Introduction
Worldwide, 94 million individuals suffer from blindness or are visually impaired, and the most common cause of blindness is cataract (1). Cataract is highly associated with a decreased quality of life and a reduced life expectancy (2). Age and diabetes are considered to the main causes of cataract (3,4). Surgery is the only viable method with which to treat cataracts, and replacing them with an artificial lens can restore vision (5). However, not all countries have sufficient resources for surgery (6); thus, alternative treatment strategies also need to be explored. Apart from age and diabetes, smoking, myopia, intraocular infection, metabolic diseases and drug use are also defined as the factors associated with cataract development (7-10). However, it is not known whether changing levels of low-density lipoprotein cholesterol (LDL-C) may cause cataract.
The level of LDL-C is considered to be related to cardiovascular disease and recurrent stroke (11). Previous dietary trials have demonstrated that a reduction in LDL-C levels is beneficial to human health (12). As regards diabetes, high levels of LDL-C are associated with the of developing diabetes (13). High LDL-C levels are also related to abnormal liver and kidney function and hypertension (14,15). LDL-C is a common index examined in clinical practice.
Mendelian randomization (MR) provides a method which can be used to assess the association between a risk factor and a clinical disease (16). By using genetic variants as instrumental variables (IVs) for the tested exposure, confounding or reverse causality can be solved in this process (17). Two-sample MR refers to the use of MR methods in a two-sample environment; in this case, the instrument-exposure and instrument-outcome associations are estimated in non-overlapping sets of individuals (18). The majority of the data used for analysis can be obtained from genome-wide association studies (GWAS) (19).
GWAS aim to reveal the genotype-phenotype associations by testing genetic variants across the genomes of different individuals (20). Data of single-nucleotide polymorphism (SNP) arrays are used for analysis in GWAS (21). The wide application of GWAS has proven to be beneficial to several complex disease genetics over the past decades (22). Moreover, novel causes of diseases and mechanisms of diseases are defined in this manner (23). Although there are some limitations of GWAS, such as the fact that it implicates a high number of loci, GWAS are a useful tool due to their capacity of understand the genetic basis of human diseases (22).
The present study employed bidirectional two-sample MR to investigate the causal association between LDL-C and cataract. By addressing the global challenge of cataract, particularly where surgical options are limited, the present study aimed to identify novel preventive strategies. By exploring genetic links, the present study aimed to unveil modifiable risk factors, providing insight into non-surgical interventions to reduce the incidence of cataract and improve public health outcomes.
Data and methods
Exposure and outcome data. The SNPs for LDL-C were used as IVs and sourced from the IEU OpenGWAS project (mrcieu.ac.uk) GWAS dataset (GWAS ID: ebi-a-GCST002222). This project provides summary statistics, publications and top associations, and does not require additional ethical approval. The data includes 94,595 European participants and 2,409,690 SNPs. To obtain result data, summary data for cataracts were also utilized from the IEU OpenGWAS project (GWAS ID: ukb-a-426), which consists of 108,817 European participants (10,475 cases and 98,342 controls), covering a total of 10,894,596 SNPs.
It is important to note that the conclusions of the present study are based on data from European populations. Therefore, the findings presented herein may not be applicable to non-European populations. Explicitly acknowledging this limitation and suggesting future research directions to explore the generalizability of this association in different population groups would be beneficial.
IV selection criteria
The SNPs for LDL-C were selected as IVs from the IEU Open GWAS project. Specifically, genome-wide significant SNPs (P<0.00000005) were used. This P-value threshold is a standard criterion in GWAS, ensuring that only variants with strong statistical evidence of association with LDL-C are included. Additionally, a linkage disequilibrium (LD) cut-off of r²<0.1 was applied within a 1 MB window. This threshold minimizes the impact of closely related genetic variants (in high LD) that could bias the results. The selected SNPs were then filtered to remove duplicates and palindromic SNPs, ensuring a robust set of instrumental variables.MR-PRESSO and MR-Egger regression tests were conducted to further evaluate the potential for horizontal pleiotropy, enhancing the credibility of the findings.
MR analysis
MR analysis was performed to determine the casual association between LDL-C and cataract. Among the five methods of MR analysis, the inverse-variance weighted (IVW) method is considered to be slightly more powerful than the others (24); thus, the IVW test was used as the main statistical method in the present study. The other methods of MR analysis (simple mode, simple median, weighted mode, MR-Egger regression and weighted median estimator) were used as complementary methods. Leave-one-out analysis refers to the observation of residual stability following the stepwise remove of SNPs, ideally with minimal change after culling, which is similar to the meta-analysis of culling. Further more, F-statistics was calculated to evaluate weak instrument bias, as an F value <10 indicates weak instrument strength. Odds ratios (ORs) and 95% confidence intervals (95% CI) were indicated as causal estimates. All data analyses were performed using tge R package (two-sample MR and MR-PRESSO).
Heterogeneity
MR-heterogeneity mainly depends on the P-value of Cochran's Q test; when the statistical Q value is <0.05, it is considered that there is heterogeneity. Heterogeneity is acceptable and allowed to exist.
Horizontal pleiotropy
The MR-Egger method was used to test the horizontal pleiotropy, In the MR-Egger analysis, the existence of an Egger-intercept was considered and used to compare with 0 and assess horizontal pleiotropy. In the case that there is no significant difference (P>0.05), no horizontal pleiotropy was considered to exist.
Results
Effect of LDL-C on cataract
In the two-way MR analysis, 75 SNPs were extracted with LDL-C as the exposure and cataract as the outcome. In the set of IVs (P<0.00000005), IVW analysis revealed that there was a positive causal association between LDL-C and cataract (OR, 1.01; P=0.036). The sample mode analysis also indicated that LDL-C was associated with cataract (OR, 1.02; P=0.013). Similarly, the sample median analysis revealed that LDL-C was causally associated with cataract (OR, 1.02; P=0.003) (Figs. 1, and 2A and C). A leave-one-out sensitivity test was performed to test the robustness of the results; following the stepwise elimination of SNPs, the results were not markedly altered (Fig. 2D).
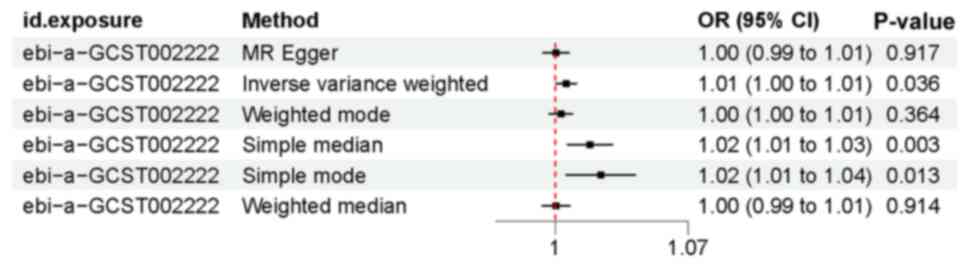 |
Figure 1
Mendelian randomization forest result. Causal effects of low-density lipoprotein cholesterol on cataracts estimated by Mendelian randomization given as ORs and 95% CIs. OR, odds ratio; CI, confidence interval.
|
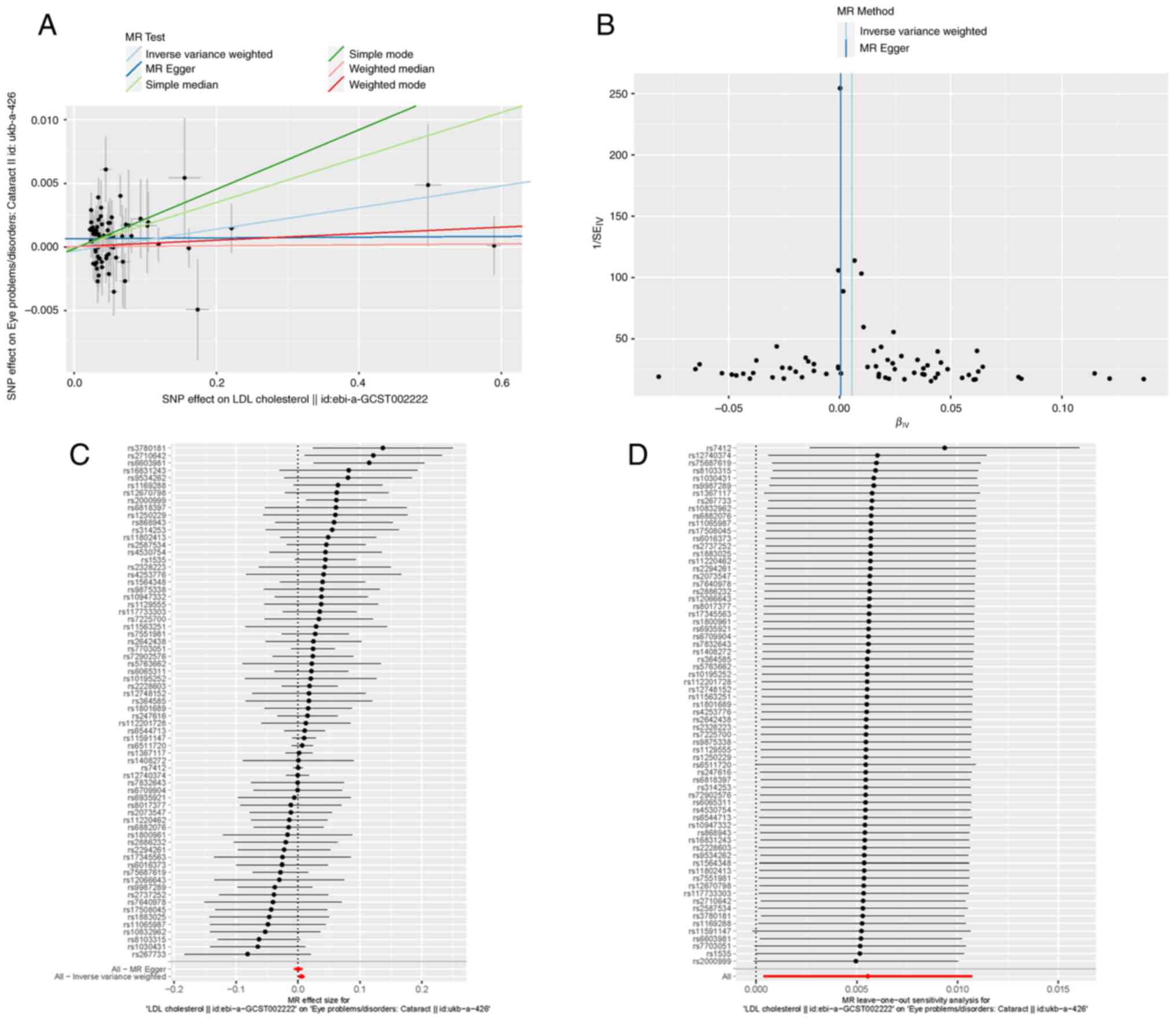 |
Figure 2
(A) Scatter plot of the association between LDL cholesterol and cataract. The six Mendelian randomization methods are presented. Lines in light blue, blue, light green, green, orange and red, respectively the inverse-variance weighted, MR-Egger, simple median, weighted median, simple mode, weighted median and weighted mode methods. (B) Funnel plot depicting whether there was significant heterogeneity in the observed associations. (C) Forest plot illustrating the MR estimate and 95% confidence intervals for each SNP; the gray line segment represents 95% confidence intervals; the inverse-variance weighted and MR-Egger results are shown at the bottom. (D) Leave-one-out sensitivity test; SNPs were removed one by one and the results of MR were calculated. LDL, low-density lipoprotein; MR, Mendelian randomization; SNP, single nucleotide polymorphism.
|
The MR-Egger regression method was applied to measure horizontal pleiotropy. It was found that there was no directional pleiotropy in the MR-Egger regression (P>0.05). At the same time, Cochran's Q test indicated that there was no evidence of heterogeneity in the present study (Q value >0.05). The funnel plot can also prove this finding (Fig. 2B).
Discussion
In the present study, a bidirectional MR was performed to investigate the causal association between LDL-C and cataract; it was found that LDL-C was causally associated with cataract. Moreover, no significant heterogeneity or pleiotropy were found in the present study, which confirmed the validity of the conclusions.
To the best of our knowledge, this is the first study to use the MR method to assess the association between LDL-C and cataract. Some previous studies have found a possible association between the LDL-C and cataract. In previous research, through a univariate and multivariate Cox model analysis, time-varying LDL-C lowering (annually assessed) was associated with a lower incident of cataract (25,26). Moreover, it was previously found that following the inhibition of plasma proprotein convertase subtilisin/kexin type 9, the decrease of LDL-C levels was associated with an increased risk of cataracts (27). Another study using a common data model revealed that LDL-C variability was associated with developing cataracts (26). However, whether patients with very low LDL-C levels are at a risk of developing cataract remains a matter of debate (26,28).
The present study was based on the methods of MR analysis, and several advantages of the study should be mentioned. Firstly, through analysis using the MR method, reverse causality and unmeasured confounding factors can be reduced (29), which can improve the accuracy of causal inference. Secondly, the GWAS database provides a large scale of significant SNPs; thus, sufficient IV strength was ensured in the present study. Thirdly, the present study found an association between LDL-C and cataract, which may provide new insight into the prevention and treatment of cataract. At the same time, the control of LDL-C levels will help to reduce the risk of developing diabetes (30), thus also preventing the occurrence of diabetes-related cataracts to a certain extent (31).
There are some limitations to the present study which should also be mentioned. The use of a European-only dataset limits the generalizability of the results to other populations. Genetic heterogeneity across different ethnic groups means that findings in European populations may not be directly applicable to non-European groups. Future studies are thus required to include diverse populations to enhance the external validity of the results and provide a more comprehensive understanding of the genetic determinants of cataract risk. Additionally, reflecting on how these limitations may influence the outcomes of the present study is vital. For instance, the exclusion of certain genetic variants could lead to an underestimation of the true causal effect of LDL-C on cataract. This underestimation may affect clinical recommendations and public health policies derived from the study. Acknowledging this limitation helps in tempering the interpretation of the findings and emphasizes the need for cautious application in clinical settings. To address these gaps, future research could adopt a more inclusive approach to IV selection, perhaps by incorporating methods that allow for the inclusion of variants with weaker associations. Furthermore, expanding the study to include diverse populations may provide a broader understanding of the genetic underpinnings of cataract. Collaborations with international cohorts and the inclusion of multi-ethnic GWAS datasets can help achieve this goal.
In conclusion, the present study leverages two-sample MR to reveal a causal link between LDL-C levels and cataract, advocating for revised lipid management strategies to mitigate cataract risk and enhance ocular health.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WO and ZH were involved in the conception of study, as well as in data curation, in the writing and reviewing of the manuscript, and in data visualization. YL and ZR were involved in data curation, in the writing of the original draft, and in data visualization. YP was involved in the writing of the original draft, in the reviewing and editing of the manuscript and in data curation and visualization. All the authors confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Lee CM and Afshari NA: The global state of cataract blindness. Curr Opin Ophthalmol. 28:98–103. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Brian G and Taylor H: Cataract blindness-challenges for the 21st century. Bull World Health Organ. 79:249–256. 2001.PubMed/NCBI
|
|
3
|
Asbell PA, Dualan I, Mindel J, Brocks D, Ahmad M and Epstein S: Age-related cataract. Lancet. 365:599–609. 2005.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Auger N, Tang T, Healy-Profitós J and Paradis G: Gestational diabetes and the long-term risk of cataract surgery: A longitudinal cohort study. J Diabetes Complications. 31:1565–1570. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Dobrzynski JM, Kostis JB, Sargsyan D, Zinonos S and Kostis WJ: Effect of cholesterol lowering with statins or proprotein convertase subtilisin/kexin type 9 antibodies on cataracts: A meta-analysis. J Clin Lipidol. 12:728–733. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Do DV, Gichuhi S, Vedula SS and Hawkins BS: Surgery for postvitrectomy cataract. Cochrane Database Syst Rev. 1(CD006366)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Pan CW, Cheng CY, Saw SM, Wang JJ and Wong TY: Myopia and age-related cataract: A systematic review and meta-analysis. Am J Ophthalmol. 156:1021–1033.e1. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ye J, He J, Wang C, Wu H, Shi X, Zhang H, Xie J and Lee SY: Smoking and risk of age-related cataract: A meta-analysis. Invest Ophthalmol Vis Sci. 53:3885–3895. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lim JC, Caballero Arredondo M, Braakhuis AJ and Donaldson PJ: Vitamin C and the lens: New insights into delaying the onset of cataract. Nutrients. 12(3142)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chan NS, Ti SE and Chee SP: Decision-making and manage-ment of uveitic cataract. Indian J Ophthalmol. 65:1329–1339. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wang A, Dai L, Zhang N, Lin J, Chen G, Zuo Y, Li H and Wang Y, Meng X and Wang Y: Oxidized low-density lipoprotein (LDL) and LDL cholesterol are associated with outcomes of minor stroke and TIA. Atherosclerosis. 297:74–80. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Silverman MG, Ference BA, Im K, Wiviott SD, Giugliano RP, Grundy SM, Braunwald E and Sabatine MS: Association between lowering LDL-C and cardiovascular risk reduction among different therapeutic interventions: A systematic review and meta-analysis. JAMA. 316:1289–1297. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Hu X, Liu Q, Guo X, Wang W, Yu B, Liang B, Zhou Y, Dong H and Lin J: The role of remnant cholesterol beyond low-density lipoprotein cholesterol in diabetes mellitus. Cardiovasc Diabetol. 21(117)2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kintu C, Soremekun O, Kamiza AB, Kalungi A, Mayanja R, Kalyesubula R, Bagaya SB, Jjingo D, Fabian J, Gill D, et al: The causal effects of lipid traits on kidney function in Africans: Bidirectional and multivariable Mendelian-randomization study. EBioMedicine. 90(104537)2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Xiong X, Wang P, Li X, Zhang Y and Li S: The effects of red yeast rice dietary supplement on blood pressure, lipid profile, and C-reactive protein in hypertension: A systematic review. Crit Rev Food Sci Nutr. 57:1831–1851. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Birney E: Mendelian randomization. Cold Spring Harb Perspect Med. 12(a041302)2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Sekula P, Del Greco MF, Pattaro C and Köttgen A: Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol. 27:3253–3265. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hartwig FP, Davies NM, Hemani G and Davey Smith G: Two-sample Mendelian randomization: Avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol. 45:1717–1726. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Boehm FJ and Zhou X: Statistical methods for Mendelian randomization in genome-wide association studies: A review. Comput Struct Biotechnol J. 20:2338–2351. 2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ishigaki K: Beyond GWAS: From simple associations to functional insights. Semin Immunopathol. 44:3–14. 2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Abdellaoui A, Yengo L, Verweij KJH and Visscher PM: 15 years of GWAS discovery: Realizing the promise. Am J Hum Genet. 110:179–194. 2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Tam V, Patel N, Turcotte M, Bossé Y, Paré G and Meyre D: Benefits and limitations of genome-wide association studies. Nat Rev Genet. 20:467–484. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al: Finding the missing heritability of complex diseases. Nature. 461:747–753. 2009.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Lin Z, Deng Y and Pan W: Combining the strengths of inverse-variance weighting and Egger regression in Mendelian randomization using a mixture of regressions model. PLoS Genet. 17(e1009922)2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Bang CN, Greve AM, La Cour M, Boman K, Gohlke-Bärwolf C, Ray S, Pedersen T, Rossebø A, Okin PM, Devereux RB and Wachtell K: Effect of randomized lipid lowering with simvastatin and ezetimibe on cataract development (from the simvastatin and ezetimibe in aortic stenosis study). Am J Cardiol. 116:1840–1844. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Park JS, Kim DH, Kim BK, Park KH, Park DH, Hwang YH and Kim CY: Effect of cholesterol variability on the incidence of cataract, dementia, and osteoporosis: A study using a common data model. Medicine (Baltimore). 102(e35548)2023.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Masson W, Lobo M, Huerín M, Molinero G, Lobo L and Nogueira JP: Plasma proprotein convertase subtilisin/kexin type 9 inhibitors and cataract risk: A systematic review and meta-analysis. Arch Soc Esp Oftalmol (Engl Ed). 94:75–8. 2019.PubMed/NCBI View Article : Google Scholar : (In English, Spanish).
|
|
28
|
Yu S, Chu Y, Li G, Ren L, Zhang Q and Wu L: Statin use and the risk of cataracts: A Systematic review and meta-analysis. J Am Heart Assoc. 6(e004180)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Bowden J and Holmes MV: Meta-analysis and Mendelian randomization: A review. Res Synth Methods. 10:486–496. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Hu X, Liu Q, Guo X, Wang W, Yu B, Liang B, Zhou Y, Dong H and Lin J: The role of remnant cholesterol beyond low-density lipoprotein cholesterol in diabetes mellitus. Cardiovasc Diabetol. 21(117)2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Alabdulwahhab KM: Senile cataract in patients with diabetes with and without diabetic retinopathy: A Community-based comparative study. J Epidemiol Glob Health. 12:56–63. 2022.PubMed/NCBI View Article : Google Scholar
|
















