huang H and Yan H:
Insulin-like growth factor 2 mRNA-binding protein 2-A potential
link between type 2 diabetes mellitus and cancer. J Clin Endocrinol
Metab. 106:2807–2818. 2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Trinh I, Gluscencova OB and Boulianne GL:
An in vivo screen for neuronal genes involved in obesity identifies
Diacylglycerol kinase as a regulator of insulin secretion. Mol
Metab. 19:13–23. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ali W, Sharaf SAE, Hamid EO and Ali AT:
Association of common variants in the IGF2BP2 gene with type
2 diabetes. Sohag Med J. 25:62–69. 2021.
|
|
41
|
Huang Z, Dong M, Li J, Qiu W and Li S:
Meta-analysis of the association of IGF2BP2 gene rs4402960
polymorphisms with T2DM in Asia. BIO Web Conf. 8(02003)2017.
|
|
42
|
Rao P, Wang H, Fang H, Gao Q, Zhang J,
Song M, Zhou Y, Wang Y and Wang W: Association between
IGF2BP2 polymorphisms and type 2 diabetes mellitus: a
case-control study and meta-analysis. Int J Environ Res Public
Health. 13(574)2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Askin L, Tibilli H, Tanriverdi O and
Turkmen S: The relationship between coronary artery disease and
SIRT1 protein. North Clin Istanb. 7:631–635. 2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Alam F, Syed H, Amjad S, Baig M, Khan TA
and Rehman R: Interplay between oxidative stress, SIRT1,
reproductive and metabolic functions. Curr Res Physiol. 4:119–124.
2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Sadeghi MB, Nakhaee A, Saravani R, Sadeghi
MH, Sargazi S and Nia MH: SIRT1 functional polymorphisms
(rs12778366, rs3758391) as genetic biomarkers of susceptibility to
type 2 diabetes mellitus in Iranians: A case-control study and
computational analysis. Int J Diabetes Dev Ctries. 41:447–455.
2021.
|
|
46
|
Kaabi YA, Abdelmola AA, Abdelwahab SI,
Alshaikh NA, Halawi MA and Kuriri HM: Common genetic variants in
SIRT1 gene promoter and type 2 diabetes mellitus in Saudi
Arabia. Clin Lab: Feb 1, 2024 (Epub ahead of print).
|
|
47
|
Mohammed NI, Alzubaidi ZF and Khudhair M:
The relevance of rs6777038 and rs6444082 of IGF2BP2 gene
polymorphism and type 2 diabetes mellitus: A case control study.
Wiad Lek. 75:2811–2816. 2022.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Liu J, Song G, Zhao G and Meng T: Lack of
association between IGF2BP2 rs4402960 polymorphism and gestational
diabetes mellitus: A case-control study, meta-analysis and trial
sequential analysis. Biosci Rep. 40(BSR20200990)2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Li W, She L, Zhang M, Yang M, Zheng W, He
H, Wang P, Dai Q and Gong Z: The associations of IGF2, IGF2R and
IGF2BP2 gene polymorphisms with gestational diabetes mellitus: A
case-control study. PLoS One. 19(e0298063)2024.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Zhu Z, Zhang Y, Bai R, Yang R, Shan Z, Ma
C, Yang J and Sun D: Association of genetic polymorphisms in
microRNAs with type 2 diabetes mellitus in a Chinese population.
Front Endocrinol (Lausanne). 11(587561)2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Pang S, Zhang Z, Zhou Y, Zhang J and Yan
B: Genetic variants of SIRT1 gene promoter in type 2
diabetes. Int J Endocrinol. 2023(6919275)2023.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Letonja J, Završnik M, Makuc J, Šeruga M,
Peterlin A, Cilenšek I and Petrovič D: Sirtuin 1 rs7069102
polymorphism is associated with diabetic nephropathy in patients
with type 2 diabetes mellitus. Bosn J Basic Med Sci. 21:642–646.
2021.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Li J, Yang Y, Xia Y, Luo S, Lin J, Xiao Y,
Li X, Huang G, Yang L, Xie Z and Zhou Z: Effect of SIRT1 gene
single-nucleotide polymorphisms on susceptibility to type 1
diabetes in a Han Chinese population. J Endocrinol Invest.
47:819–826. 2024.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Deeksha C, Kumar SG, Sangeeta S, Kumar SS
and Kaleem AM: Unraveling epigenetic signatures for early detection
of diabetes nephropathy in type 2 diabetes: A case-control
investigation. Biomed Biotechnol Res J. 8:108–116. 2024.
|
|
55
|
Canto-Cetina T, Silva-Nicanor D,
Coral-Vázquez RM, Cano-Martínez LJ and Canto P: RS3480 polymorphism
of FNDC5/Irisin is associated with type 2 diabetes mellitus
in Maya-Mestizo women. Metab Syndr Relat Disord. 21:503–508.
2023.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Atere AD, Chukwuemeka CE, Oluwatuyi KO and
Olupeka BT: Serum amyloid A as acute phase protein and its
association with dyslipidemia in type 2 diabetes. Biomed Biotechnol
Res J. 7:195–200. 2023.
|
|
57
|
Huang T, Wang L, Bai M, Zheng J, Yuan D,
He Y, Wang Y, Jin T and Cui W: Influence of IGF2BP2, HMG20A, and
HNF1B genetic polymorphisms on the susceptibility to type 2
diabetes mellitus in Chinese Han population. Biosci Rep.
40(BSR20193955)2020.PubMed/NCBI View Article : Google Scholar
|