Introduction
A considerable percentage of head and neck
malignancies are invasive and aggressive oral squamous cell
carcinoma (OSCC). OSCC is a type of cancerous growth that arises
from the stratified squamous epithelium of the oral cavity. It
continues to pose a significant worldwide health concern and causes
>90% of all oral cancers (1).
In areas, such as Southeast Asia and portions of Europe where the
disease is more common, OSCC has a major influence on world health.
It poses a serious public health concern in these areas, since, for
instance, OSCC accounts for ~45% of cases worldwide (2). As the population of the world ages
and exposure to established risk factors, including alcohol,
tobacco and chewing betel quid increases, the incidence of OSCC is
expected to increase further. According to current statistics, OSCC
is the sixth most prevalent type of cancer among females and the
fourth most common among males (3). The condition can appear on the
tongue, floor of the mouth, or the buccal mucosa, the latter of
which is more frequently affected due to long-term irritation and
exposure to cigarette smoke. The usual regulatory systems
regulating cell growth and differentiation are disrupted by a
complex multi-step process of genetic and epigenetic modifications
that comprise the pathogenesis of OSCC. Currently, there is no
effective treatment strategy for OSCC. Even though improvements
have been made in surgical techniques, radiotherapy and
chemotherapy, as these methods have limited success in improving
long-term survival rates and reducing recurrence. Thus, in order to
combat this condition, the primary objective is to develop novel
therapeutic strategies that can effectively manage the diverse gene
disturbances, as well as molecular abnormalities associated with
the development of OSCC.
Human papillomavirus (HPV), in particular high-risk
HPV-16, has also been linked to the etiology of OSCC in a growing
number of patients, particularly those that do not have typical
risk factors. As a result of the integration of HPV-16 into the
host genome, tumor suppressor proteins become inactive and a
buildup of genetic abnormalities that accelerate the development of
cancer occurs (4). The process of
field cancerization, in which a number of distinct regions of
dysplasia develop inside the oral cavity and frequently result in
the development of carcinoma in situ and invasive carcinoma,
is another critical part of the pathophysiology of OSCC (5). The chance of tumor recurrence and
treatment resistance is increased by this process, which is
suggestive of extensive genetic instability in the afflicted
tissue. The invasion of underlying connective tissues, angiogenesis
stimulation and metastasis, which frequently involves nearby lymph
nodes and distant organs, are the types of aggressive behavior
displayed by OSCC as it progresses. Advanced-stage OSCC is
associated with a poor prognosis due to its tendency for early
metastases and local invasion (6).
Moreover, among its counterparts implicated in OSCC, the
phosphatase and tensin homolog (PTEN) gene stands out as it plays a
crucial role in maintaining cellular homeostasis hence preventing
malignant transformation. In the majority of cases of OSCC, the
expression of PTEN, as a well-established tumor suppressor, is lost
or frequently downregulated. Upon the loss of PTEN, patients
exhibit accelerated cell growth rates and invasiveness that lead to
resistance against standard care interventions, which expose them
to poorer clinical outcomes (7).
Recent studies have demonstrated the importance of epigenetic
alterations, such as the methylation of the PTEN promoter, as major
factors contributing to PTEN expression reduction, which further
accelerates the progression of OSCC (8).
In addition to genetic alterations, the regulation
of PTEN is also influenced by microRNAs (miRNAs/miRNAs), which are
small non-coding RNAs that modulate gene expression at the
post-transcriptional level (9).
miRNAs exert their effects by binding to complementary sequences on
target mRNAs, leading to their degradation or inhibition of
translation (10). Recent studies
have identified several miRNAs that directly target PTEN, affecting
its expression and contributing to tumor progression in various
cancers, including OSCC (9). These
findings suggest that miRNAs may serve as valuable tools for
elucidating the molecular mechanisms underlying OSCC and for the
development of novel diagnostic and therapeutic strategies
(11).
The present study aimed to integrate these insights
by employing a two-pronged approach. First, the present study
utilized computational methods to identify miRNAs that target PTEN,
focusing on those that exhibit significant regulatory interactions.
Subsequently, the present study validated these miRNAs through
experimental analyses, assessing their expression profiles in OSCC
samples and evaluating their functional impact on PTEN. By
combining computational predictions with empirical validation, the
present study aimed to uncover miRNAs that may serve as biomarkers,
thereby enhancing the accuracy of early OSCC diagnosis. These
miRNAs may also aid in the exploration of novel therapeutic avenues
by understanding the interplay between PTEN and its regulatory
miRNAs.
Materials and methods
Identification of a key tumor
suppressor in OSCC
The pathophysiology of OSCC was investigated through
an extensive literature review, which included identifying key
genes involved in the disease. Among the various candidates, a
well-established tumor suppressor of interest was selected for
further research due to its critical role in maintaining cellular
homeostasis and its frequent downregulation in OSCC. Although not a
novel protein, the importance of this tumor suppressor (PTEN) in
OSCC underscores its relevance for the study, aiming to deepen the
understanding of its regulatory mechanisms and potential as a
therapeutic target.
Selection and characterization of
miRNAs
After identifying the gene, the present study
concentrated on identifying possible miRNAs that target this gene,
which may be critical for the progression of OSCC. TargetScan
(https://www.targetscan.org/vert_80/)
was used to anticipate miRNAs that could control PTEN
computationally (12). A thorough
list of potential miRNAs was supplied by this database, and
particular miRNAs of interest were selected.
Sequence retrieval and structural
analysis
Reputable miRNA databases, namely miRBase
(https://www.mirbase.org/), provided the sequence
of the selected miRNA. The present study utilized RNAfold
(http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi)
to analyze the secondary structure of the miRNA to gain a deeper
understanding of its functional characteristics (13). The evaluation of the stability and
possible binding abilities of the miRNA, two essential components
of its regulatory roles, was made possible.
Sample collection
The present study received approval from the
Institutional Ethics Committee, Department of Medicine, Saveetha
Medical College (IHEC/SDC/PhD/O-PATH-1916/19/432) and all samples
were collected during the time from period September, 2023 to
January, 2024 in strict adherence to the Declaration of Helsinki.
The sample size for the study was calculated using Gpower and a set
of 30 tissue samples, including OSCC and adjacent normal tissues,
were acquired from patients who provided informed consent through
the Department of Medicine at Saveetha Medical College and
Hospitals (Chennai, India) to validate the experiment. Informed
consent was obtained from each individual (Table I). Patients >18 years of age
with a confirmed diagnosis of OSCC and no notable medical
disorders, such as hypertension or hypothyroidism were included in
the present study. The diagnosis of OSCC was validated by the
Saveetha Medical College and Hospitals Department of Biochemistry.
For further examination, the materials were stored at -20˚C after
being washed with PBS.
 | Table ICharacteristics of the patients in the
present study. |
Table I
Characteristics of the patients in the
present study.
| Characteristic | Description |
|---|
| Age | 30-75 years (mean,
55±10 years) |
| Sex | Males, 20; Females,
10 |
| Tumor Stage | Stage I, 5; stage II,
10; stage III, 10; stage IV, 5 |
| Metastasis | Yes, 8; no, 22 |
| Additional
factors | Smoking, 15 (yes);
alcohol consumption, 12 (yes) |
RNA extraction and quantification
Utilizing TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), total RNA was isolated from the
tissue samples in accordance with the manufacturer's
recommendations. A Thermo Fisher Scientific NanoDrop 2000 Lite
spectrophotometer (Thermo Fisher Scientific, Inc.) was used to
evaluate the amount and caliber of the isolated RNA (14). Before being used again, the RNA
samples were kept at -20˚C.
Reverse transcription
Reverse transcription was performed on the isolated
RNA to create complementary DNA (cDNA). For genes, an oligo(dT)18
primer (Promega Corporation, 50 µM) was employed, and for miRNAs, a
universal adapter. Nuclease-free water and dNTPs (10 mM each) from
New England Biolabs Inc. were added to the mixture. Following 5 min
of incubation at 65˚C, this mixture was rapidly chilled.
Nuclease-free water, 5X prime buffer, reverse transcriptase (New
England Biolabs Inc.), and murine RNase inhibitor comprised the
final reaction mixture. The following temperatures were used for
the reverse transcription process in a MiniAmp Plus heat cycler
(Thermo Fisher Scientific, Inc.): 30˚C for 10 min, 42˚C for 30 min,
and 95˚C for 5 min, with a pause at 4˚C in between (14). Using a Nanodrop Lite
spectrophotometer, the cDNA was measured and stored.
Expression analysis using quantitative
PCR (qPCR)
The expression levels of the selected miRNA and PTEN
gene were measured using qPCR with SYBR-Green (Takara Bio, Inc.).
U6 and β-actin were employed as housekeeping controls for the
expression of miRNA and genes, respectively. The primers were
provided by Eurofins Genomics LLC, and the Bio-Rad CFX96 Realtime
System was utilized to perform the expression analysis (Table II). Following a 30-sec initial
denaturation phase at 95˚C, there were 35-40 cycles of 5 sec at
95˚C and 30 sec at the annealing temperature throughout the PCR
cycling conditions. After the PCR cycles were completed, a melt
curve analysis was carried out (15). Each test was run in duplicate, and
the relative expression levels were determined using the
2-∆∆Cq technique (16).
 | Table IImiRNAs and reference gene primer
sequences used in the gene expression analysis. |
Table II
miRNAs and reference gene primer
sequences used in the gene expression analysis.
| miRNA/reference gene
name | Forward primer | Reverse primer |
|---|
| β-actin |
5'-GCACCACACCTTCTACAATG-3' |
5'-TGCTTGCTGATCCACATCTG-3' |
| PTEN |
5'-TGAGTTCCCTCAGCCGTTACCT-3' |
5'-GAGGTTTCCTCTGGTCCTGGTA-3' |
| U6 |
5'-CTCGCTTCGGCAGCACA-3' |
5'-ACGCTTCACGAATTTGC-3' |
| miR-141 |
5'-GCGGAAAGAGGCCCCG-3' |
5'-AGTGCAGGGTCCGAGGTATT-3' |
Statistical analysis
The means of the repeated experiments, along with
the standard error of the mean (SEM) were used to display the
results.. For comparisons between two groups, the Student's t-test
(paired) was used with GraphPad Prism 10.1.0 statistical software
(15). All comparisons in the
present study were made between two groups. To analyze the
correlation between variables in the control and patient groups,
Pearson's correlation was used. A value of P<0.05 was considered
to indicate a statistically significant difference.
Results
miRNA selection and analysis
Computational predictions were made once PTEN was
identified as a major gene of interest to find potential miRNAs
that may control PTEN. TargetScan was utilized to build an
extensive list of potential miRNAs (Fig. 1). The candidate miRNA, miR-141 was
chosen based on their anticipated interaction with PTEN and their
biological importance in OSCC.
miRNA sequence and structural
analysis
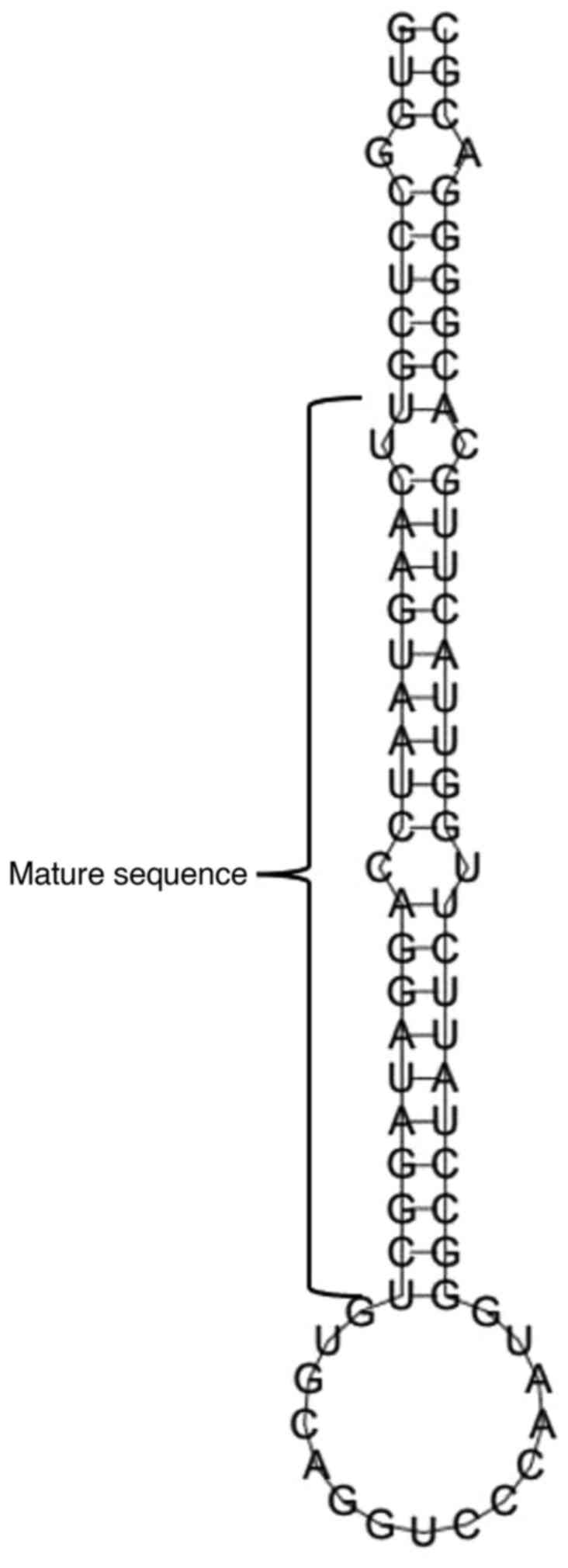
The sequence of miR-141 was extracted from miRBase
and a stable stem-loop shape was predicted by further analysis
using RNAfold, which is necessary for the correct processing and
operation of the miRNA (Fig. 2). A
minimal free energy of -37.30 kcal/mol was found, exhibiting a high
structural stability. These properties underline the possible
involvement of miR-141 in OSCC (Table III).
 | Table IIIMinimum free energy, mature sequence,
match extent and A+U content of hsa-miR-141. |
Table III
Minimum free energy, mature sequence,
match extent and A+U content of hsa-miR-141.
| Source miRNA | Source organism | Minimum free
energy | Mature sequence | Match extension | Strand | A+U% |
|---|
| miR-141 | Homo
sapiens | -37.30 kcal |
UAACACUGUCUGGUAAAGAUGG | 22/22 | 3p | 45.33 |
Expression profiling of PTEN and
miR-141
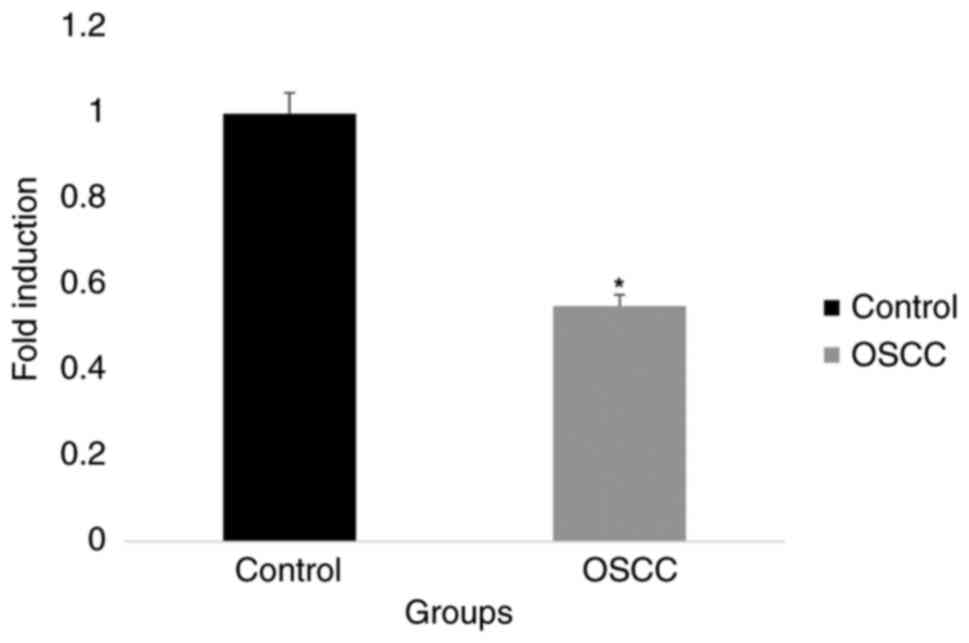
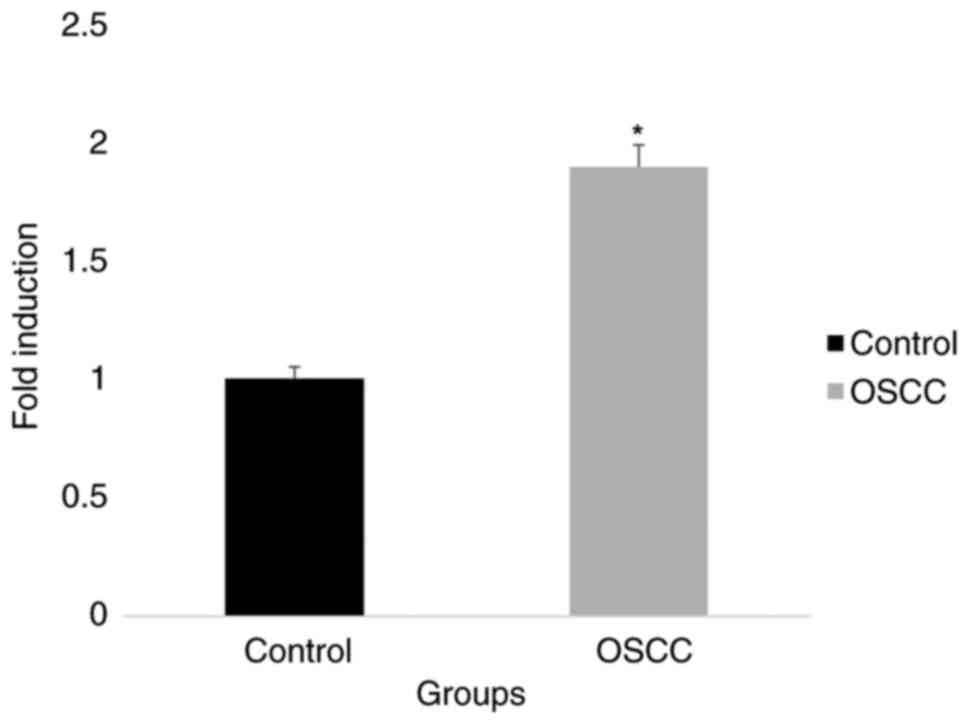
When comparing tissue samples from OSCC to those
from normal tissues the gene expression analysis revealed different
patterns of PTEN and miR-141 levels. PTEN expression was
significantly downregulated, although miR-141 levels were
noticeably upregulated in OSCC samples. Based on these data, it
appears that PTEN and miR-141 may be involved in the development of
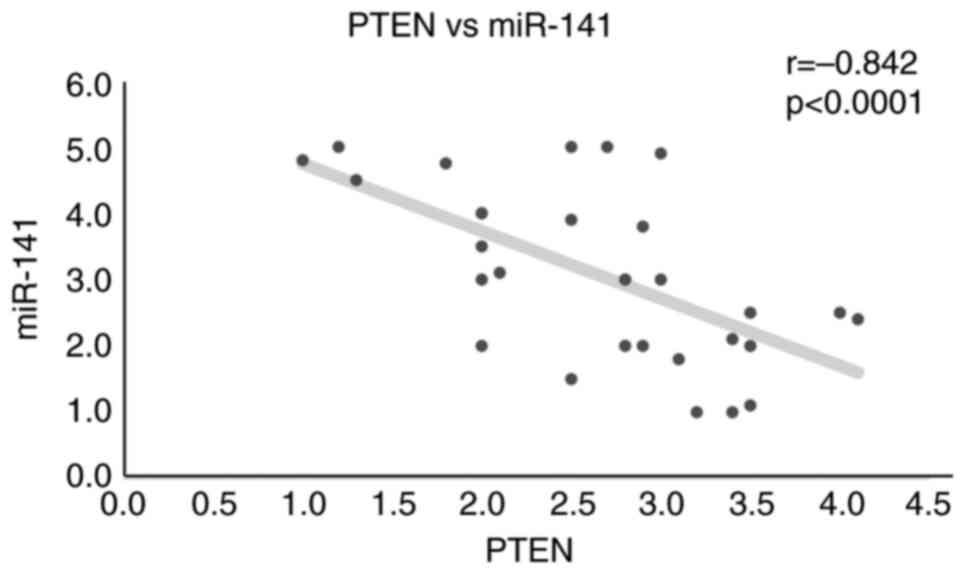
OSCC, which calls for further research (Figs. 3 and 4). An inverse correlation between PTEN
levels and miR-141 was also observed, suggesting that in OSCC, an
increase in the expression of miR-141 is correlated with a decrease
in PTEN expression (Fig. 5). The
Pearson's correlation coefficient (r) for this corrleation was
-0.842, indicating a strong negative correlation, while the P-value
was <0.0001, demonstrating that this correlation is
statistically significant.
Discussion
OSCC poses a major worldwide health burden, mostly
due to its high mortality rates and rapid development (17). Known for its intricate interactions
between genetic and epigenetic elements, OSCC frequently entails
the suppression of key tumor suppressor genes, such as PTEN, which
are essential for preserving cellular homeostasis and averting
malignant transformation (18).
The present study centered on the function of miRNAs in regulating
the expression of PTEN, specifically focusing on miR-141, a pivotal
regulator of PTEN found in several malignancies, including OSCC. In
OSCC tissues, the data revealed a strong negative association
between miR-141 and PTEN expression, with an increased expression
of miR-141 corresponding to downregulated levels of PTEN. miR-141
has also been demonstrated to target and suppress PTEN in other
types of cancer, including gastric and prostate cancer, which has
resulted in increased tumor development and treatment resistance
(19,20). These findings are in line with
these studies. These comparative findings demonstrate the wider
function that miR-141 plays in the oncogenesis of a number of
cancer types and point to its potential as a therapeutic target
that may be used universally.
This discovery also coincides with studies on
hepatocellular carcinoma (HCC), where miR-141 has been linked to
the downregulation of PTEN, hence increasing carcinogenesis
(21). On the other hand, these
findings suggest that, in OSCC, miR-141 may potentially enhance
invasiveness and metastatic potential by interfering with
PTEN-mediated pathways, in contrast to HCC, where miR-141 primarily
addresses cell proliferation. As the functional impact varies
throughout cancer types, different treatment approaches are
required, highlighting the intricacy of miRNA regulation. The
results presented herein have consequences that go beyond simple
molecular understanding. In OSCC, the deregulation of the
miR-141/PTEN axis may be a useful biomarker for prognosis and early
identification. Comparative research on breast cancer has
demonstrated that diagnostic accuracy is increased when miRNA
profiling is combined with conventional markers. Comparably, early
identification rates for OSCC may be increased by including PTEN
expression analysis and miR-141 into the existing diagnostic
methods, particularly for individuals lacking conventional risk
factors. Furthermore, a viable treatment approach is to target the
miR-141/PTEN pathway. Preclinical research on lung cancer has
demonstrated that miRNA inhibitors can render tumors more sensitive
to chemotherapy and restore PTEN expression (22). By reactivating tumor suppressor
pathways, innovative medicines that overcome resistance to current
medications may be developed if this technique is used to OSCC
(23).
In conclusion, the present study statistically
analyzed the expression of PTEN and miR-141 in OSCC in comparison
to normal tissue samples. The findings presented herein provide a
deeper understanding of the molecular interactions between these
critical regulators, providing insight which may aid in the
development of novel therapeutic approaches. miR-141 plays a
crucial role in regulating PTEN expression in OSCC, with
significant implications for both diagnosis and treatment. The
present study highlights the potential of miR-141 as a universal
therapeutic target by contrasting the results with research in
other malignancies. Further studies are required however, to
explore the therapeutic potential of targeting the miR-141/PTEN
pathway in OSCC, particularly in combination with conventional
therapies, in order to improve patient outcomes and reduce
mortality rates.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
DS conceptualized the study, and was also involved
in the study supervision, formal analysis and reviewing of the
manuscript. AA was involved in the writing of the original draft of
the manuscript, as well as in data analysis and interpretation.
ASUPP was involved in the design of the study, in data curation and
assisted with data analysis. AKP was involved in data collection
and revised the manuscript. DS and AKP confirm the authenticity of
all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was conducted in accordance with
the Declaration of Helsinki and was approved by the Institutional
Ethics Committee of Department of Medicine at Saveetha Medical
College and Hospitals (IHEC/SDC/PhD/O-PATH-1916/19/432). Written
informed consent was obtained from all patients prior to the
collection of tissue samples for research purposes.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Arumugam P, M SM and Jayaseelan VP: A
novel m6A reader RBFOX2 expression is increased in oral squamous
cell carcinoma and promotes tumorigenesis. J Stomatol Oral
Maxillofac Surg. 102041:2024.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Murugesan D, Kannan B, As SG, Jayaseelan
VP and Arumugam P: Alteration of SERPINH1 is associated with
elevated expression in head and neck squamous cell carcinomas and
its clinicopathological significance. J Stomatol Oral Maxillofac
Surg. 125(101811)2024.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Miguelanez-Medran BC, Pozo-Kreilinger JJ,
Cebrian-Carretero JL, Martinez-Garcia MA and Lopez-Sanchez AF: Oral
squamous cell carcinoma of tongue: Histological risk assessment. A
pilot study. Med Oral Patol Oral Cir Bucal. 24:e603–e609.
2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sri S, Ramani P, Premkumar P, Ramshankar
V, Ramasubramanian A and Krishnan R: Prevalence of Human
Papillomavirus (HPV) 16 and 18 in oral malignant and potentially
malignant disorders: A polymerase chain reaction analysis-A
comparative study. Ann Maxillofac Surg. 11:6–11. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tan Y, Wang Z, Xu M, Li B, Huang Z, Qin S,
Nice EC, Tang J and Huang C: Oral squamous cell carcinomas: state
of the field and emerging directions. Int J Oral Sci.
15(44)2023.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Shetty SS, Sharma M, Fonseca FP, Jayaram
P, Tanwar AS, Kabekkodu SP, Kapaettu S and Radhakrishnan R:
Signaling pathways promoting epithelial mesenchymal transition in
oral submucous fibrosis and oral squamous cell carcinoma. Jpn Dent
Sci Rev. 56:97–108. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Liu M, Song H, Xing Z, Lu G, Li J and Chen
D: Correlation between PTEN gene polymorphism and oral squamous
cell carcinoma. Oncol Lett. 18:1755–1760. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Fusco N, Sajjadi E, Venetis K, Gaudioso G,
Lopez G, Corti C, Rocco EG, Criscitiello C, Malapelle U and
Invernizzi M: PTEN alterations and their role in cancer management:
Are we making headway on precision medicine? Genes (Basel).
11(719)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Travis G, McGowan EM, Simpson AM, Marsh DJ
and Nassif NT: PTEN, PTENP1, microRNAs, and ceRNA Networks:
Precision targeting in cancer therapeutics. Cancers (Basel).
15(4954)2023.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Diener C, Keller A and Meese E: The
miRNA-target interactions: An underestimated intricacy. Nucleic
Acids Res. 52:1544–1557. 2024.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chakrabortty A, Patton DJ, Smith BF and
Agarwal P: miRNAs: Potential as biomarkers and therapeutic targets
for cancer. Genes (Basel). 14(1375)2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Gu Y, Tang S, Wang Z, Cai L, Shen Y and
Zhou Y: Identification of key miRNAs and targeted genes involved in
the progression of oral squamous cell carcinoma. J Dent Sci.
17:666–676. 2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ahsan M, K P A, Usman P P AS and Sekar D:
Computational and expression analysis of microRNA-149-5p and its
target, interleukin-6, in chronic kidney disease. Biomed Res Ther.
10:6103–6109. 2023.
|
|
14
|
Suthagar P, Purayil ASUP, Parambath AK and
Sekar D: In silico identification and gene expression of
miR-148a-5p and IL-6 in hypothyroid patient blood samples. Biomed
Res Ther. 11:6379–6386. 2024.
|
|
15
|
Narayan RN, Parambath AK, Purayil ASUP and
Sekar D: microRNA-9-5p and its target nuclear factor kappa B are
differentially expressed in type-2 diabetes patients. Biomed Res
Ther. 11:6183–6190. 2024.
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Oral cancer - the fight must go on against
all odds. Evid Based Dent. 23:4–5. 2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Mesgari H, Esmaelian S, Nasiri K,
Ghasemzadeh S, Doroudgar P and Payandeh Z: Epigenetic regulation in
oral squamous cell carcinoma microenvironment: A comprehensive
review. Cancers (Basel). 15(5600)2023.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zedan AH, Osther PJS, Assenholt J, Madsen
JS and Hansen TF: Circulating miR-141 and miR-375 are associated
with treatment outcome in metastatic castration resistant prostate
cancer. Sci Rep. 10(227)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Guz M, Jeleniewicz W and Cybulski M:
Interactions between circRNAs and miR-141 in Cancer: From
pathogenesis to diagnosis and therapy. Int J Mol Sci.
24(11861)2023.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Khare S, Khare T, Ramanathan R and Ibdah
JA: Hepatocellular Carcinoma: The Role of MicroRNAs. Biomolecules.
12(645)2022.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yang H, Liu Y, Chen L, Zhao J, Guo M, Zhao
X, Wen Z, He Z, Chen C and Xu L: MiRNA-based therapies for lung
cancer: Opportunities and challenges? Biomolecules.
13(877)2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Bou Antoun N and Chioni AM: Dysregulated
signalling pathways driving anticancer drug resistance. Int J Mol
Sci. 24(12222)2023.PubMed/NCBI View Article : Google Scholar
|