Introduction
Rheumatoid arthritis (RA) is the most common
autoimmune inflammatory disease worldwide, primarily affecting the
joints. It is marked by a progressive course that can result in
pannus formation and joint destruction if left untreated (1,2).
Rheumatoid arthritis affects 0.5 to 1.0% of the adult population,
with the incidence varying by region (3). The disease is more prevalent among
females than males and the incidence tends to increase with age
(4). The main symptoms of RA
include pain, stiffness and swelling, primarily affecting the
joints of the hands and feet, although other joints can also become
inflamed. Additionally, RA may affect other organs (5). Research suggests that multiple
genetic and environmental factors contribute to the development of
RA (6,7). Despite advancements in more effective
treatments over the past two decades, achieving disease remission
in RA remains uncommon (8).
Previous studies have demonstrated that cytokine
networks play a crucial role in the pathogenesis of RA (9). Interleukin (IL)-15, a cytokine
produced by myeloid cells, plays a critical role in the
proliferation and survival of natural killer cells, memory
CD8+ T-cells and invariant natural killer T-cells
(10). It is also implicated in
the pathogenesis of RA and may be considered a potential biomarker
for the progression of RA. Furthermore, elevated levels of IL-15,
rheumatoid factor (RF) and anti-citrullinated protein antibodies
(ACPAs) have been reported in patients with RA (11,12).
IL-29 is a key member of the type III interferon family and the
most active cytokine in this group. It plays a role in modulating
autoimmune inflammation and protection against viral infections.
Previous studies have indicated that IL-29 is expressed in the
lining layers of RA synovium by CD68+ macrophages and
FGF-2+ fibroblasts. Moreover, the production of IL-29 is
significantly elevated in the serum, peripheral blood mononuclear
cells (PBMCs) and synovial tissue of patients with RA compared to
healthy controls. However, the precise molecular mechanisms
underlying the role of IL-29 in RA are not yet fully understood
(13,14).
Epigenetics is the alteration in gene expression and
function without changes in the DNA sequences, leading to heritable
phenotypes. Epigenetic modifications involve DNA methylation, the
post-translational modifications of proteins and the
post-transcriptional regulation of genes. Epigenetics plays a key
role in autoimmune diseases, including RA (15,16).
DNA methylation is a process in which a CH3 group is
added to cytosine at cytosine-guanine (CG) dinucleotide sites,
known as CpG islands. Under normal conditions, the DNA methylation
of CpG islands prevents transcription factors from binding to the
DNA through various mechanisms, leading to the inhibition of gene
expression (17,18). Hydroxymethylcytosine (5hmC) is an
epigenetic modifier formed through the active demethylation of DNA
via oxidation by ten-eleven-translocation (TET) enzymes. Previous
studies have reported that patients with RA exhibit differential
methylation patterns in peripheral blood mononuclear cells,
fibroblast-like synoviocytes and synovial T-cells. As a result, DNA
methylation in peripheral blood, or the methylome, has been
proposed as a biomarker for predicting the response of patients
with RA to treatment (19,20). Furthermore, DNA methylation can be
considered as a biomarker for RA susceptibility. Detectable DNA
methylation signatures at the earliest stages of the disease
provide the optimal opportunity to initiate early treatment and
potentially achieve remission (21).
The present study aimed to assess the potential
roles of global DNA methylation levels, along with certain
physiological and immunological markers, in the development of
RA.
Materials and methods
Subjects and blood collection
A total of 60 patients with RA (45 females and 15
males) visiting the Department of Rheumatology in Private Nursing
Home Hospital in Medical City, Baghdad, Iraq, from October, 2023 to
January, 2024 were recruited for the study. All patients were
diagnosed according to the 2010 American College of
Rheumatology/European League Against Rheumatism (ACR/EULAR)
classification criteria. The patients with RA were aged between 30
and 70 years, and were newly diagnosed. Patients with prolonged
disease duration, those on extended treatment and other autoimmune
diseases, active infections, or who were pregnant or breastfeeding,
were excluded from the study. Additionally, 40 healthy control
samples (10 males and 30 females; aged 30 to 66 years) with no
history of autoimmune or chronic diseases and who appeared to be in
good health were enrolled in the study. The present study was
approved by the Ethics Committee of the Department of Biology,
College of Science, University of Baghdad, under reference no.
CSEC/0923/0105, on September 25, 2023. Written informed consent was
obtained from all patients in the study. The research adhered to
the standards set by the latest revision of the Declaration of
Helsinki.
Sample collection
A total of 5 ml venous blood was drawn from the
radial vein of the participants using disposable syringes. In
total, 3 ml were slowly transferred into disposable serum tubes
containing separating gel, allowed to clot at room temperature for
10 to 15 min, and then centrifuged at 1,508.3 x g for 10 to 15 min
at room temperature. The serum was then distributed into Eppendorf
tubes (EPPENDORF) in equal amounts and stored at -20˚C for later
use in serological tests. The remaining 2 ml blood were transferred
to ethylenediaminetetraacetic acid (EDTA)-containing tubes
(Hangzhou Ciping Medical Devices Co., Ltd.) for complete blood
count (CBC), erythrocyte sedimentation rate (ESR), and the
measurements of 5mC and 5hmC.
Measurement of immunological markers
using enzyme-linked immunosorbent assay (ELISA)
The quantification of C-reactive protein (CRP) and
anti-cyclic citrullinated peptides (anti-CCPs) was performed using
the human CRP ELISA kit (cat. no. MBS564038, MyBioSource) and the
human anti-CCP antibody ELISA kit (cat. no. MBS7235871,
MyBioSource) all materials mentioned hereafter are components of
these kits. These kits are based on ELISA, a diagnostic method
widely used in medicine. Additionally, ELISA is used in biomedical
research as an analytical tool for the detection and measurement of
specific antigens or antibodies in selected samples. The principle
of ELISA relies on the fundamental immunological concept of
antigens binding to specific antibodies, allowing for the detection
of small quantities of antigens, such as proteins, peptides,
hormones, or antibodies in fluid samples. This process was
conducted according to the manufacturer's instructions. All
reagents and samples were allowed to reach room temperature prior
to use. A total of 100 µl of standards and samples were added to
their respective wells, followed by incubation for 30 min at 37˚C
and washing with 300 µl of wash buffer. Subsequently, 100 µl HRP
conjugate were added to each well followed by incubation for 30 min
at 37˚C, then washed again. Subsequently, 50 µl Substrates A and B
were added followed by incubation for 10 min at 37˚C. Finally, 50
µl stop solution were added before measuring the absorbance at 450
nm using an ELISA reader (BioTek Instruments, Inc.).
Detection of IL-15 and IL-29 levels
using ELISA
The levels of IL-15 and IL-29 were evaluated using
human IL ELISA kits (cat. nos. E0097Hu and E0040Hu, respectively,
BT LAB) all materials mentioned hereafter are components of these
kits. Following the manufacturer's instructions, all reagents and
samples were brought to room temperature prior to use. A total of
50 µl of standard and 40 µl of sample were added to their
respective wells. Subsequently, 10 µl anti-IL-15 antibody and human
IL-29 were added to the sample wells, followed by the addition of
50 µl streptavidin-HRP to both the sample and standard wells (but
not the blank well). The wells were then incubated for 60 min at
37˚C and washed with 300 µl wash buffer. Following this, 50 µl
substrate solutions A and B were added to each well followed by
incubation for 10 min at 37˚C in the dark. Finally, 50 µl stop
solution were added to each well, and the absorbance was read at
450 nm using an ELISA reader (BioTek Instruments, Inc.).
Measurement of RF levels
The RF-latex slide agglutination test (cat. no.
SL003, Bioresearch) was performed to determine the RF levels all
materials mentioned hereafter are components of this kit. First, 40
µl of the sample and one drop each of the negative (usually
contains animal serum, which not contain RF) and positive controls
(Human serum with a RF concentration >30 IU/ml) were placed into
separate circles on the test slide. Subsequently, 40 µl RF latex
reagent was added adjacent to the sample and the negative and
positive controls for testing. Thirdly, the RF reagent was mixed
with the sample, positive and negative controls, and spread over
the entire surface of each circle using a stirrer. Finally, the
slide was placed on a mechanical rotator at 80 to 100 rpm for 2
min. The presence or absence of visible agglutination was assessed
immediately after removing the slide from the rotator.
DNA extraction and assessment of
global DNA methylation
Whole DNA was extracted from the blood samples of
all the studied cases and healthy controls using the
gSYNC™ DNA extraction kit (cat. no. GS100, Geneaid
Biotech Ltd.). The purity and concentration of the genomic DNA were
assessed by measuring the A260/A280
absorbance using a NanoDrop spectrophotometer (Thermo Fisher
Scientific, Inc.). A ratio between 1.8 and 2.0 indicates that the
genomic DNA is of high quality. The evaluation of global
methylation was performed using the MethylFlash™ Global
DNA Methylation (5mC) and MethylFlash™ Hydroxymethylated
DNA (5hmC) Quantification kits (cat. nos. P-1034 and P-1036,
EpigenTek). A total of 100 ng of the extracted genomic DNA from
each test sample were diluted in the supplied binding solution
provided with the 8-well assay strip kit. In brief, the DNA
methylation fraction that bound to the monoclonal antibodies of the
well-assay strip was captured for detection in the subsequent assay
steps. This process included the addition of wash solution,
detection antibody, enhancer solution, developer and stop reaction
solution. Ultimately, the quantification of 5mC and 5hmC was
calculated as proportional to the optical density (OD) intensity
read at 450 nm using a microplate reader (BioTek Instruments,
Inc.). The kit demonstrates excellent sensitivity, with a detection
limit of 0.02 ng for 5mC and 0.04 ng for 5hmC DNA. Additionally, it
has high specificity, as the antibody selectively detects only 5mC
and 5hmC.
Statistical analysis
Graph Pad Prism (Version 8) was employed to create
various graphical representations of the data. The Statistical
Analysis System (SAS) program (2018) was used to assess the effects
of different groups (patients and controls) on the study
parameters. The mean ± standard error (SE) were calculated for each
group. An unpaired t-test was employed to evaluate the significance
of differences between the means. One-way ANOVA was used to assess
the differences among the age groups. Tukey's Honest Significant
Difference (HSD) post hoc test was performed for pairwise
comparisons. Cohen's d was calculated to assess the effect size of
the difference between the two group means. A value of P<0.05
was considered to indicate a statistically significant
difference.
Results
A total of 45 females (75%) and 15 males (25%) with
RA participated in the present study, while the control group
comprised 30 females (75%) and 10 males (25%), as presented in
Table I. The mean age of the
patients with RA was 48.72±1.32 years, compared to 45.60±1.63 years
for the control group. The body mass index (BMI) results for both
the patients with RA and the control group are presented in
Table I. The mean BMI of the
patients with RA was 28.52±0.46, while the control group had a mean
BMI of 27.81±0.44, indicating that both groups were classified as
overweight. However, no significant differences were observed in
age or sex based on the obtained P-values. Additionally, the
present study demonstrated a significant increase in the ESR among
patients with RA compared with the control group, with values of
39.67±1.91 and 8.69±0.59 mm/h, respectively. The results of the CBC
indicated that the mean red blood cell (RBC) count of patients with
RA was 4.49±0.07x106/µl, compared to
4.97±0.10x106/µl in the control group, exhibiting a
significant decrease (P-value=0.0002). However, the white blood
cell (WBC) and platelet (PLT) counts did not exhibit significant
differences between the patients with RA and the control group.
Additionally, there was a significant increase in the mean
corpuscular volume in patients with RA compared with the control
group (Table I). A significant
increase in hematocrit (HCT) levels was observed in the control
group compared with the patients with RA. The mean corpuscular
hemoglobin concentration, mean corpuscular hemoglobin and
hemoglobin levels did not exhibit any significant differences
between the patients with RA and the control group. The results of
the WBC count indicated that the levels of neutrophils (57.96%),
lymphocytes (32.02%), monocytes (8.17%) and eosinophils (5.37%) in
the patients with RA were higher compared to those in the control
group, which had levels of 51.06, 26.09, 6.13 and 3.21%,
respectively. These results indicated a significant increase in the
levels of neutrophils (P-value=0.0003), lymphocytes
(P-value=0.0022), monocytes (P-value=0.0001) and eosinophils
(P-value=0.0001) in the patients with RA compared to the levels in
the control group (Table I).
 | Table IComparison of age, BMI, ESR and CBC
parameters between patients with and the control group. |
Table I
Comparison of age, BMI, ESR and CBC
parameters between patients with and the control group.
| | Mean ± SE | |
|---|
| Parameter | Patients with
RA | Controls | t-test | P-value | Cohen's d |
|---|
| Sex | Females (n=45)
(75%) | Females (n=30)
(75%) | - | 0.0087a | - |
| | Males (n=15)
(25%) | Males (n=10)
(25%) | - | 0.0087a | - |
| Age (years) | 48.72±1.32 | 45.60±1.63 | - | 0.141 NS | 0.303 |
| BMI
(kg/m2) | 28.52±0.46 | 27.81±0.44 | - | 0.294 NS | 0.215 |
| ESR (mm/h) | 39.67±1.91 | 8.69±0.59 | 4.752b | 0.0001 | 2.641 |
| RBC
(x106/µl) | 4.49±0.07 | 4.97±0.10 | 0.239b | 0.0002 | -0.808 |
| WBC
(103/µl) | 6.97±0.27 | 6.35±0.21 | 0.745 NS | 0.102 | 0.339 |
| PLT
(103/µl) | 265.20±9.82 | 261.00±9.73 | 28.462 NS | 0.769 | 0.060 |
| MCV (fl) | 86.19±0.79 | 83.98±0.60 | 2.132a | 0.0424 | 0.423 |
| MCHC (g/dl) | 33.49±0.37 | 33.04±0.15 | 0.926 NS | 0.333 | 0.200 |
| MCH (pg) | 29.03±0.53 | 28.95±0.39 | 1.425 NS | 0.913 | 0.022 |
| HCT (%) | 39.35±0.92 | 41.79±0.50 | 2.308a | 0.0386 | -0.513 |
| HGB (g/dl) | 13.06±0.23 | 13.57±0.20 | 0.647 NS | 0.117 | -0.325 |
| Neutrophils
(%) | 57.96±1.38 | 51.06±0.93 | 3.652b | 0.0003 | 0.771 |
| Lymphocytes
(%) | 32.02±1.24 | 26.09±1.38 | 3.745b | 0.0022 | 0.646 |
| Monocytes (%) | 8.17±0.34 | 6.13±0.28 | 0.941b | 0.0001 | 0.886 |
| Eosinophils
(%) | 5.37±0.31 | 3.21±0.18 | 0.824b | 0.0001 | 1.066 |
The results presented in Table II indicated that the anti-CCP
levels in the patients with RA were significantly higher
(P-value=0.0001) than those in the control group. A significant
difference (P-value=0.0003) was also observed in the CRP levels
between the patients with RA and the controls. The RF levels in the
patients with RA were 73.33%, which differed significantly
(P-value=0.0001) from those in the control group. Furthermore, the
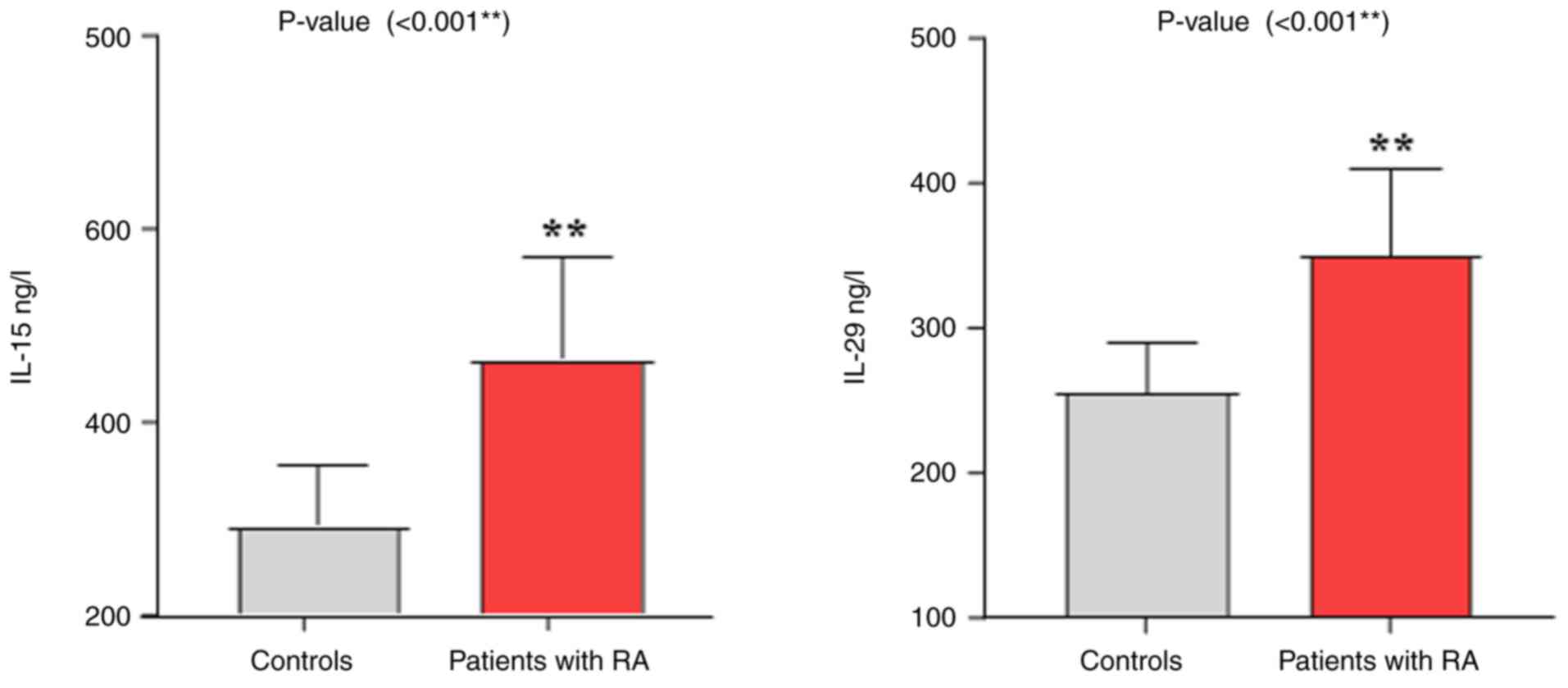
results from the analysis of the levels of IL-15 and IL-29
indicated that the patients with RA exhibited higher levels of both
ILs compared with the control group, with a P-value of 0.001,
demonstrating a highly significant difference between the two
groups (Table III and Fig. 1). Additionally, the results
presented in Table IV and
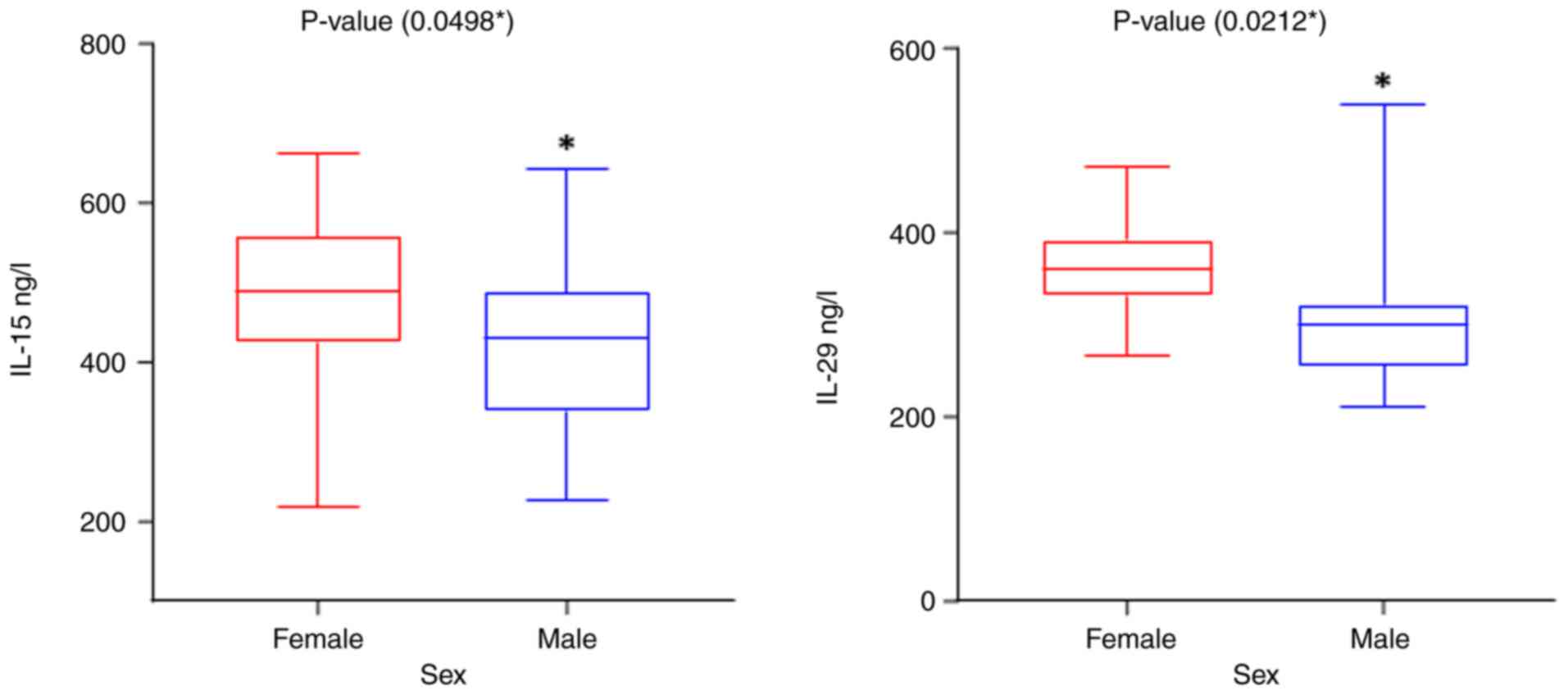
Fig. 2 indicate a significant
difference in IL-15 and IL-29 levels among the patients based on
sex, with the levels being higher in females compared to males
(Fig. 2). The results regarding
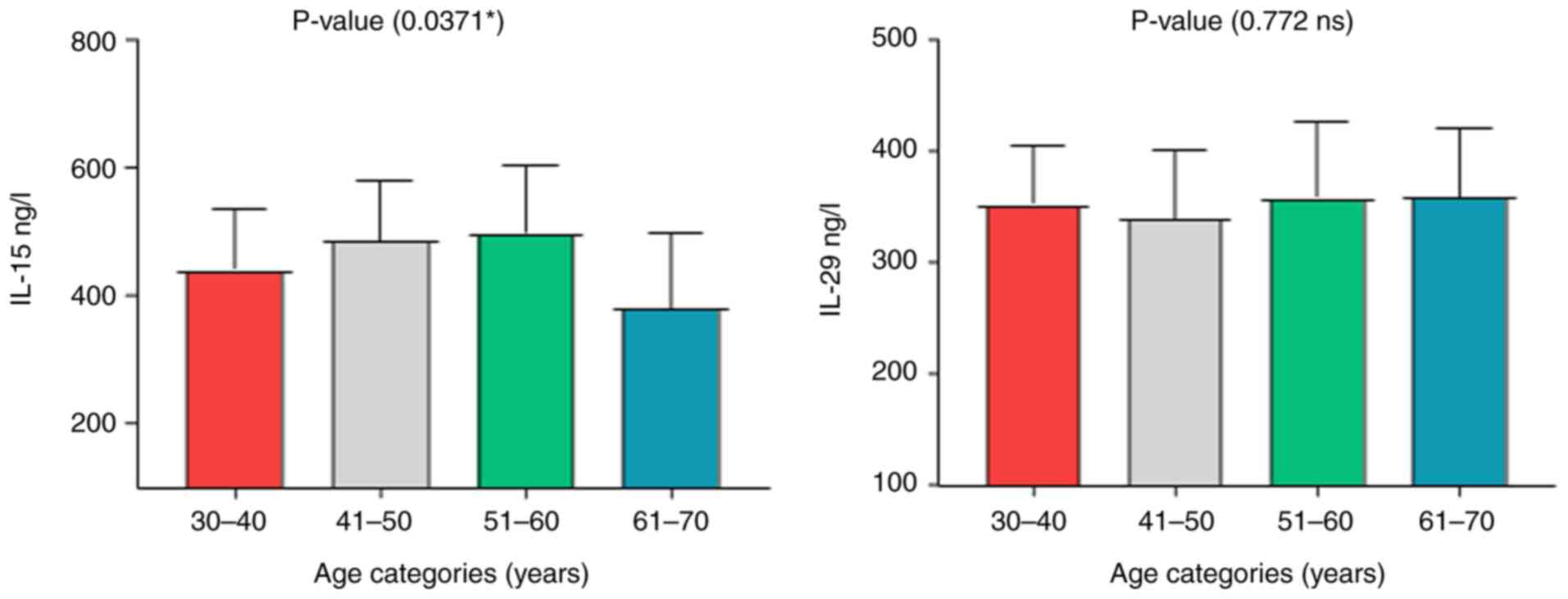
the age groups indicated that the P-value for the IL-15 levels was
0.037, suggesting a significant difference across the age groups.
By contrast, the P-value for IL-29 was 0.772, indicating no
significant difference in its levels among the age groups (Table IV and Fig. 3). The results of the analysis of
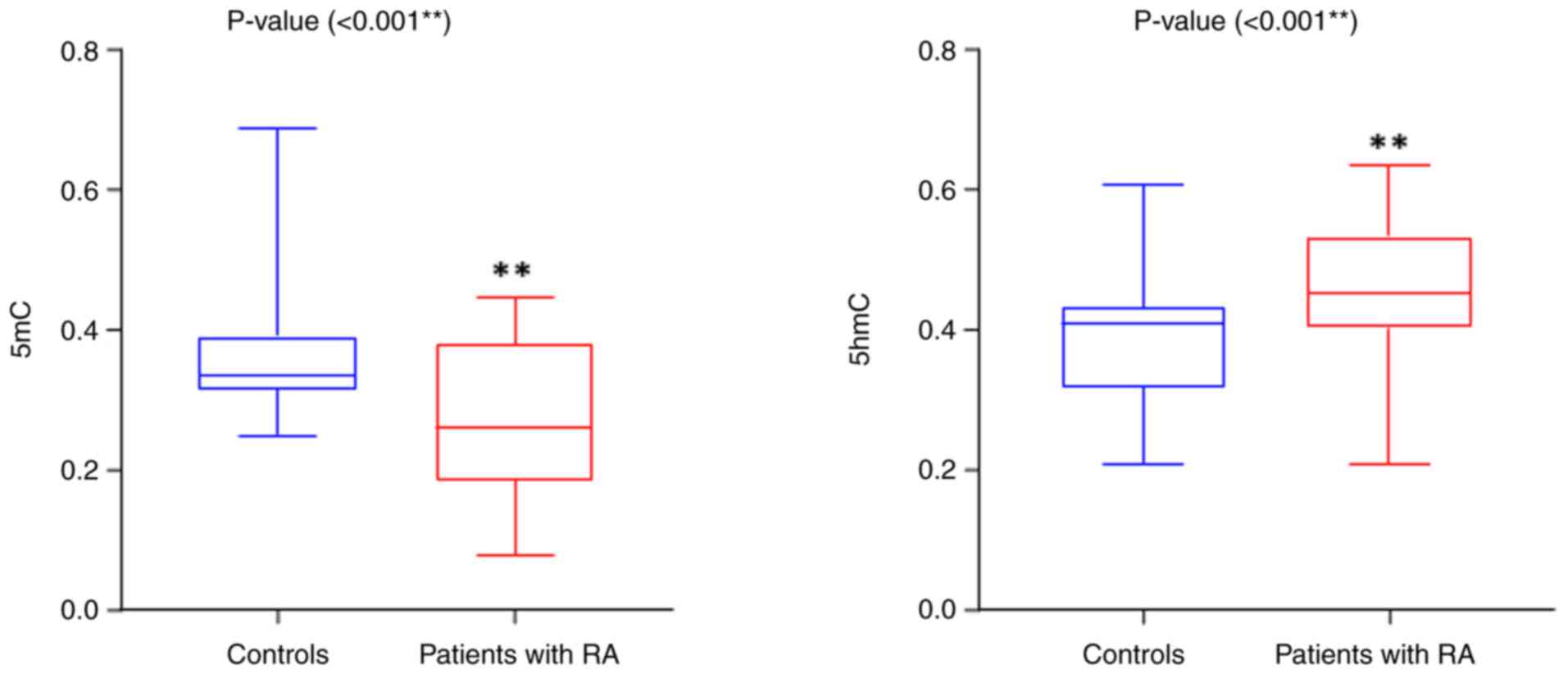
global DNA methylation (5mC and 5hmC; Table V) revealed a highly significant
decrease in 5mC levels in the patients with RA compared with the
control group, with a P-value of 0.0001. Conversely, the 5hmC
levels were elevated in the patients with RA compared with the
control group, exhibiting a significant difference
(P-value=0.0004), as illustrated in Fig. 4. Additionally, the results
indicated no significant differences in the 5hmC levels based on
age groups or sex among the patients with RA, as presented in
Table VI.
 | Table IIComparison of CRP, anti-CCP and RF
between patients with RA and the controls. |
Table II
Comparison of CRP, anti-CCP and RF
between patients with RA and the controls.
| | Mean ± SE | |
|---|
| Parameter | Patients with
RA | Controls | P-value | Cohen's d |
|---|
| CRP | 11.50±1.12 | 4.83±0.38 | 0.0003 | 0.985 |
| Anti-CCP | 0.629±0.03 | 0.307±0.01 | 0.0001 | 1.962 |
| RF % | 73.33% | 0% | 0.0001 | - |
 | Table IIIComparison of IL-15 and IL-29 levels
between patients with RA and the controls. |
Table III
Comparison of IL-15 and IL-29 levels
between patients with RA and the controls.
| | Mean ± SE |
|---|
| Group | IL-15 (ng/l) | IL-29 (ng/l) |
|---|
| Patients with
RA | 464.61±13.89 | 349.45±7.87 |
| Control | 291.47±10.32 | 255.37±5.45 |
| P-value | 0.001 | 0.001 |
| Cohen's d | 1.859 | 1.807 |
 | Table IVDistribution of IL-15 and IL-29 in
patients with RA according to age groups and sex. |
Table IV
Distribution of IL-15 and IL-29 in
patients with RA according to age groups and sex.
| | Parameters Mean ±
SE | |
|---|
| Descriptive
data | | IL-15 (ng/l) | IL-29 (ng/l) |
|---|
| Sex | Female | 479.35±15.86 | 359.68±7.38 |
| | Male | 420.40±26.43 | 318.73±20.98 |
| | P-value | 0.0498 | 0.0212 |
| | Cohen's d | 0.6 | 0.7 |
| Age categories
(years) | 30-40 | 440.29±25.524 | 351.21±14.267 |
| | 41-50 | 485.67±21.174 | 338.43±13.530 |
| | 51-60 | 498.71±25.813 | 357.53±16.520 |
| | 61-70 | 379.50±42.338 | 358.13±21.915 |
| | P-value | 0.037a | 0.772 |
| | Eta-squared | 0.139 | 0.020 |
 | Table VComparison of 5mC and 5hmC levels
between patients with and the controls. |
Table V
Comparison of 5mC and 5hmC levels
between patients with and the controls.
| | Mean ± SE |
|---|
| Group | 5mC | 5hmC |
|---|
| Patients with
RA | 0.269±0.01 | 0.227±0.01 |
| Control | 0.376±0.02 | 0.193±0.01 |
| t-test | 0.0438a | 0.0187a |
| P-value | 0.0001 | 0.0001 |
| Cohen's d | 0.984 | 0.754 |
 | Table VIDistribution of 5mC and 5hmC in
patients with RA according to age groups and sex. |
Table VI
Distribution of 5mC and 5hmC in
patients with RA according to age groups and sex.
| | Parameter (mean ±
SE) | |
|---|
| Descriptive
data | | 5mC | 5hmC |
|---|
| Sex | Female | 0.270±0.02 | 0.231±0.01 |
| | Male | 0.268±0.03 | 0.217±0.01 |
| | P-value | 0.939 NS | 0.328 NS |
| | Cohen's d | 0.023 | 0.294 |
| Age categories
(years) | 30-40 | 0.296±0.03 | 0.231±0.011 |
| | 41-50 | 0.255±0.023 | 0.222±0.01 |
| | 51-60 | 0.260±0.024 | 0.228±0.012 |
| | 61-70 | 0.283±0.04 | 0.237±0.019 |
| | P-value | 0.699 NS | 0.877 NS |
| | Eta-squared | 0.025 | 0.012 |
Discussion
The results of the present study indicated no
significant differences between the patients with RA and the
control group as regards age and sex. However, clinical and
laboratory data from previous research suggest that demographic
factors, such as age, sex, disease duration and clinical
parameters, including ESR and anti-CCP levels, do not differ
significantly among patients with RA with mild, moderate and severe
disease (22). The serum marker
findings from the present study align with those observed in
previous research (23),
particularly the increased levels of the inflammatory marker, CRP,
in the patients with RA compared with the controls. Furthermore,
the levels of autoantibodies, specifically anti-CCP, were
significantly elevated in the patient group compared with the
controls. RF autoantibodies were detected in 73.33% of the patients
with RA. These findings are consistent with those in the studies by
Bagdi et al (24) and
Hashiam and Aldahhan (25).
Elevated CRP levels are associated with higher disease activity in
RA and play a role in bone damage and disease progression. Anti-CCP
and RF autoantibodies have been utilized as diagnostic markers for
RA for several years. However, ~25% of patients with RA do not
produce RF (26), which is
consistent with the findings of the current study. Anti-CCP
exhibited a specificity of 95-99% and a sensitivity of 60-75%,
rendering them a valuable tool for the early diagnosis of RA
(27).
Another notable finding of the present study is the
elevated concentrations of IL-15 and IL-29 in patients with RA,
indicating a highly significant difference compared with the
control group. These findings align with those of previous studies
(14,28). Age groups influence IL-15 levels in
RA patients, while IL-29 levels remain unaffected by age.
Additionally, the levels of both ILs are not affected by sex.
Elevated IL-15 and IL-29 levels could be implicated in the
pathogenesis of RA by several mechanisms. IL-15 plays a critical
role in the pathogenesis and progression of RA. It activates and
promotes the proliferation of T-cells and natural killer cells,
which produce pro-inflammatory cytokines, leading to increased
inflammation and joint damage. Additionally, IL-15 is involved in
the activation of neutrophils and macrophages, which play a crucial
role in modulating inflammation, as well as in the survival and
proliferation of synovial fibroblasts, leading to pro-inflammatory
cytokine production, contributing to synovial hyperplasia.
Furthermore, this cytokine stimulates the production of TNF-α and
IL-17, exacerbating joint inflammation and osteoclastogenesis,
which is associated with the progression of joint destruction
(10,29). Furthermore, IL-15 plays a critical
role in the early stages of RA development (11), and has been identified as a
potential therapeutic target (30). In patients with undifferentiated
arthritis (UA), IL-15 levels were more pronounced in those who
later developed RA compared to those with RF or anti-CCP
antibodies. Consequently, IL-15 has been proposed as a serum marker
for RA, demonstrating greater sensitivity and specificity than the
aforementioned autoantibodies (31). Furthermore, the role of IL-29 in
the development of RA has recently been discovered. IL-29
contributes to neutrophil chemotaxis, triggering inflammatory
responses and tissue damage, inhibiting the differentiation of
T-follicular helper cells and stimulating fibroblast-like
synoviocytes, which leads to synovial hyperplasia and contributes
to joint bone destruction; it also induces the production of
pro-inflammatory cytokines involved in the pathogenesis of RA, such
as IL-6, IL-8 and IL-10. This cascade promotes inflammation,
exacerbating joint inflammation and damage. Additionally, IL-29
stimulates the activation and differentiation of B-cells, which are
responsible for producing RF and anti-CCP antibodies; these
autoantibodies play a crucial role in the development and
progression of RA by triggering inflammation, the formation of the
immune complex and tissue damage (32,33).
The use of IL-29 biologics in patients with RA blocks the
production of T-follicular helper cells, which may help suppress
disease progression and provide novel targets for clinical
treatment (14,34). Overall, the findings of the present
study suggest that the levels of IL-15 and IL-29 in patients with
RA could serve as prognostic risk factors and may be utilized in
the diagnosis of the disease.
The results of the present study also revealed a
decreased level of 5mC in patients with RA, indicating DNA
hypomethylation in PBMCs, which aligns with findings from previous
research (35). In addition, there
was no significant effect of age or sex on 5mC levels. The
dysregulation of DNA methylation affects gene expression in immune
cells by coordinating the control of immune cell differentiation
and function. This dysregulation affects the activation of immune
responses and inflammatory pathways, thereby contributing to the
pathogenesis and development of RA (36,37).
There are several potential explanations for the decrease in DNA
methylation, leading to DNA hypomethylation in patients with RA.
These include the action of pro-inflammatory cytokines, such as
TNF-α, which can stimulate alterations in DNA methylation.
Additionally, the inflammatory environment plays a critical role in
influencing DNA methylation. Elevated levels of oxidative stress
and environmental factors, such as smoking, can also induce changes
in DNA methylation (38,39).
DNA hypomethylation has been observed in the PBMCs
of patients with RA (40).
Differential methylation patterns have also been reported in RA
synovial fibroblasts, which correspond to the aggressive phenotype
acquired by these fibroblasts and contribute to the development of
RA. Abnormal gene expression resulting from DNA hypomethylation has
been associated with increased levels of various genes, including
growth factors and receptors, extracellular matrix proteins,
adhesion molecules and matrix-degrading enzymes (41,42).
By contrast, the present study found that levels of 5hmC were
increased in patients with RA compared with the control group.
These findings align with those of previous research (43), indicating that 5hmC levels are
higher in patients with both RA and osteoarthritis. It has been
suggested that the TET3 enzyme functions as an epigenetic
gatekeeper at the point of no return in the progression and
chronicity of RA. It has been established that patients with RA
exhibit distinct methylation patterns in their genomes compared to
healthy individuals, as well as different DNA methylation profiles
between treatment responders and non-responders (43). These findings suggest the potential
for using DNA methylation as a predictive biomarker (44). The analysis of CpG sites within the
promoters of genes in patients with RA indicates that changes in
DNA methylation occur at a very early stage of the disease, with
numerous genes displaying significant hypermethylation at their
promoter sites. Thus, the identified epigenetic modifications in
genes may serve as valuable prognostic biomarkers for the
progression of RA (45). The
findings of the present study also indicated no association between
age and sex as regards DNA methylation and hydroxymethylation in
patients with RA. However, previous studies (46,47)
have revealed a positive association between age and the decline in
global DNA methylation (5mC). One possible explanation is the
age-related changes in the expression of enzymes involved in DNA
methylation and demethylation. The findings of the present study
have notable clinical implications. The elevated levels of IL-15
and IL-29, and the alteration of DNA methylation levels in the
patient with RA indicates that these factors contribute to the
pathogenesis of RA. Furthermore, this indicates that DNA
methylation may serve as early diagnostic tool for the disease, and
may also function as a target in novel treatment strategies.
Increased DNA hydroxymethylation is associated with a decrease in
DNA methylation levels (hypomethylation) in patients. This
connection reflects a dynamic epigenetic backdrop in RA.
The present study is an extension of previous
research in the field. examining the levels of IL-15, IL-29 and
global DNA methylation modifications on the pathogenesis of RA.
While previous studies (11,35,48)
have examined the roles of cytokines and epigenetic changes
separately, the present study provides novel insight by combining
these cytokine levels with epigenetic changes. This integrated
analysis provides a more comprehensive understanding of the
interplay of these biomarkers in RA pathogenesis and may aid in the
development of novel therapeutic strategies targeting both
inflammatory and epigenetic pathways.
However, future research with a larger sample size
could enable a more robust analysis. Further studies are required
in the future to compare the results obtained in RA with those of
other autoimmune diseases. Moreover, longitudinal follow-up studies
that evaluate the association between epigenetic and immunological
variations and disease progression, as well as assess changes in
these markers with treatment, could provide valuable insight into
the potential applications of these findings.
In conclusion, the present study indicated
statistically significant differences in the levels of 5mC and 5hmC
between patients with RA and healthy controls. However, further
research is required to extensively explore the association between
DNA methylation and demethylation patterns in genes associated with
RA. Additionally, focusing on IL-15 levels in seronegative patients
and targeting IL-29 in new therapies may provide insight into novel
aspects of the pathogenesis of the disease and may lead to the
identification of novel therapeutic targets.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
Both authors (NAH and RMKAJ) contributed to the
conception and design of the study, as well as in the preparation
of materials, including the ELISA kits. Sample and data collection,
analysis, and manuscript writing were carried out by NAH. RMKAJ was
involved in the reviewing and editing of the manuscript, and
supervised the study. Both authors have read and approved the final
manuscript. Additionally, NAH and RMKAJ confirm the authenticity of
all the raw data
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Department of Biology, College of Science,
University of Baghdad (Baghdad, Iraq), under reference no.
CSEC/0923/0105, on September 25, 2023. Written informed consent was
obtained from all patients for their participation in the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sokolova MV, Schett G and Steffen U:
Autoantibodies in rheumatoid arthritis: Historical background and
novel findings. Clin Rev Allergy Immunol. 63:138–151.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhang HG, McDermott G, Seyok T, Huang S,
Dahal K, L'Yi S, Lea-Bonzel C, Stratton J, Weisenfeld D, Monach P,
et al: Identifying shared genetic architecture between rheumatoid
arthritis and other conditions: A phenome-wide association study
with genetic risk scores. eBioMedicine. 92(104581)2023.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Rija FF, Hussein SZ and Abdalla MA:
Physiological and immunological disturbance in rheumatoid arthritis
patients. Baghdad Sci J. 18(0247)2021.
|
|
4
|
Li Z and Wang XQ: Clinical effect and
biological mechanism of exercise for rheumatoid arthritis: A mini
review. Front Immunol. 13(1089621)2023.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Poniewierska-Baran A, Bochniak O, Warias P
and Pawlik A: Role of sirtuins in the pathogenesis of rheumatoid
arthritis. Int J Mol Sci. 24(1532)2023.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Edilova MI, Akram A and Abdul-Sater AA:
Innate immunity drives pathogenesis of rheumatoid arthritis. Biomed
J. 44:172–182. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Mutlak QM and Kasim AA: Impact of MTHFR
gene polymorphism on the outcome of methotrexate treatment in a
sample of Iraqi rheumatoid arthritis patients. Sci Rep.
14(15119)2024.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Laragione T, Harris C, Azizgolshani N,
Beeton C, Bongers G and Gulko PS: Magnesium increases numbers of
Foxp3+ Treg cells and reduces arthritis severity and joint damage
in an IL-10-dependent manner mediated by the intestinal microbiome.
EBioMedicine. 92(104603)2023.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Matsumoto H, Fujita Y, Asano T, Matsuoka
N, Temmoku J, Sato S, Yashiro-Furuya M, Yokose K, Yoshida S, Suzuki
E, et al: Association between inflammatory cytokines and
immune-checkpoint molecule in rheumatoid arthritis. PLoS One.
16(e0260254)2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Reyes-Pérez IV, Sánchez-Hernández PE,
Muñoz-Valle JF, Martínez-Bonilla GE, García-Iglesias T,
González-Díaz V, García-Arellano S, Cerpa-Cruz S, Polanco-Cruz J
and Ramírez-Dueñas MG: Cytokines (IL-15, IL-21, and IFN-γ) in
rheumatoid arthritis: Association with positivity to autoantibodies
(RF, anti-CCP, anti-MCV, and anti-PADI4) and clinical activity.
Clin Rheumatol. 38:3061–3071. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kurowska W, Przygodzka M, Jakubaszek M,
Kwiatkowska B and Maslinski W: Interleukin-15 as a biomarker
candidate of rheumatoid arthritis development. J Clin Med.
9(1555)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Koper-Lenkiewicz OM, Sutkowska K,
Wawrusiewicz-Kurylonek N, Kowalewska E and Matowicka-Karna J:
Proinflammatory cytokines (IL-1, -6, -8, -15, -17, -18, -23, TNF-α)
single nucleotide polymorphisms in rheumatoid arthritis-a
literature review. Int J Mol Sci. 23(2106)2022.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang JM, Huang AF, Xu WD and Su LC:
Insights into IL-29: Emerging role in inflammatory autoimmune
diseases. J Cell Mol Med. 23:7926–7932. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xu T, Yan T and Li P: Interleukin-29
regulates T follicular helper cells by repressing BCL6 in
rheumatoid arthritis patients. Clin Rheumatol. 39:3797–3804.
2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yang C, Li D, Teng D, Zhou Y, Zhang L,
Zhong Z and Yang GJ: Epigenetic regulation in the pathogenesis of
rheumatoid arthritis. Front Immunol. 13(859400)2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Al-Attar MM, Al-Awadi SJA and Abdulfattah
SY: Gene expression and methylation levels of PCSK9 gene in iraqi
patients with coronary artery disease. Baghdad Sci J. 20:2124–2133.
2023.
|
|
17
|
Zhao J, Wei K, Chang C, Xu L, Jiang P, Guo
S, Schrodi SJ and He D: DNA methylation of T lymphocytes as a
therapeutic target: Implications for rheumatoid arthritis etiology.
Front Immunol. 13(863703)2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ahmed DF and AL-Jumaily RMK: Role of
hematopoietic growth factors as immune modulators (GM-CSF &
IL-3) in newly diagnosed colorectal cancer patients and their
correlation with P53 expression and global DNA methylation. Iraqi J
Sci. 65:3047–3056. 2024.
|
|
19
|
Gosselt HR, Griffioen PH, van Zelst BD,
Oosterom N, de Jonge R and Heil SG: Global DNA (hydroxy)methylation
is stable over time under several storage conditions and
temperatures. Epigenetics. 16:45–53. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hageman I, Mol F, Atiqi S, Joustra V,
Sengul H, Henneman P, Visman I, Hakvoort T, Nurmohamed M, Wolbink
G, et al: Novel DNA methylome biomarkers associated with adalimumab
response in rheumatoid arthritis patients. Front Immunol.
14(1303231)2023.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Nair N and Wilson AG: Assessing the
potential of epigenetic targets as biomarkers in the diagnosis and
treatment of rheumatoid arthritis. Expert Rev Clin Immunol.
19:483–488. 2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hajizadeh MR, Moslemizadeh N, Rezaeian M,
Ranjbar FM and Abbasifard M: Expression of genes involved in
epigenetic modifications in patients with rheumatoid arthritis. Hum
Gene. 33(201054)2022.
|
|
23
|
Ibrahim S and Sarhat E: EvaluatIon of
serum levels of interleukin-6, fetuin-a, lipocalin-2, and
C-reactive protein in rheumatoid arthritis patients. Georgian Med
News. 42–45. 2022.PubMed/NCBI
|
|
24
|
Bagdi R, Aswani P, Singh VK and Verma MK:
C-reactive protein as a disease activity marker in rheumatoid
arthritis. Int J Health Sci. 6:10587–10593. 2022.
|
|
25
|
Hashiam MD and Aldahhan HAA: Assessment of
auto-antibodies (RF, Anti-CCP, and anti-RA33) in rheumatoid
arthritis patients: Comparative study. Int J Health Sci.
6:5434–5444. 2022.
|
|
26
|
Zheng Z, Mergaert AM, Fahmy LM, Bawadekar
M, Holmes CL, Ong IM, Bridges AJ, Newton MA and Shelef MA:
Disordered antigens and epitope overlap between anti-citrullinated
protein antibodies and rheumatoid factor in rheumatoid arthritis.
Arthritis Rheumatol. 72:262–272. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ma L, Wang W, Li L, Chen Y, Chen B, Shao
M, Cheng Y and Zhou R: Comparison of different assays for the
detection of anticyclic citrullinated peptide antibodies in
patients with rheumatoid arthritis. Front Immunol.
13(940713)2022.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Lin SJ, Hsu CY, Kuo ML, Lee PT, Hsiao HS
and Chen JY: Phenotypic and functional characterization of natural
killer cells in rheumatoid arthritis-regulation with
interleukin-15. Sci Rep. 10(5858)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Allard-Chamard H, Mishra HK, Nandi M,
Mayhue M, Menendez A, Ilangumaran S and Ramanathan S:
Interleukin-15 in autoimmunity. Cytokine.
136(155258)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Shah P, Siddique A, Thakkar A, Gharat S,
Godad A, Kale P and Doshi G: An update on novel therapeutic
intervention in rheumatoid arthritis. Int Immunopharmacol.
109(108794)2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Fonseca Peixoto R, Ewerton Maia Rodrigues
C, Henrique de Sousa Palmeira P, Cézar Comberlang Queiroz Davis Dos
Santos F, Keesen de Souza Lima T and de Sousa Braz A: Immune
hallmarks of rheumatoid arthritis management: A brief review.
Cytokine. 158(156007)2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Xu TS, Jia SY and Li P: Interleukin-29 and
interleukin-28A induce migration of neutrophils in rheumatoid
arthritis. Clin Rheumatol. 40:369–375. 2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhao J, Guo S, Schrodi SJ and He D:
Molecular and cellular heterogeneity in rheumatoid arthritis:
Mechanisms and clinical implications. Front Immunol.
12(790122)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Mohamed MMH and Abdelsattar AB:
Interleukin-29 in autoimmune diseases: Review article. Egypt J Hosp
Med. 91:4614–4618. 2023.
|
|
35
|
Liebold I, Grützkau A, Göckeritz A, Gerl
V, Lindquist R, Feist E, Zänker M, Häupl T, Poddubnyy D, Zernicke
J, et al: Peripheral blood mononuclear cells are hypomethylated in
active rheumatoid arthritis and methylation correlates with disease
activity. Rheumatology (Oxford). 60:1984–1995. 2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhang R, Chang C, Jin Y, Xu L, Jiang P,
Wei K, Xu L, Guo S, Sun S and He D: Identification of DNA
methylation-regulated differentially expressed genes in RA by
integrated analysis of DNA methylation and RNA-Seq data. J Transl
Med. 20(481)2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Guo S, Xu L, Chang C, Zhang R, Jin Y and
He D: Epigenetic regulation mediated by methylation in the
pathogenesis and precision medicine of rheumatoid arthritis. Front
Genet. 11(811)2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Webster AP, Plant D, Ecker S, Zufferey F,
Bell JT, Feber A, Paul DS, Beck S, Barton A, Williams FMK and
Worthington J: Increased DNA methylation variability in rheumatoid
arthritis-discordant monozygotic twins. Genome Med.
10(64)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Rodríguez-Ubreva J, de la Calle-Fabregat
C, Li T, Ciudad L, Ballestar ML, Català-Moll F, Morante-Palacios O,
Garcia-Gomez A, Celis R, Humby F, et al: Inflammatory cytokines
shape a changing DNA methylome in monocytes mirroring disease
activity in rheumatoid arthritis. Ann Rheum Dis. 78:1505–1516.
2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Hoang TT, Lee Y, McCartney DL, Kersten
ETG, Page CM, Hulls PM, Lee M, Walker RM, Breeze CE, Bennett BD, et
al: Comprehensive evaluation of smoking exposures and their
interactions on DNA methylation. EBioMedicine.
100(104956)2024.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Anteneh H, Fang J and Song J: Structural
basis for impairment of DNA methylation by the DNMT3A R882H
mutation. Nat Commun. 11(2294)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Nemtsova MV, Zaletaev DV, Bure IV,
Mikhaylenko DS, Kuznetsova EB, Alekseeva EA, Beloukhova MI,
Deviatkin AA, Lukashev AN and Zamyatnin AA Jr: Epigenetic changes
in the pathogenesis of rheumatoid arthritis. Front Genet.
10(570)2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Kawabe A, Yamagata K, Kato S, Nakano K,
Sakata K, Tsukada YI, Ohmura K, Nakayamada S and Tanaka Y: Role of
DNA dioxygenase Ten-Eleven translocation 3 (TET3) in rheumatoid
arthritis progression. Arthritis Res Ther. 24(222)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Cribbs A, Feldmann M and Oppermann U:
Towards an understanding of the role of DNA methylation in
rheumatoid arthritis: Therapeutic and diagnostic implications. Ther
Adv Musculoskelet Dis. 7:206–219. 2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Karouzakis E, Raza K, Kolling C, Buckley
CD, Gay S, Filer A and Ospelt C: Analysis of early changes in DNA
methylation in synovial fibroblasts of RA patients before
diagnosis. Sci Rep. 8(7370)2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Morgan AE, Davies TJ and Mc Auley MT: The
role of DNA methylation in ageing and cancer. Proc Nutr Soc.
77:412–422. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Sahar BR and Al-Jumaily RMK: The impact of
global DNA methylation and hypoxia-inducible factor 1 alpha levels
in the progression of breast cancer. Opera Med Physiol. 11:28–39.
2024.
|
|
48
|
Wang F, Xu L, Feng X, Guo D, Tan W and
Zhang M: Interleukin-29 modulates proinflammatory cytokine
production in synovial inflammation of rheumatoid arthritis.
Arthritis Res Ther. 14(R228)2012.PubMed/NCBI View
Article : Google Scholar
|