Introduction
The liver is the largest organ of the mammalian
body, with a highly varied and complex function. The role of the
liver is demonstrated by the fact that despite intensive efforts,
its action cannot be replaced by artificial technology. The liver
contributes to the maintenance of the homeostasis of an organism as
an active bidirectional biofilter. It is essential for protein,
carbohydrate and fat metabolism in the human body (1). Liver cancer is associated with the
third-highest mortality rate worldwide. The global mortality rate
from liver cancer has reached 8.2%. The proper diagnosis of benign
and malignant tumors is essential for effective treatment (2). Liver cancer is the fifth- and
ninth-most prevalent malignancy among both males and females. It is
the second-highest cause of cancer-related mortality worldwide.
Furthermore, secondary liver cancer is more prevalent than primary
liver cancer (3). Hepatocellular
carcinoma (HCC) is a type of primary liver tumor that develops in a
setting of chronic liver disease or cirrhosis. It has been
responsible for the majority of liver cancer diagnosis and deaths.
HCC is associated with a poor prognosis, with a 5-year survival
rate of 20-40%, particularly in individuals with advanced-stage
disease (4).
In Egypt, liver cancer is the fourth most frequent
type of cancer. Numerous hospital researches have revealed an
elevation in the incidence of HCC. This increased incidence may be
due to the advanced technologies used for screening and diagnosis.
These technologies increase the rate of survival of patients
suffering from cirrhosis, those at risk of developing HCC, and
hepatitis C virus (HCV)-related complications. The most common risk
factor for developing liver cancer in Egypt, including HCC is HCV
(5). Several liver function tests
are performed on patients to assess and treat hepatic dysfunction.
Alpha-fetoprotein (AFP) is the most commonly tested biomarker for
monitoring HCC. Other benign or malignant illnesses can cause an
increase in serum AFP levels. The specificity of AFP is hampered by
its presence in other illnesses, such as hepatitis B virus (HBV),
intrahepatic cholangiocarcinoma, and acute and chronic hepatitis
(6).
The most effective techniques for identifying
genomic areas involved in the control of traits of interest are
molecular markers. They also make it easier to select target
genomic regions based on marker genotype rather than concerned
trait phenotype (7). For primer
design, the random amplified polymorphic DNA (RAPD) approach does
not require DNA probes or prior sequence information. As random
primers are widely available, such sequence data information is
irrelevant (8). However, RAPD
analysis has certain limitations and disadvantages, such as the
fact that almost all RAPD markers are dominant, i.e., it is not
possible to distinguish whether a DNA segment is amplified from a
locus that is heterozygous (1 copy) or homozygous (2 copies). The
RAPD technique needs carefully developed laboratory protocols to be
reproducible. In addition, the results from RAPD can be difficult
to interpret. It lacks prior knowledge on the identity of the
amplification products (9).
The use of RAPD has been shown to be successful in
studying genetic polymorphisms of the specific breast cancer
related genes (10). The genomic
alteration of renal tumors was previously illustrated by RAPD
analysis (11). RAPD-PCR analysis
is used to identify genetic alterations and polymorphisms in human
tumors, such as neck and head squamous cell carcinoma, lung cancer,
ovarian cancer, lymphoma, brain tumors, HCC and leukemia (12). RAPD-PCR provides the potential to
generate diagnostic markers for studying genomic instability in
bladder tumors (13). RAPD has
been used to identify a unique pattern of amplified DNA fragments
in the genomic DNA of patients with acute lymphoblastic leukemia
(14). Thus, the present study
aimed to use RAPD to perform the biochemical and molecular
detection of genetic instability in Egyptian patients with liver
cancer, in order to aid in the medical diagnostic sector.
Patients and methods
Clinical specimens
After obtaining approval from the Medical Ethics
Committee of Mansoura University (Mansoura, Egypt; approval no.
MS.22.11.2211), the present prospective study was carried out on 20
patients with liver cancer. Informed consent was obtained for the
publication of their data. Their ages were 45-80 years and they
included both sexes (16 males and 4 females). They were selected
randomly from the Gastroenterology Surgical Center, Mansoura
University. The selected patients had no other malignant tumors.
From April, 2022 to January, 2023, tissue true cut samples and
blood samples were collected from patients with different grades
and stages of liver cancer. From each liver cancer patient, tumor
and normal tissues (as a self-negative control) were collected for
the molecular analysis, while the blood samples were collected for
the biochemical analysis.
Data collection
Patient data, including age and sex were collected
from the files of the patients through the internal network of
Gastroenterology Surgical Center, Mansoura University.
Histopathological
characterization
Liver cancer histopathology of tissues biopsies
including, tumor type, staging and grading was performed.
Etiological data
Data on viral infections, liver inflammation types
and tumor classification were combined through virology
reports.
Radiological examination
Patients with liver cancer underwent an abdominal
ultrasound and a computerized tomography (CT) scan.
Biochemical analysis
Blood was collected from all the patients into
different metal-free safety vacutainer blood-collecting tubes
containing heparin (Kemico) for blood chemistry analysis, including
serum creatinine (Creat), sodium (Na+), potassium
(K+), glucose (Gluc), albumin (Alb), alanine
transaminase (ALT), aspartate aminotransferase (AST), total protein
(TP), total bilirubin (T.Bil) and AFP. The values of these tests
were determined using a Cobas c311 autoanalyzer (Roche Diagnostics)
and reagent cassettes were purchased from Roche Diagnostics Egypt.
The second type of vacutainer blood-collecting tubes containing
K2EDTA (Kemico) were used for the analysis of complete blood count
(CBC), including red blood cells (RBCs), white blood cells (WBCs),
platelets (PLTs) and hemoglobin level (Hb); these were measured
using a Sysmex cell counter (Bioline 6500; Sysmex America, Inc.)
and all the reagents were purchased from Sysmex, Egypt LLC. The
third type of vacutainer blood-collecting tubes contained sodium
citrate (3.2%) as an anti-coagulant (Greiener Bio-One) for the
international normalized ratio (INR) test, and was measured using
the Stago compact coagulation analyzer (Diagnostica Stago, Inc.)
and all the reagents were purchased from Stago, Etico, Egypt. The
normal references of all the biochemical tests were determined from
the data sheet that was provided with the kit of each test.
Molecular analysis
Frozen true cut samples from tumor and normal
tissues were weighed exactly and were grounded homogenously using a
tissue homogenizer (Mechanika Precyzyjna, Poland; https://www.tachografy.kalisz.pl/kontakt/) and DNA was
extracted by Phenol-chloroform isoamylalcohol manual method
(15). The extracted genomic DNA
concentration and purity were assessed by nanodrop
spectrophotometer (Thermo Scientific 2000, USA). The optimum purity
ratio was 1.8-2.0. Extracted pure genomic DNA integrity was
assembled through gel electrophoresis in 0.7% agarose' (Industrias
Roko S.A.). Agarose was boiled in a microwave for 2 min in 1X Tris
Acetate EDTA (TAE) and mixed with 5 µl Midori Green Advance DNA
stain (Nippon Corp.). The extracted genomic DNA was loaded in the
prepared 0.7% agarose lanes and one lane (the first or last lane)
was loaded with lambda/DNA HindIII marker (Sigma-Aldrich;
Merck KGaA). Electrophoresis was performed in a gel electrophoresis
apparatus (Labnet) for 45 min at 100 Vin 1XTAE, and demonstrated on
ultraviolet light box (Fotodyne).
DNA amplification was carried out with five primers
(A-01 to A-05) (Eurofins Genomics) to examine the efficiency of the
primer kit (Table I). The PCR
mixture was prepared in a 200 µl sterile PCR Eppendorf tube by
mixing 1 µg pure genomic DNA, 2 U Taq DNA polymerase enzyme
(Viviantis, Malizia; https://www.vivantechnologies.com/), 30 pm of each
primer, one PCR bead (Enzynomics) and sterile (free of any salts
and microorganisms) and distilled H2O was added to reach
a final volume of 20 µl. The PCR Eppendorf was shaken and was spun
at 13,000 rpm in an Eppendorf centrifuge (Beckman Coulter, Inc.). A
total of 20 µl mineral oil (Merck KGaA) was dropped over the PCR
mixture. The PCR Eppendorf containing the PCR components was
transferred to the PCR machine (Proflex Industries Pte. Ltd.) which
was programmed into three programs. The first program was cycled
for 40 cycles; each cycle was programmed as four steps. The first
step was set at 95˚C for 1 min to denature the DNA (initial), the
second step was set at 95˚C for 1 min to denature the DNA (final),
the third step was set at 30˚C for 1 min to anneal the primer with
DNA strands, and the fourth step was set at 72˚C for 2 min for the
new DNA extension. The first program was linked to the second
program, which was set as one cycle at 72˚C for 7 min for the final
extension of the new DNA copies. The second program was linked to
the third program, which was programmed to terminate the reaction
by keeping the PCR Eppendorf tube at 4˚C in the PCR machine. The
results of RAPD-PCR were analyzed through gel electrophoresis as
previously described (highlighted green) with some modifications:
The agarose gel was prepared as 2%, and the DNA marker (GeneDirex)
was a 1-kb DNA ladder. DNA bands molecular weights were calculated
using GellApp software 1.2.7 android app.
 | Table IKit-A primers sequences. |
Table I
Kit-A primers sequences.
| Name | Sequence |
|---|
| A-01 | 5'-CAGGCCCTTC-3' |
| A-02 | 5'-TGCCGAGCTG-3' |
| A-03 | 5'-AGTCAGCCAC-3' |
| A-04 | 5'-AATCGGGCTG-3' |
| A-05 | 5'-AGGGGTCTTG-3' |
Statistical analysis
GraphPad Prism 5.0 software (Dotmatics) was used for
all statistical analyses. The results were presented as the mean ±
the standard error of the mean (SEM) (n=6), where n is the number
of patients in each group. One-way analysis of variance (ANOVA) was
used to perform statistical comparisons followed by the
Student-Newman-Keuls post hoc test (16). The Chi-squared test and Fisher's
exact test (<20 cases) were used to analyze the association
between genomic instability, histological type and the grade of
liver cancer. A value of P<0.05 was considered to indicate a
statistically significant difference. The GellApp analyzer program
was used to determine the molecular weight of the amplified DNA
fragments including normal and tumor DNA bands by plotting a curve
illustrating the detected fragment molecular weight against the
molecular weight of the marker fragments.
Results
According to the ages and sex of the patients, the
incidence of liver cancer among the male patients was ~4-fold
higher compared with that among the female patients and the age
range of the patients was 45-80 years (Table II). A histopathological
examination of the tumor tissues biopsies was performed, revealing
that HCC comprised the highest frequency (85%) followed by
fibrolamellar carcinoma (10%), while hepatic adenoma (rare type)
exhibited the lowest percentage (5%) (Table III). As regards tumor grading and
staging, grade II exhibited the highest frequency (80%) followed by
grade III (15%) and grade I (5%) (Table IV), while the majority of patients
(40%) had stage I disease followed by stage III (30%) and stage II
(20%); only a minority of patients (10%) had stage IV disease
(Table V). As regards the liver
cancer etiology of the patients, ~50% of patients had fatty liver
cirrhosis, 10% of patients had HBV and 40% of patients had HCV.
According to radiological analyses, when patients underwent
radiological investigations, it was found that the majority of the
patients (85%) had large-sized tumors and the remaining (15%) of
patients had smaller-sized tumors. On the other hand, the majority
of the patients (50%) were suffering from fatty liver cirrhosis,
while in 10% of patients, the lymph nodes were invaded by the tumor
(metastasis) and the remaining 90% of the patients did not have
lymph node metastasis (Table
VI).
 | Table IIAge and sex of the patients. |
Table II
Age and sex of the patients.
| Characteristic | Statistics |
|---|
| Number | 20 |
| Age in years, mean ±
SD (range) | 63.3 (45-80) |
| Sex
(male/female) | 16/4 |
 | Table IIIHistopathological subtypes. |
Table III
Histopathological subtypes.
| Histopathological
subtypes | No. of patients | % |
|---|
| Hepatocellular
carcinoma | 17 | 85 |
| Fibrolamellar
carcinoma | 2 | 10 |
| Hepatic
adenoma | 1 | 5 |
 | Table IVTumor grade. |
Table IV
Tumor grade.
| Tumor grades | No. of
patients | % |
|---|
| Grade I | 1 | 5 |
| Grade II | 16 | 80 |
| Grade III | 3 | 15 |
 | Table VTumor stage. |
Table V
Tumor stage.
| Tumor stage | No. of
patients | % |
|---|
| Stage I | 8 | 40 |
| Stage II | 4 | 20 |
| Stage III | 6 | 30 |
| Stage IV | 2 | 10% |
 | Table VIRadiology results. |
Table VI
Radiology results.
| Parameter | No. of patients
(%) |
|---|
| Tumor size | |
|
Small (≤3
cm) | 3(15) |
|
Large (>3
cm) | 17(85) |
| Lymph node
invasion | |
|
Negative | 18(90) |
|
Positive | 2(10) |
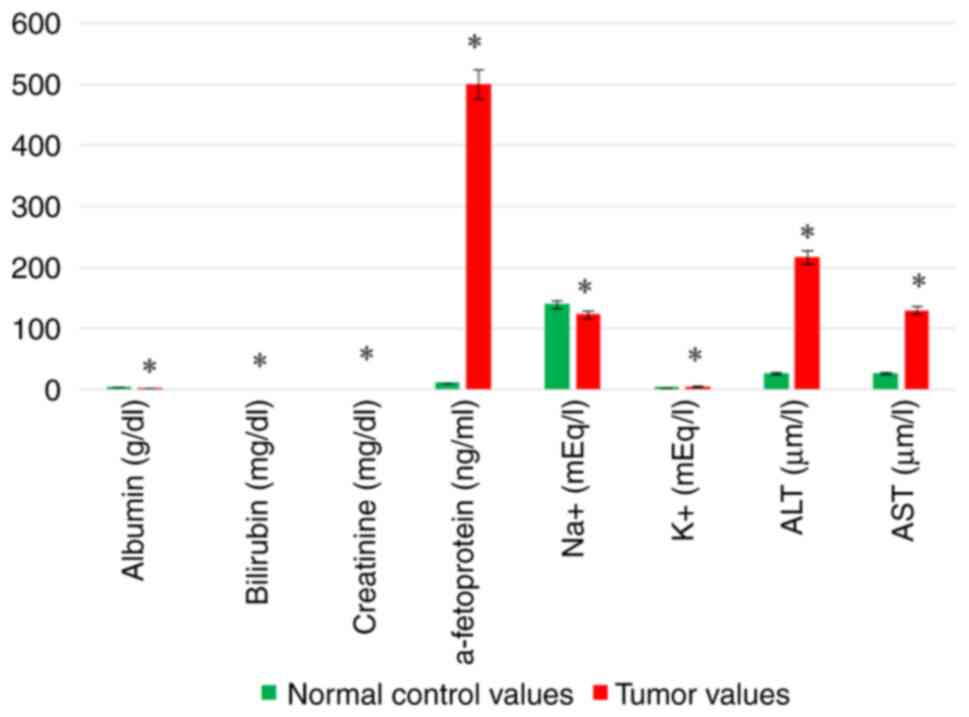
Biochemical analysis of the patients with liver
cancer included chemical and hematological analyses. Chemical
analysis revealed a significant decrease in both serum Alb and
Na+ levels. A significant increase in serum Bil, Creat,
K+, AFP, ALT and AST levels was recorded in the tumor
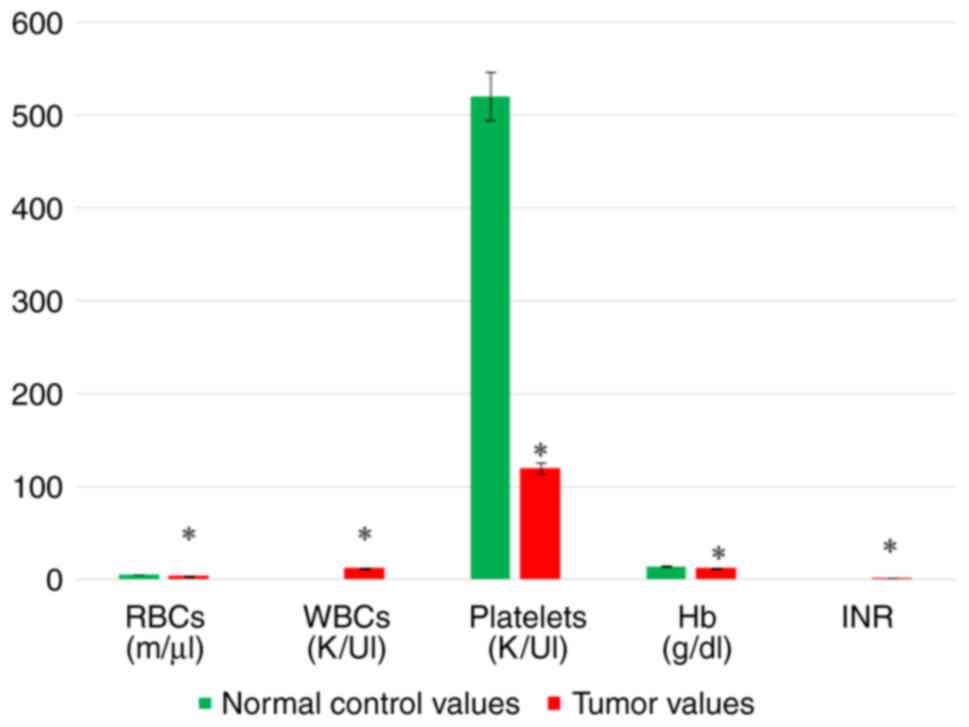
tissues compared with the normal tissues (Table VII and Fig. 1). Hematological parameters included
CBC and INR analyses of the patients with liver cancer. The CBC
analyses exhibited a significant decrease in the RBC count, PLT
count and Hb content in the tumor tissues compared with the normal
control values, whereas the WBC count was significantly increased
in the tumor tissues compared with the normal tissues. Moreover,
the INR level exhibited a significant increase in the tumor tissues
(Table VII and Fig. 2).
 | Table VIIResults of biochemical (hematologic
and chemical) analyses. |
Table VII
Results of biochemical (hematologic
and chemical) analyses.
| Variable | Normal control
values (n=20) | Tumor values
(n=20) | P-value |
|---|
| RBCs | 5.253±0.5446 | 3.421±1.070 | 0.0001 |
| WBCs | 5.11 2±0.4996 | 12.30±2.577 |
<0.0001 |
| PLTs | 520.6±71.99 | 119.8±34.79 |
<0.0001 |
| Hb | 13.90±1.148 | 11.71±0.7876 |
<0.0001 |
| INR | 1.009±0.07549 | 1.470±0.2214 | 0.0001 |
| Alb | 4.440±0.4037 | 2.720±0.7463 | 0.0019 |
| Bil | 0.8167±0.2229 | 1.60±0.3633 | 0.0011 |
| Creat | 0.7067±0.04274 | 1.563±0.3275 |
<0.0001 |
| AFP | 10.98±2.217 | 500.4±132.7 |
<0.0001 |
| Na+ | 139.8±3.435 | 123.8±7.997 |
<0.0001 |
| K+ | 3.600±0.8380 | 5.740±0.7397 |
<0.0001 |
| ALT | 27.60±10.21 | 217.0±44.18 |
<0.0001 |
| AST | 27.20±5.263 | 130.4±42.00 | 0.0006 |
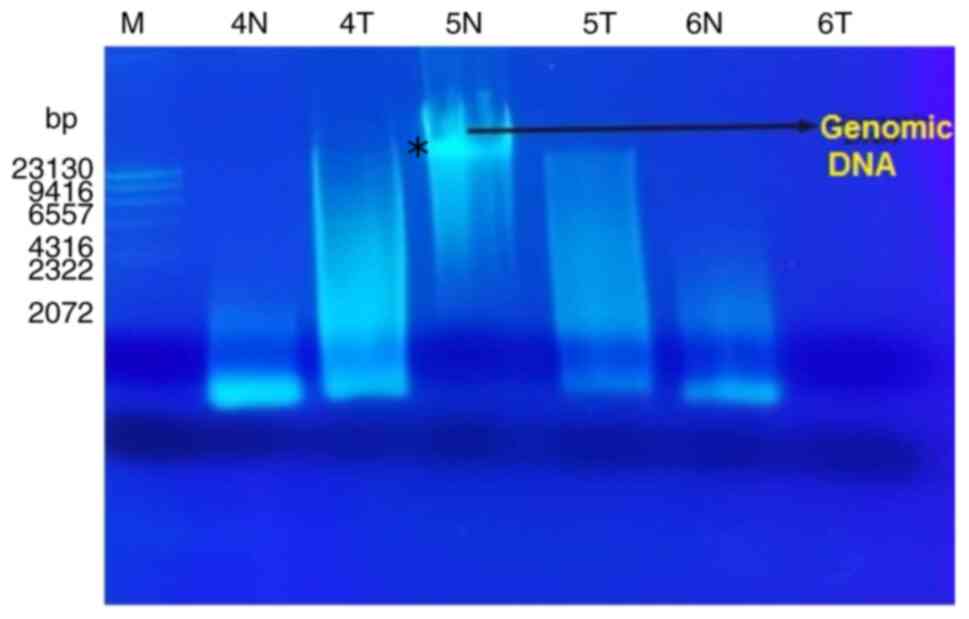
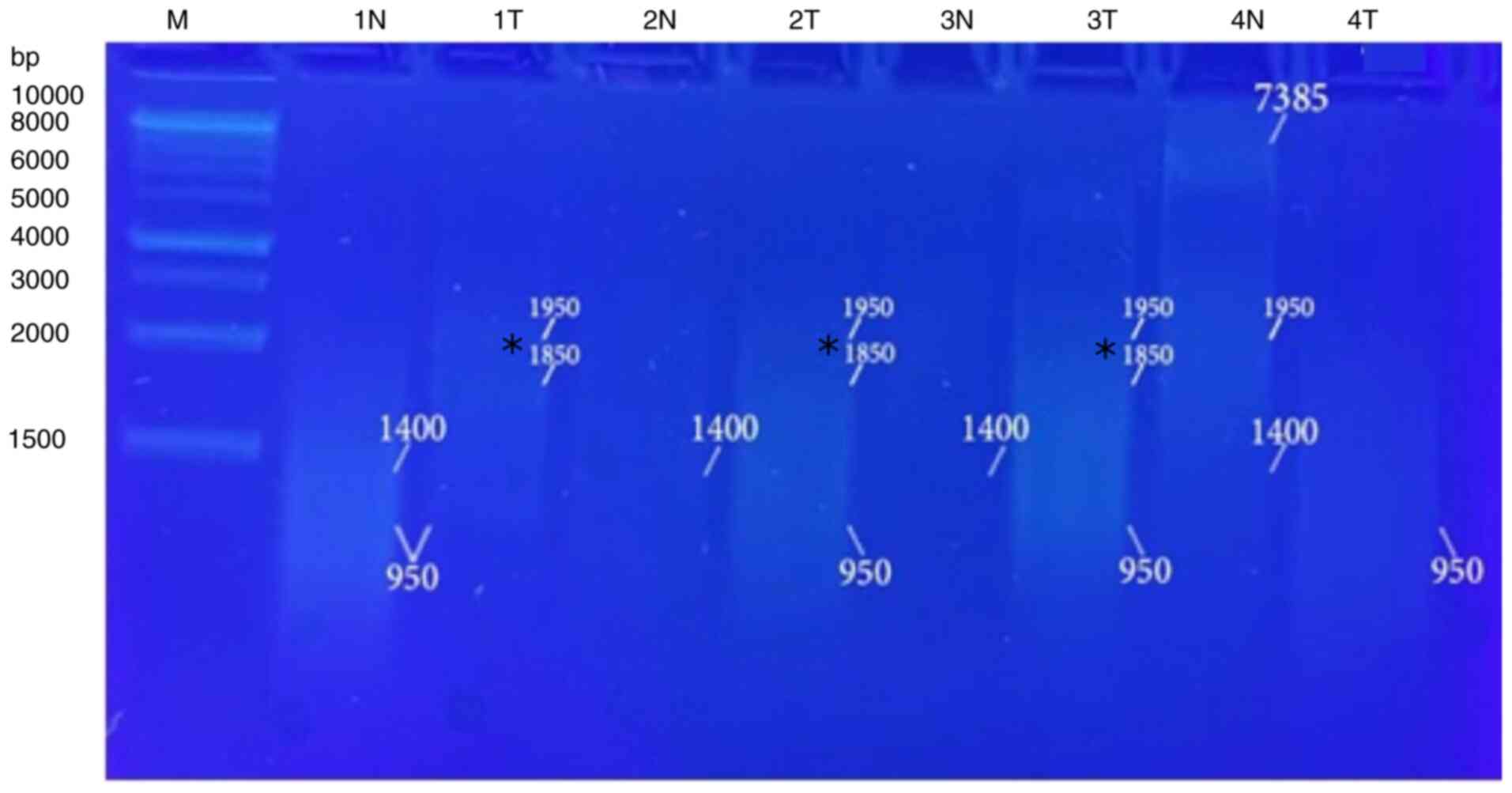
The results of molecular analysis included extracted
genomic DNA profile and RAPD-PCR pattern profile. Purified
extracted DNA of the patients with liver cancer was run on 0.7%
agarose gels (Fig. 3). RAPD-PCR
was used for analysis with five distinct primers of A-01 to A-05
(Table I). Two primers (A-01 and
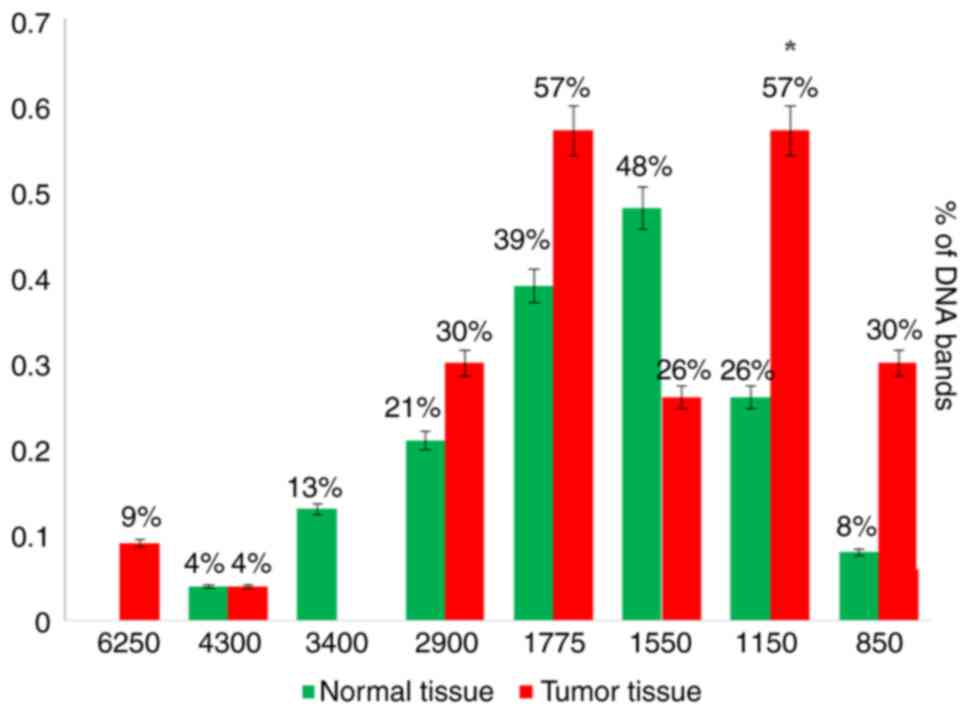
A-03) were amplified and presented clear and firm bands with the
majority of the tested samples. The A-01 primer revealed several
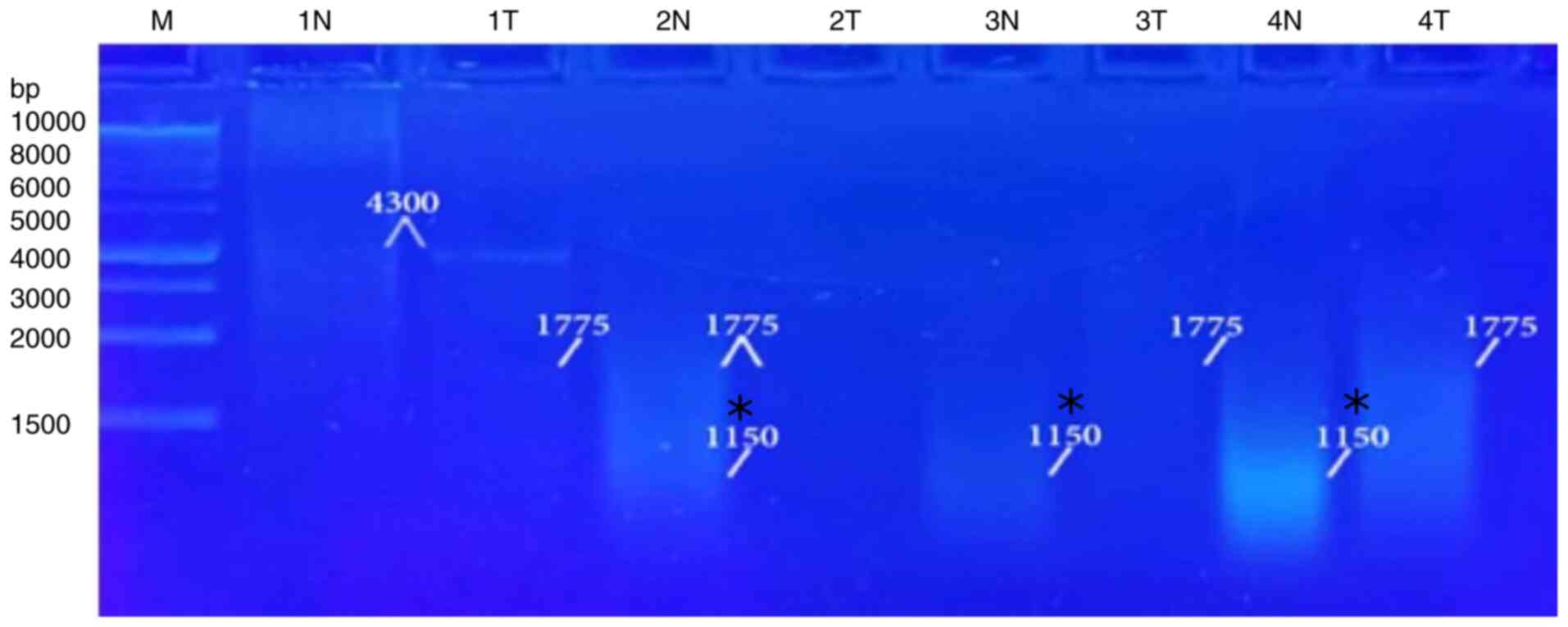
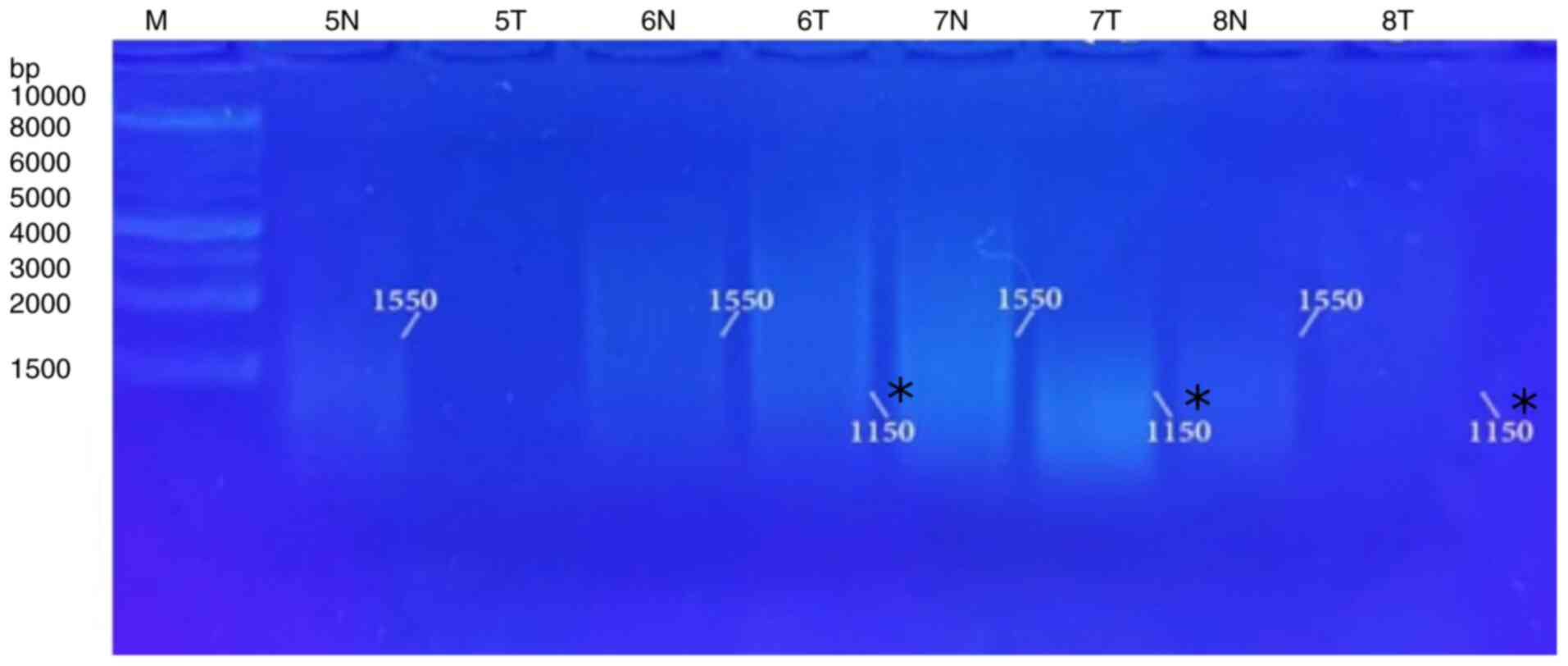
bands and eliminated others when used in the RAPD-PCR approach. A
6,250-bp band was observed in 9% of the tumor tissue samples and
was completely absent in all the normal tissue samples. While it
was presented in 13% of all the normal tissue samples, the 3,400-bp
band was completely absent from all the malignant tissue samples.
In 4, 21, 39, 48 and 8%, respectively of the normal tissue samples,
and in 4, 30, 57, 26 and 30%, respectively of the tumor tissue
samples, five common bands 4,300, 2,900, 1,775, 1,550 and 850 bp
were observed. A distinctive significant appearance (P=0.036071) of
a tumor marker band of 1,150 bp in 26% of the normal tissue
samples, and in 57% of the tumor tissue samples was noted (Fig. 4, Fig.
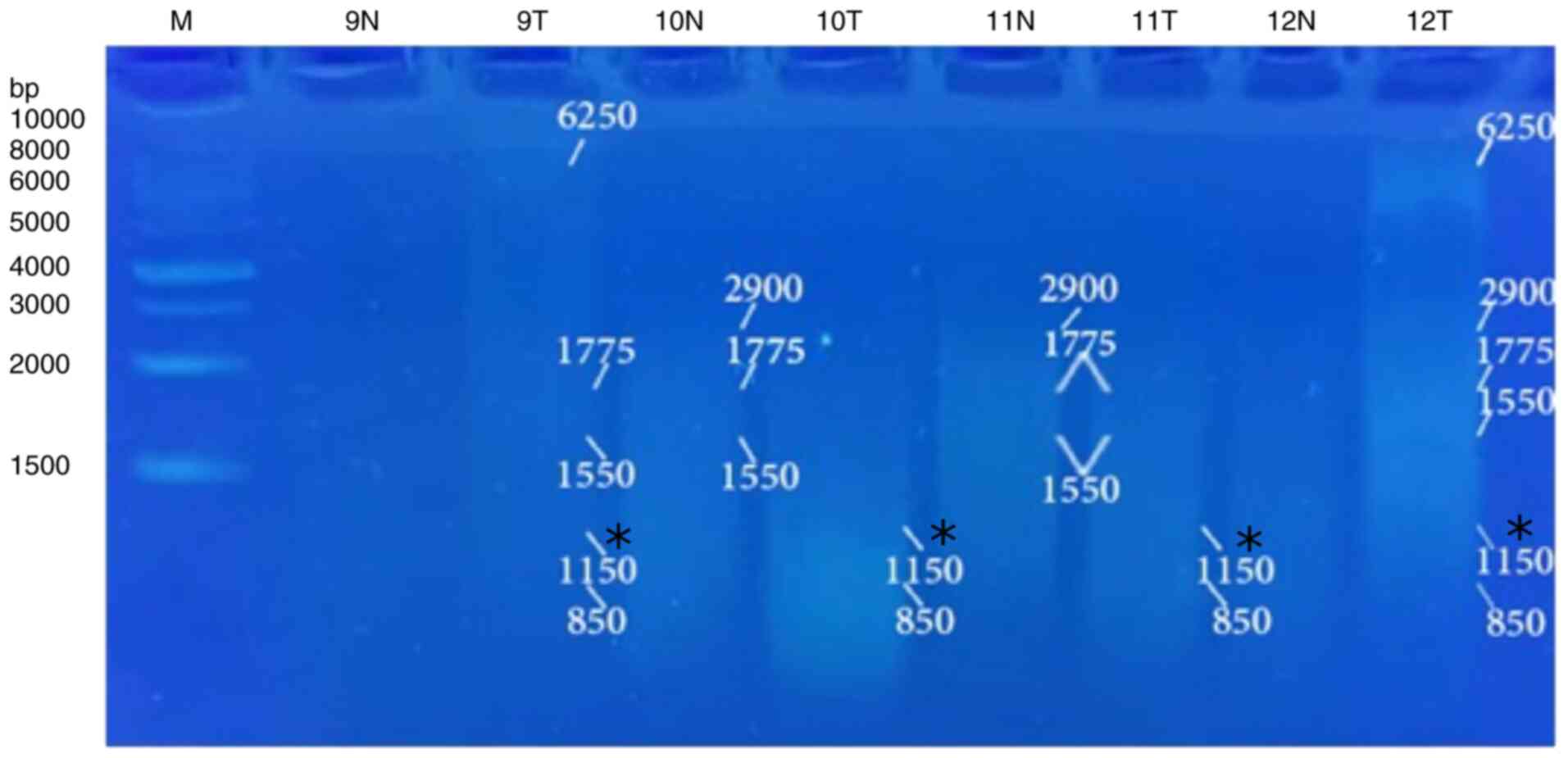
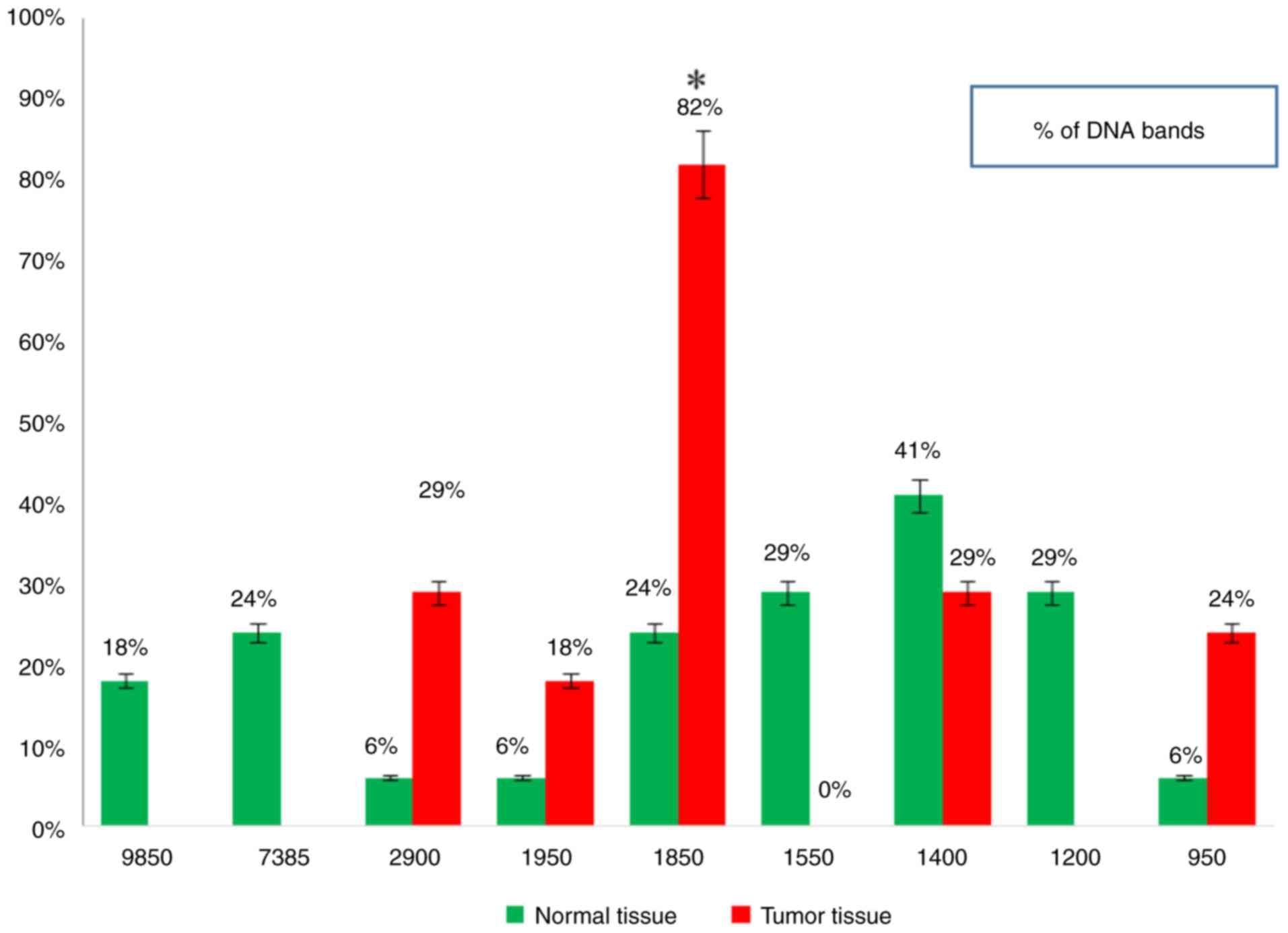
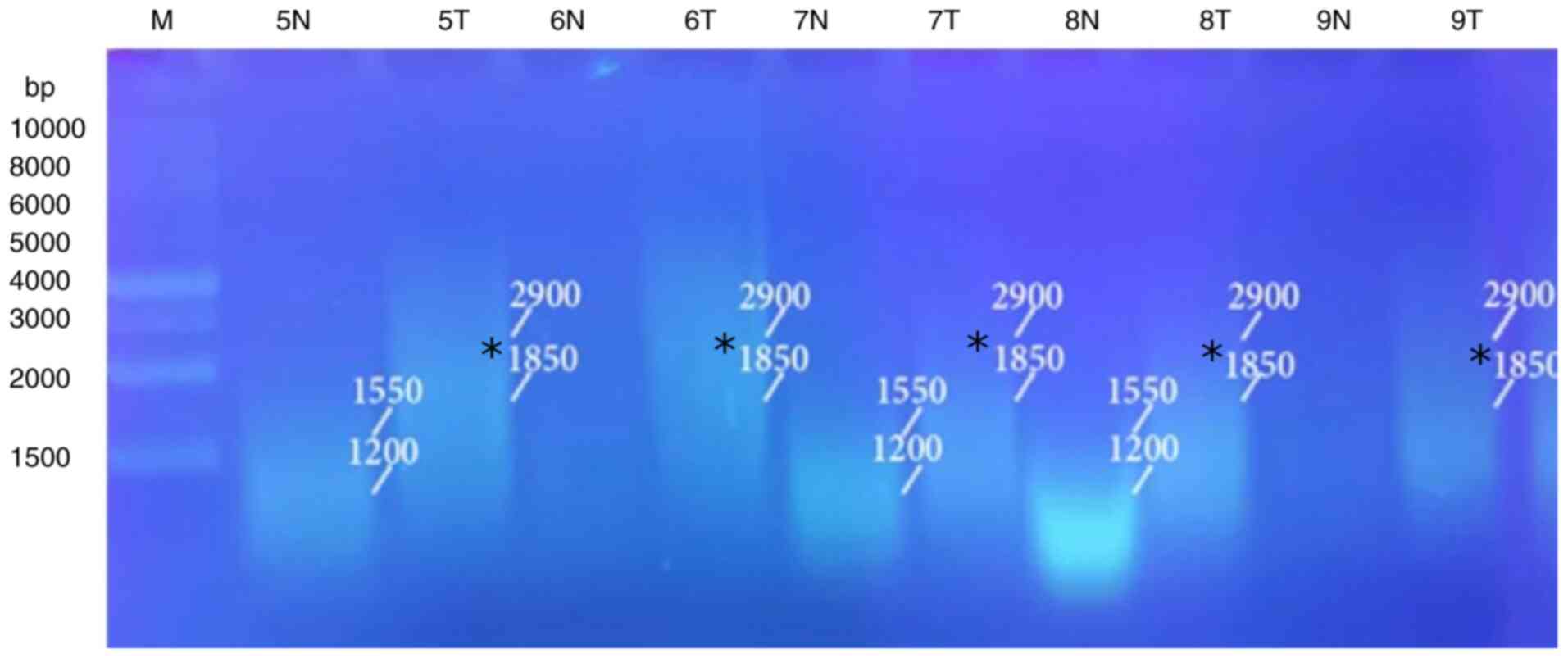
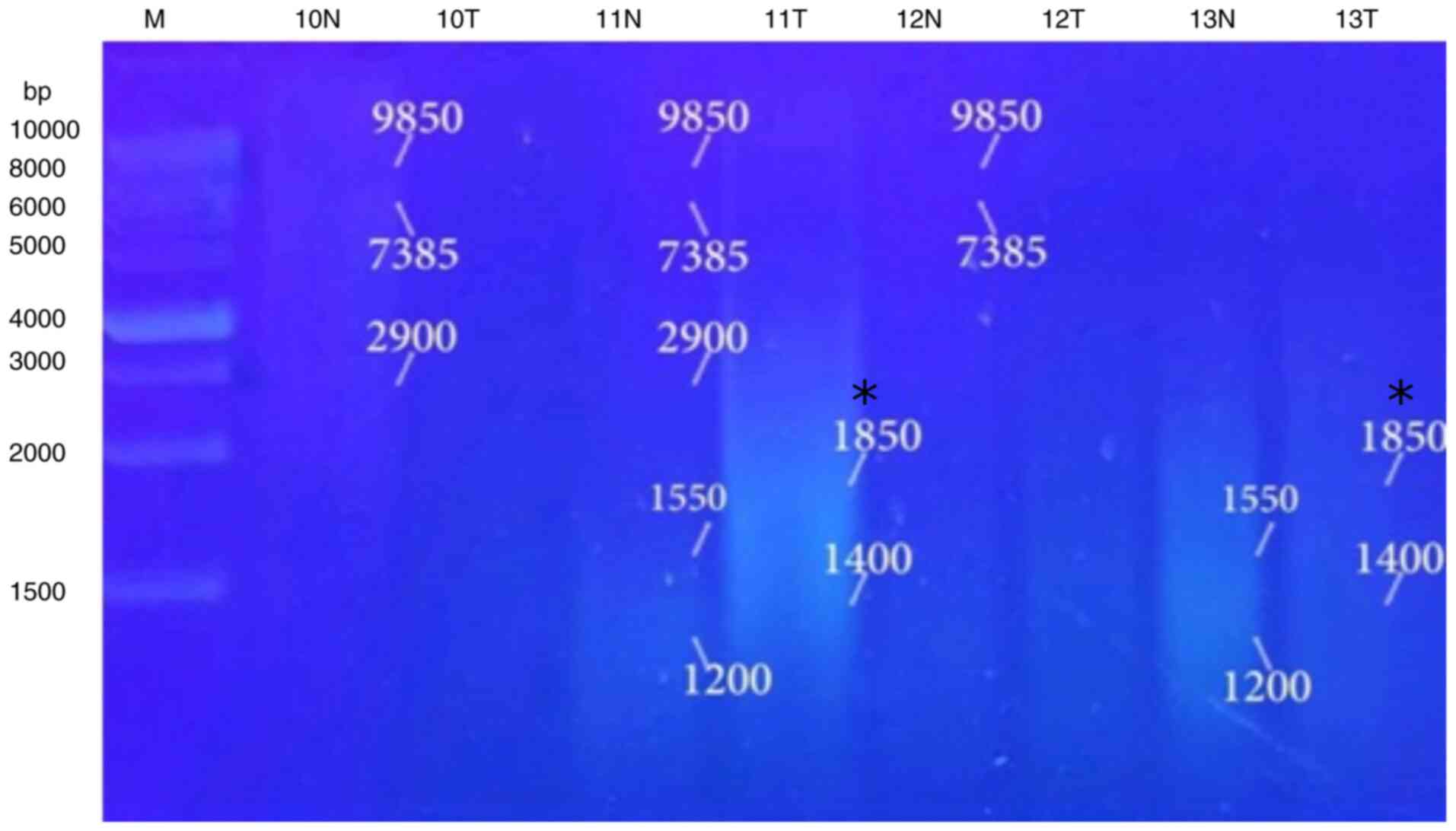
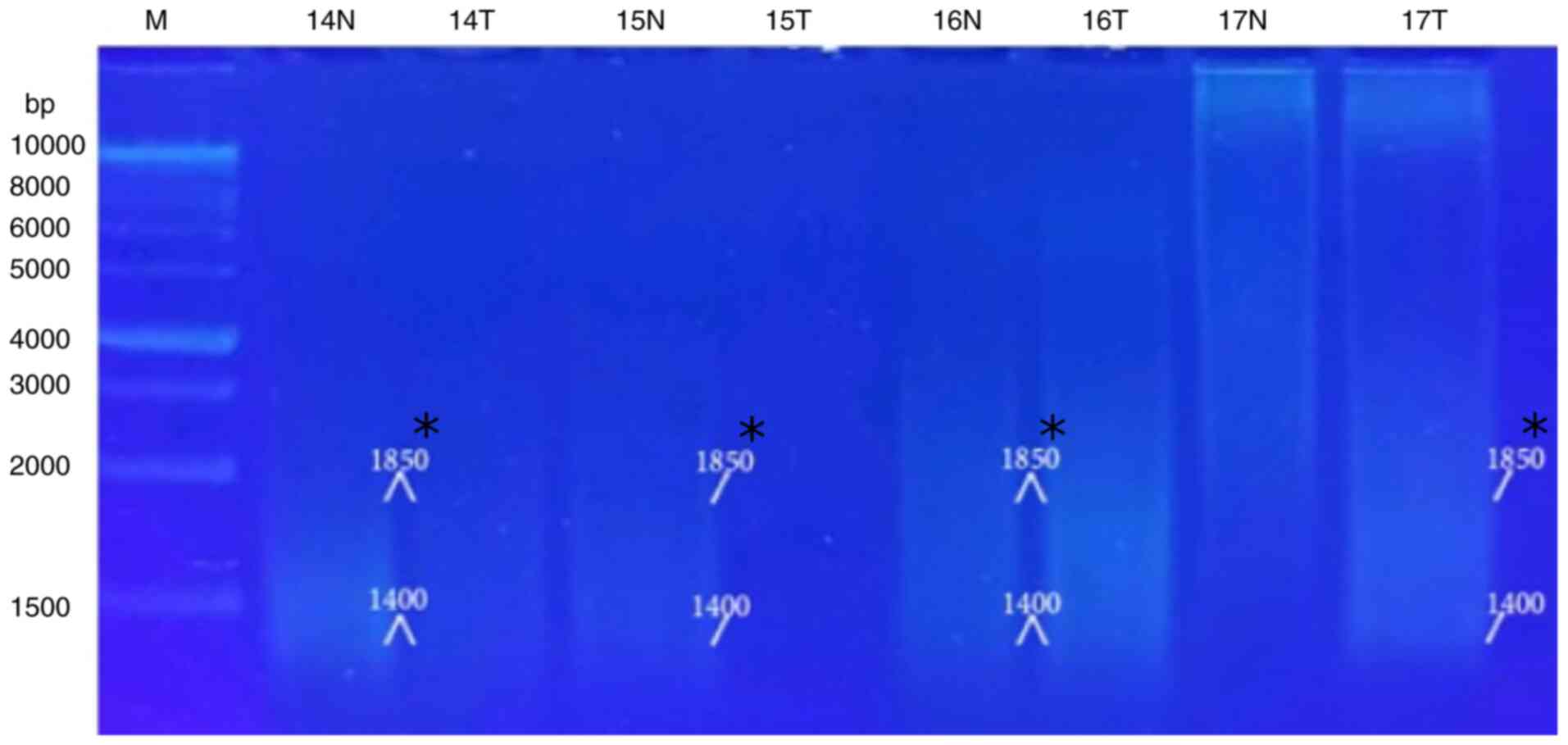
5, Fig. 6, Fig. 7 and Fig. 8 and Table VIII). When primer the A-03 was
employed in the RAPD-PCR approach, four bands of 9,850, 7,385,
1,550 and 1,200 bp were detected in 18, 24, 29 and 29%,
respectively of all the normal tissue samples however; they were
completely absent from all malignant tissue samples. The four
common bands 2,900, 1,950, 1,400 and 950 bp were found in 6, 6, 41
and 6 of the normal tissue samples, respectively, and in 29, 18, 29
and 24% of the tumor tissue samples, respectively. A highly
significant appearance (P=0.000591) of a tumor marker band of 1,850
bp was observed in 24% of the normal tissue samples, and in 82% of
the tumor tissue samples (Fig. 9,
Fig. 10, Fig. 11, Fig. 12 and Fig. 13 and Table IX).
 | Table VIIIStatistical analysis of amplified DNA
fragments generated using the A-01 primer for liver tumor and
normal tissues. |
Table VIII
Statistical analysis of amplified DNA
fragments generated using the A-01 primer for liver tumor and
normal tissues.
| DNA band molecular
weight (bp) | Tumor tissue | Normal tissue | |
|---|
| | No. of
patients | % | No. | % | χ² | P-value |
|---|
| 6,250 | 2 | 9 | 0 | 0.0 | 0.3566 | 0.550407 |
| 4,300 | 1 | 4 | 1 | 4 | 0.0 | 0.999 |
| 3,400 | 0 | 0.0 | 3 | 13 | 1.0952 | 0.295314 |
| 2,900 | 7 | 30 | 5 | 21 | 0.451 | 0.50187 |
| 1,775 | 13 | 57 | 9 | 39 | 1.3939 | 0.237741 |
| 1,550 | 6 | 26 | 11 | 48 | 2.3327 | 0.126685 |
| 1,150 | 13 | 57 | 6 | 26 | 4.3938 |
0.036071 |
| 850 | 7 | 30 | 2 | 8 | 3.4535 | 0.06312 |
 | Table IXStatistical analysis of amplified DNA
fragments which generated by A-03 primer of liver tumor and normal
tissues. |
Table IX
Statistical analysis of amplified DNA
fragments which generated by A-03 primer of liver tumor and normal
tissues.
| DNA band molecular
weight (bp) | Tumor tissue | Normal tissue | |
|---|
| | No. of
patients | % | No. | % | χ² | P-value |
|---|
| 9,850 | 0 | 0.0 | 3 | 18 | 1.1333 | 0.287065 |
| 7,385 | 0 | 0.0 | 4 | 24 | 2.1103 | 0.146306 |
| 2,900 | 5 | 29 | 1 | 6 | 3.2381 | 0.071944 |
| 1,950 | 3 | 18 | 1 | 6 | 1.1333 | 0.287065 |
| 1,850 | 14 | 82 | 4 | 24 | 11.8056 |
0.000591 |
| 1,550 | 0 | 0.0 | 5 | 29 | 3.2381 | 0.071944 |
| 1,400 | 5 | 29 | 7 | 41 | 0.5152 | 0.472917 |
| 1,200 | 0 | 0.0 | 5 | 29 | 3.2381 | 0.071944 |
| 950 | 4 | 24 | 1 | 6 | 2.1103 | 0.146306 |
Discussion
In the present study, the ages of the patients
ranged between 45-80 years and included both sexes, where the male
sex was ~4-fold more common than females combined with a previous
hypothesis (17,18). In addition, the present study
demonstrated that the HCC type comprised the highest frequency
(85%) followed by the mixed type (15%) and fibrolamellar carcinoma
(10%), while hepatic adenoma (rare type) was the lowest percentage
(5%). Another study indicated that 75-85% of cases were HCC,
followed by intrahepatic cholangiocarcinoma and the remaining cases
were less common tumors; grade II liver cancer comprised the
majority of cases (80%), while grade I cancer comprised the
minority (5%) (19). On the
contrary, previous research has revealed a significantly high
percentage of grade I and a low percentage of grade III liver
cancer tumors (20).
Compared to previous findings, the present study
demonstrated that 40% of the patients were in stage I, while in
another study, only 4.6% of patients had stage I disease (21). The present study conducted the
radiological analysis of patients with liver cancer using CT scan
and abdominal ultrasound. It was revealed that 85% of the patients
had large-sized tumors and the remaining 15% had smaller-sized
tumor. To a certain extent, these results are in accordance with
the findings of another study where 25% of the liver cancer lesions
are <3 cm and 50% of the tumors were >3 cm in diameter
(22). The present study found
that fatty liver cirrhosis comprised 50% of the cases, HBV
comprised 10% and HCV comprised 40% of the cases. Of note, in
another study, 95.7% of the cases had HCV or HBV infection, with
the incidence of HCV predominating (91.4%) (23). Egypt has the highest HCV prevalence
rate worldwide, with rates of 6 to 28% (23). The hematological findings of
patients with liver cancer have been demonstrated in previous
studies (24-26);
in these patients, RBCs, the PLT count and Hb content were
significantly decreased compared to normal values, while the number
of WBCs was significantly increased (24-26).
Previous studies (27,28) have confirmed the biochemical
observations of the present study where, serum Alb and sodium
levels were significantly decreased in the serum of patients with
liver cancer compared to normal values. However, the values of
serum T.Bil, serum Creat, potassium, serum AFP, serum ALT, AST and
the plasma INR were all significantly elevated and this finding has
been corroborated by previous studies (29-34).
The present study used the RAPD-PCR approach to
demonstrate the genomic instability of Egyptian patients suffering
from liver cancer by losing normal bands and revealing new
tumor-related bands. Additionally, other researchers have supported
these findings, where the RAPD investigation of patients with HCC
revealed loss or gain of bands, change in intensity and shifting in
DNA band/s of the tumor lane compared to the corresponding healthy
DNA (35). Genetic alterations
were revealed in HCC that were discovered by one primer at least,
and the genetic mutations detectability was ranged from 20 to 70%
in each instance and 17.9 to 50% of each primer (12). In a previous study on CCA, the RAPD
analysis of chromosomes 2p25.3 and 7q11.23 appeared consistent DNA
fingerprints of tumor and normal tissue from the same patient.
Normal and tumor bands intensities were compared to examine changes
in DNA fingerprints (36).
Furthermore, that study discovered that the amplified CCA accounted
for >70% of the alterations in the genome. The significant
prevalence gain of the CCA sequence may suggest that changes in
gene copy numbers play a crucial role in this type of cancer
(36).
In the present study, the RAPD-PCR analysis of the
tumors of patients with liver cancer and normal tissues by using
the A-01 primer revealed promising results, when a distinctive
significant appearance (P=0.036071) of a tumor marker band of 1,150
bp appeared in 26% of the normal tissue samples and in 57% of tumor
tissue samples. Another notable result was observed with the A-03
primer, where a highly significant appearance (P=0.000591) of a
tumor marker band of 1,850 bp was observed in 24% of the normal
tissue samples and in 82% of the tumor tissue samples. A number of
DNA bands appeared in the tumor and/or normal tissues; however,
none of these produced any significant findings.
In conclusion, the present study performed
biochemical, hematological, histopathological and radiological
analyses examine the phenotype of the cell. Thus, sharp and
accurate technology acting on the genotype of the cancer cell, such
as RAPD-PCR, is required to discover genetic instability. The
present study indicated that RAPD-PCR analysis succeeded in
producing diagnostic marker primers (A-01 and A-03) and tumor
marker bands specific for liver cancer. The present study provides
evidence of the potential promise of RAPD-PCR analysis as a
diagnostic molecular genetic marker technology, apart from the
traditional diagnostic approaches. However, the present study was a
preliminary study, and thus further studies are required in the
future to confirm these findings.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AZM conceived the study, managed the manuscript and
performed the RAPD-PCR technique. EKA performed the hematological
and biochemical analyses. AMA provided the tissue samples and
analyzed the medical reports. SFE performed the statistical
analysis and confirmed the authenticity of the raw data. All
authors have read and approved the final manuscript. AZM and EKA
confirm the authenticity of the raw data.
Ethics approval and consent to
participants
On April 8, 2023, informed consent was obtained from
all participants and/or their legal guardians, while approval was
obtained from the Ethics Committee of the Faculty of Medicine,
Mansoura University, Mansoura, Egypt (Approval no. MS.22.11.2211).
All methods were performed in accordance with the relevant
guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Arias IM, Alter HJ, Boyer JL, Cohen DE,
Shafritz DA, Thorgeirsson SS and Wolkoff AW: The Liver: Biology and
Pathobiology. 6th edition. Wiley-Blackwell, Hoboken, NJ,
2020.
|
|
2
|
Pan F, Huang Q and Li X: Classification of
liver tumors with CEUS based on 3D-CNN. In: 2019 IEEE 4th
international conference on advanced robotics and mechatronics
(ICARM). IEEE 845-849, 2019.
|
|
3
|
Wang ZG, He ZY, Chen YY, Gao H and Du XL:
Incidence and survival outcomes of secondary liver cancer: A
surveillance epidemiology and end results database analysis. Transl
Cancer Res. 10:1273–1283. 2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Shannon AH, Ruff SM and Pawlik TM: Expert
insights on current treatments for hepatocellular carcinoma.
Clinical and molecular approaches and bottlenecks to progress. J
Hepatocell Carcinoma. 9:1247–1261. 2022.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Rashed WM, Kandeil MAM, Mahmoud MO and
Ezzat S: Hepatocellular carcinoma (HCC) in Egypt. A comprehensive
overview. J Egypt Natl Can Inst. 32(5)2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Piñero FF, Dirchwolf M and Pessôa MG:
Biomarkers in hepatocellular carcinoma: Diagnosis, prognosis and
treatment response assessment. Cells. 9(1370)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Jahnke G, Smidla J and Poczai P:
MolMarker: A simple tool for DNA fingerprinting studies and
polymorphic information content calculation. Diversity.
14(497)2022.
|
|
8
|
Al-Samarai FR and Al-Kazaz AA: Molecular
markers: An introduction and applications. Eur J Mol Biotechnol.
9:118–130. 2015.
|
|
9
|
Senthil N and Gurusubramanian G: Random
amplified polymorphic DNA (RAPD) markers and its applications. Sci
Vis. 11:116–124. 2011.
|
|
10
|
Allami ZZG and Dragh MA: Identification of
some breast cancer related genes by RAPD technique in Maysan
Province Iraq. Revis Bionatura. 7:20–28. 2022.
|
|
11
|
El-Far M, Abol-Enein H, Zakaria A and
El-Gedamy M: Detection of genomic instability in renal cancer by
random amplified polymorphic DNA analysis from urine samples as a
non-invasive method: Potential use in diagnosis. Cancer Sci Res.
1(7)2013.
|
|
12
|
Xian ZH, Cong WM, Zhang SH and Wu MC:
Genetic alterations of hepatocellular carcinoma by random amplified
polymorphic DNA analysis and cloning sequencing of tumor
differential DNA fragment. World J Gastroenterol. 11:4102–4107.
2005.PubMed/NCBI View Article : Google Scholar
|
|
13
|
El-Far M, Abol-Enein H, Zakaria A and
El-Gedamy M: Application of random amplified polymorphic DNA-PCR
technique in early diagnosis of bladder cancer using urine samples.
Curr Top Bio Res. 15:23–34. 2013.
|
|
14
|
Ibrahim MA, Saleh NA, Archoukieh E,
Al-Obaide HW, Al-Obaidi MM and Said HM: Detection of novel genomic
polymorphism in acute lymphoblastic leukemia by random amplified
polymorphic DNA analysis. Int J Cancer Res. 6:19–26. 2010.
|
|
15
|
Zakaria A and Mahmoud AZ: Molecular
characterization of TP53 gene mutations in human bladder cancer.
Pharm Biotechnol. Curr Res. 2:1–7. 2017.
|
|
16
|
Armitage P, Berry G and Matthews JNS:
Statistical Methods in Medical Research. 4th edition.
Wiley-Blackwell, Hoboken, NJ, 2002.
|
|
17
|
Bale R, Schullian P, Eberle G, Putzer D,
Zoller H, Schneeberger S, Manzl C, Moser P and Oberhuber G:
Stereotactic radiofrequency ablation of hepatocellular carcinoma: A
histopathological study in explanted livers. Hepatology.
70:840–850. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Suresh D, Srinivas AN and Kumar DP:
Etiology of hepatocellular carcinoma: Special focus on fatty liver
disease. Front Oncol. 10(601710)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Loy LM, Low HM, Choi JY, Rhee H, Wong CF
and Tan CH: Variant hepatocellular carcinoma subtypes according to
the 2019 WHO classification: An imaging-focused review. AJR Am J
Roentgenol. 219:212–223. 2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Martins-Filho SN, Paiva C, Azevedo RS and
Alves VAF: Histological grading of hepatocellular carcinoma-A
systematic review of literature. Front Med. 4(193)2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Szklaruk J, Silverman PM and Charnsangavej
C: Imaging in the diagnosis, staging, treatment, and surveillance
of hepatocellular carcinoma. AJR Am J Roentgenol. 180:441–454.
2003.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Elgamal S, Ghafar AA, Ghoneem E, Elshaer
M, Alrefai H and Elemshaty W: Characterization of patients with
hepatocellular carcinoma on the way for early detection: One center
experience. Egypt J Int Med. 30:231–238. 2018.
|
|
23
|
Omar M, Elazab T, Abdelrahman A and
Mohamed E: Clinical significance of serum midkine level as a
biomarker in diagnosis of hepatocellular carcinoma. Benha Med J
(Internal Med Hepatol) Special Issue. 37:37–46. 2020.
|
|
24
|
Fang Y, Sun X, Zhang L, Xu Y and Zhu W:
Hemoglobin/red blood cell distribution width ratio in peripheral
blood is positively associated with prognosis of patients with
primary hepatocellular carcinoma. Med Sci Monit. 28:e937146–1.
2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Newsome PN, Cramb R, Davison SM, Dillon
JF, Foulerton M, Godfrey EM, Hall R, Harrower U, Hudson M, Langford
A, et al: Guidelines on the management of abnormal liver blood
tests. Gut. 67:6–19. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Mohamed BF, Serag WM, Abdelal RM and
Elsergany HF: S100A14 protein as diagnostic and prognostic marker
in hepatocellular carcinoma. Egypt Liver J. 9:1–6. 2019.
|
|
27
|
Carr BI and Guerra V: Serum albumin levels
in relation to tumor parameters in hepatocellular carcinoma
patients. Int J Biol Markers. 32:e391–e396. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Nishikawa H, Kita R, Kimura T, Ohara Y,
Sakamoto A, Saito S and Osaki Y: Hyponatremia in hepatocellular
carcinoma complicating with cirrhosis. J Cancer. 6:482–489.
2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Guerra Ruiz AR, Crespo J, LópezMartínez
RM, Iruzubieta P, Casals Mercadal G, Lalana Garcés M and Morales
Ruiz M: Measurement and clinical usefulness of bilirubin in liver
disease. Adv Lab Med. 2:352–361. 2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Watanabe M, Yokomori H, Takahashi Y, Okada
T, Shibuya A and Koizumi W: Assessing the characteristics and
feasibility of preventing early mortality in patients with
hepatocellular carcinoma. The Turkish J Gastroenterol. 30:541–548.
2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Singh Y, Nagar D, Singh M and Maroof M:
Study of electrolyte disturbance in chronic liver disease patients
attending a hospital in Kumaon region. J Family Med Primary Care.
11:4479–4482. 2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhang J, Chen G, Zhang P, Zhang J, Li X,
Gan DN and Ye YA: The threshold of alpha-fetoprotein (AFP) for the
diagnosis of hepatocellular carcinoma: A systematic review and
meta-analysis. PLoS One. 15(e0228857)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Chen W, Wang W, Zhou L, Zhou J, He L, Li
J, Xu X, Wang J and Wang L: Elevated AST/ALT ratio is associated
with all-cause mortality and cancer incident. J Clin Lab Anal.
36(e24356)2022.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhang H, Gao C, Fang L and Yao SK:
Increased international normalized ratio level in hepatocellular
carcinoma patients with diabetes mellitus. World J Gastroenterol.
19:2395–2403. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang SH, Cong WM, Xian ZH, Dong H and Wu
MC: Genomic instability in hepatocellular carcinoma revealed by
using the random amplified polymorphic DNA method. J Cancer Res
Clin Oncol. 130:757–761. 2004.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Chariyalertsak S, Khuhaprema T,
Bhudisawasdi V, Sripa B, Wongkham S and Petmitr S: Novel DNA
amplification on chromosomes 2p25.3 and 7q11.23 in
cholangiocarcinoma identified by arbitrarily primed polymerase
chain reaction. J Cancer Res Clin Oncol. 131:821–828.
2005.PubMed/NCBI View Article : Google Scholar
|