Introduction
Although effective pharmacologic therapies for
mental illnesses are available, their efficacy is restricted,
largely due to the underlying genetic variability across
psychiatric patients and the subsequent low compliance due to the
frequency of side-effects (1).
Specifically, as regards depression, one third of patients
receiving antidepressant medications do respond to treatment and
two thirds of patients do not achieve remission (2). Additionally, ~42% of patients
discontinue their prescribed antidepressant medication in the 1st
month and 70% of patients discontinue during the first 3 months of
treatment, while 45% of patients do not take their medication as
indicated by their doctor (3,4).
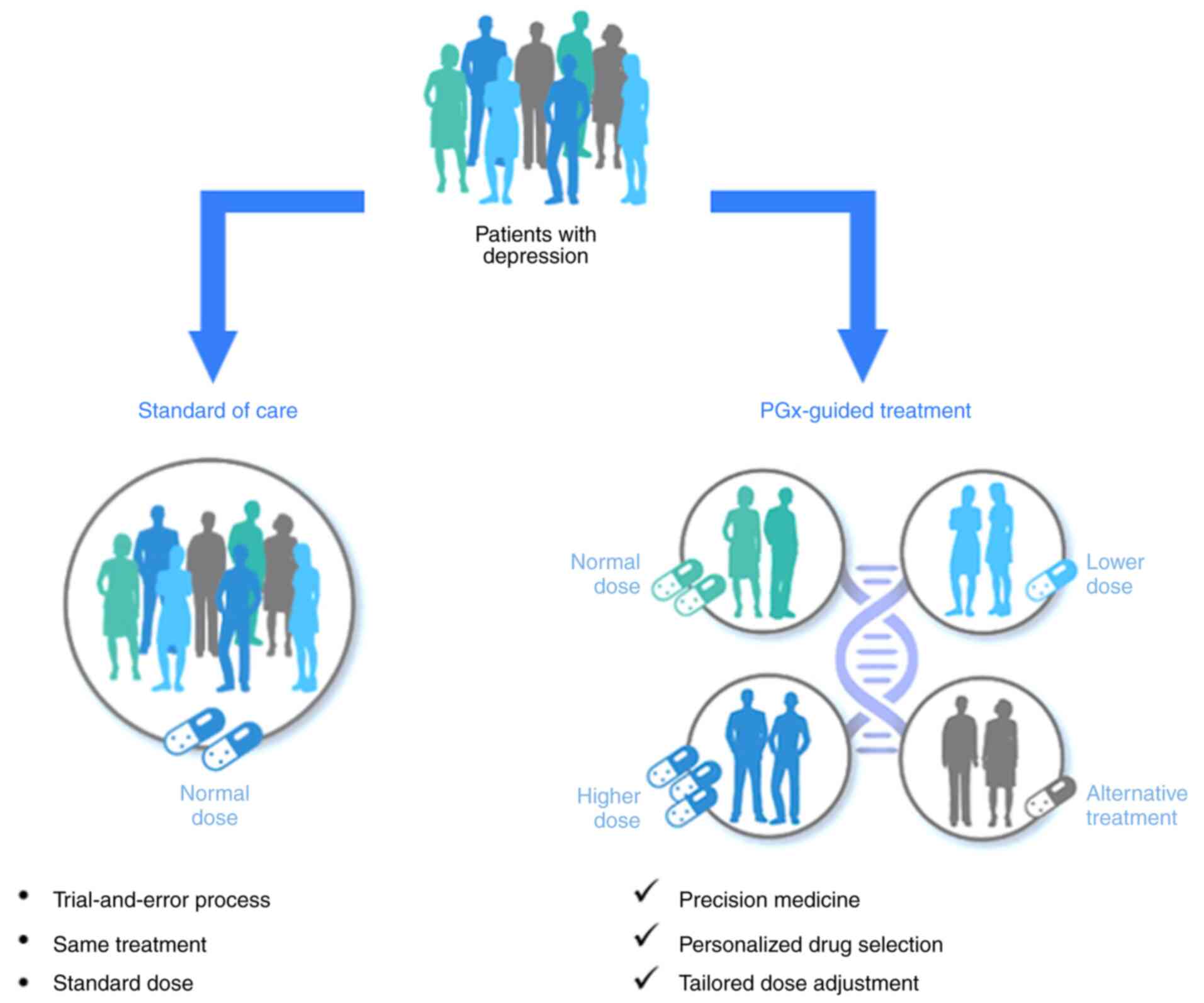
The current standard of care in psychiatric practice
relies on a trial-and-error approach that combines the experience
of a physician with specific clinical indicators. For patients
sharing the same diagnosis, most commonly, the same medication is
prescribed, usually at the recommended dose as per the drug label.
Due to interindividual differences with regards to genetics and
lifestyle, this process can take weeks, months, or even years
before the right medication is found. Pharmacogenetic (PGx)
testing, however, can personalize treatment and dosage selection,
and may thus help to avoid these long periods of trial-and-error,
empowering a precision medicine approach in mental health care
(Fig. 1). PGx testing can
identify, at a personalized level, genetic characteristics that may
predict the clinical response of a patient to a certain medication
and the probability of adverse events being developed, based on
specific genetic variants that they carry, which, among other
things, can alter the activity of enzymes that metabolize central
nervous system (CNS) medications, such as antidepressants (5,6).
Antidepressant medications are mainly metabolized by
cytochrome P450 (CYP450) enzymes, including CYP2D6 and
CYP2C19(1). Genetic variants,
specifically single nucleotide polymorphisms (SNPs), in these genes
can alter the activity levels of these enzymes, resulting in a
reduced or no activity, or even increased or significantly
increased activity. The resulting predicted phenotypes are defined
as poor, intermediate, rapid and ultra-rapid metabolizer,
respectively. Psychiatric patients with the poor or intermediate
metabolizer phenotypes may be at an increased risk of developing
adverse events when treated with medications metabolized by these
enzymes, due to the slower degradation of the drugs that results in
higher concentrations in the plasma for prolonged periods of time.
These patients, once identified by PGx testing, would be
recommended to initiate therapy on a lower dose, slow titration, or
even alternative medication. Patients with a rapid or ultra-rapid
metabolizer genotype, on the other hand, could experience either
side-effects or treatment ineffectiveness as a result of the drugs
being metabolized too rapidly and failing to act in time (7). These patients, once identified by PGx
testing, would be recommended fast titration, initiate therapy on a
higher dose, or even alternative medication not metabolized as
rapidly.
The most recent meta-analyses on major depressive
disorder (MDD) indicate that PGx-guided treatment for
moderate-to-severe depression is significantly more likely to
result in response and remission than treatment as usual (TAU)
(8,9). Despite the low confidence about the
extent, due to study limitations, there is confidence in the
direction. More precisely, the positive effects of PGx guidance for
individuals suffering from moderate-to-severe MDD may have
significant benefits for both the patients and the healthcare
system, due to the scope and magnitude of the disorder and its
consequences (8,9).
The present study aimed to examine the extent of
genetic variants in a psychiatric population, with regards to genes
implicated in antidepressant drugs metabolism, comprising CYP2D6
and CYP2C19, and response, including FKBP prolyl isomerase 5
(FKBP5). FKBP5 is a gene that encodes a protein involved in the
regulation of the stress response and has been implicated in the
pathophysiology of MDD. Variants in FKBP5 have been shown to
influence the efficacy of antidepressant treatments, thus rendering
it a key factor in pharmacogenetic studies. Specifically, the SNP
rs4713916 in FKBP5 has been shown to be associated with varying
responses to antidepressant medications, categorizing individuals
into groups with a good, increased or reduced response to treatment
(10,11). Furthermore, the present study
employed a questionnaire to assess the clinical utility of an in
vitro diagnostic PGx test, the iDNA PGx CNS (12,13),
in a population of patients suffering from MDD. Specifically,
patient responses were collected, following PGx testing and
personalized drug and dosage selection, in terms of response to
treatment, severe adverse events, changes in medication, and
frequency of doctor visits and communications.
Materials and methods
Patient data and analyses
Of note, the present study used pre-existing
anonymized data that does not allow for the identification of
individuals. The present study did not involve any invasive
procedures or direct interaction with patients. The present study
only utilized pre-existing anonymized genetic data. These factors
place the research outside the scope of requiring formal ethical
review, as it poses no foreseeable risks to individuals.
Nevertheless, it was ensured that the study adhered to the highest
ethical standards available and was carried out in accordance with
the Declaration of Helsinki. All participants were able to and
provided informed consent, allowing for the use of their anonymized
genetic data for research and statistical purposes. This also
applies to the questionnaire survey for which all participants were
capable of and provided informed consent, allowing for the use of
their answers for research and statistical purposes. Moreover, all
participants had the right to withdraw their data at any time,
ensuring their autonomy and control over their information.
The distribution of CYP2D6, CYP2C19 and FKBP5
genotypes was analyzed in a psychiatric patient population of 1,387
individuals in Greece. The genetic material of patients was
collected from buccal swab samples using the iDNA PGx CNS kit. Α
minimum DNA concentration of 50 ng in 1 ml sterile water was
required to ensure sufficient DNA yield for genotyping. DNA
extraction and purification were performed employing the PureLink
genomic DNA mini kit and genotyping was performed using PCR on
custom made iDNA PGx CNS OpenArrays on a QuantStudio 12K Flex
Real-Time PCR (Thermo Fisher Scientific, Inc.). A bioinformatics
platform, including an internal iDNA PGx CNS database and an
interface, was employed for reporting the genotypes and PGx-based
recommendations. Specifically, the iDNA PGx CNS panel, evaluates 24
SNPs across 13 genes (CYP2C19, CYP2C9, CYP2D6, DRD2, DRD3, EPHX1,
FKBP5, GRIK1, MC4R, SCN1A, ANKK1/DRD2, HTR2C and UGT2B7) and their
interactions with 30 CNS drugs, comprising antidepressants
(amitriptyline, citalopram, clomipramine, duloxetine, escitalopram,
fluoxetine, fluvoxamine, mirtazapine, paroxetine, sertraline,
venlafaxine and vortioxetine), antipsychotics (amisulpride,
aripiprazole, clozapine, haloperidol, olanzapine, paliperidone,
quetiapine, risperidone and ziprasidone), antiepileptics
(carbamazepine, lamotrigine, phenytoin, topiramate and valproic
acid) and others (clobazam, diazepam, donepezil and galantamine).
The findings concerning the total 24 SNPs of the panel across the
aforementioned 13 genes have been previously published (12).
Following genotyping analysis, patients were
categorized into a series of different predicted metabolizer status
phenotypes, including normal, intermediate, and poor metabolizers,
based on their CYPD2D6 and CYP2C19 genotypes, as well as rapid and
ultra-rapid metabolizers for CYP2C19. Patients were also
categorized based on their predicted phenotype with regards to
response to antidepressant medications, specifically into three
categories, comprising good, reduced, and increased response,
according to their FKBP5 SNP rs4713916 genotype.
A summary of the genetic analysis in steps is
presented as follows: i) DNA extraction and purification from
buccal swab samples were performed using the PureLink genomic DNA
mini kit; ii) genotyping with PCR was performed using custom made
iDNA PGx CNS OpenArrays on QuantStudio 12K Flex Real-Time PCR; iii)
bioinformatics analysis assigning genotypes to metabolizer and
response predicted phenotypes.
Finally, a sub-population of 132 patients diagnosed
with MDD was requested to answer a short questionnaire on the
following topics: i) Treatment response; ii) severe adverse events;
iii) treatment modifications; and iv) patient-physician
interactions.
Statistical analysis
Data are presented as number and percentage and
statistical analysis was performed using IBM SPSS (version 30; IBM
Corp.) and Minitab (version 22) (https://www.minitab.com/en-us/products/minitab/)
statistical software. Statistical significance was calculated using
the Chi-squared test. A value of P<0.05 was considered to
indicate a statistically significant difference.
Results
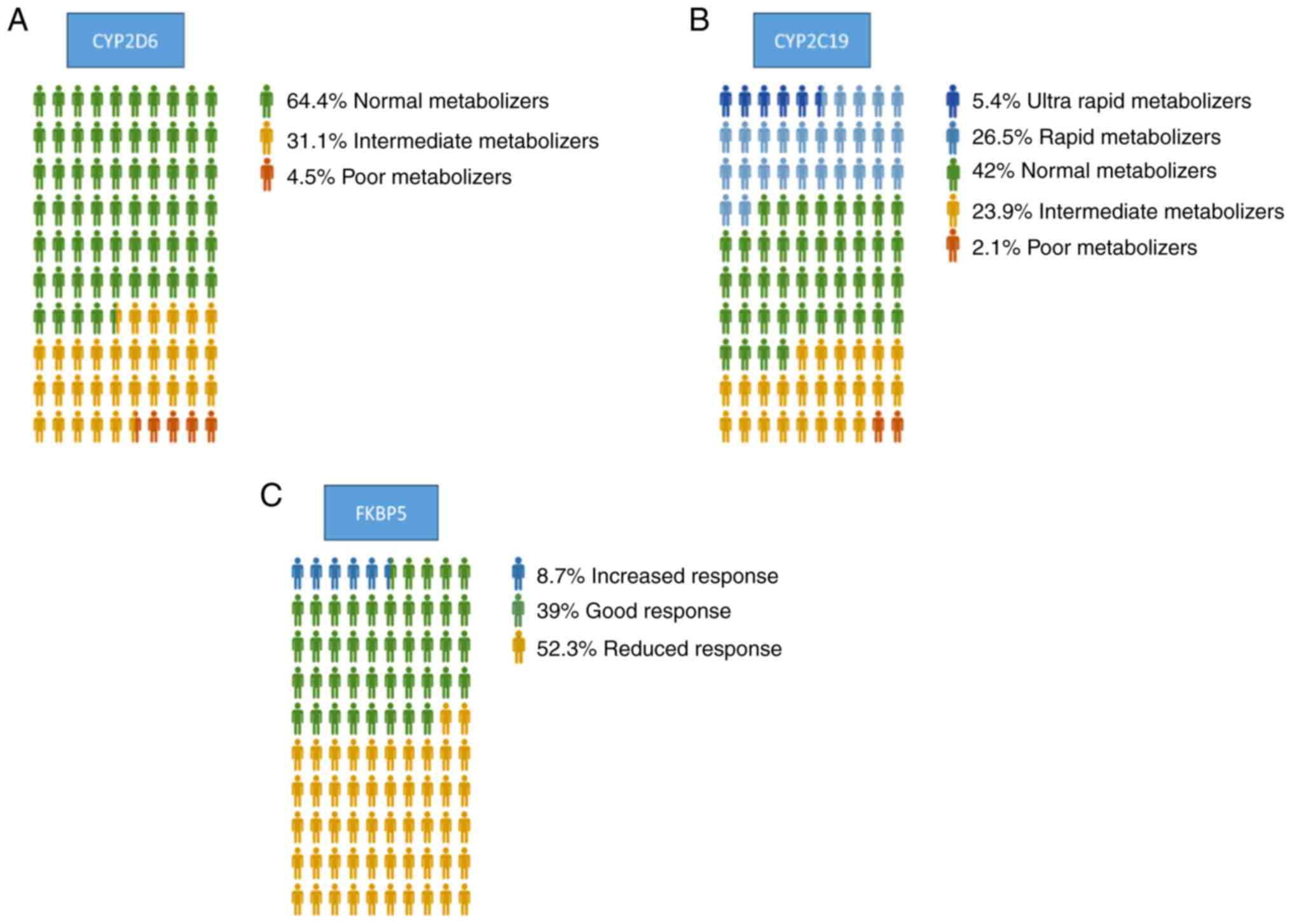
Regarding CYP2D6, 64.4% of the patients were
identified as normal metabolizers, 31.1% of the patients were
identified as intermediate metabolizers, and 4.5% of the patients
were identified as poor metabolizers (Fig. 2A). For CYP2C19, 42% of the
population exhibited normal metabolizer status, while 5.4% were
ultra-rapid metabolizers, 26.5% were rapid metabolizers, 23.9% were
intermediate metabolizers and 2.1% were poor metabolizers (Fig. 2B). The analysis of the genetic
distribution in SNP rs4713916, in FKBP5, revealed that 39% of the
subjects were expected to exhibit a good response, 8.7% an
increased response, while 52.3% were identified to carry a reduced
response genotype (Fig. 2C).
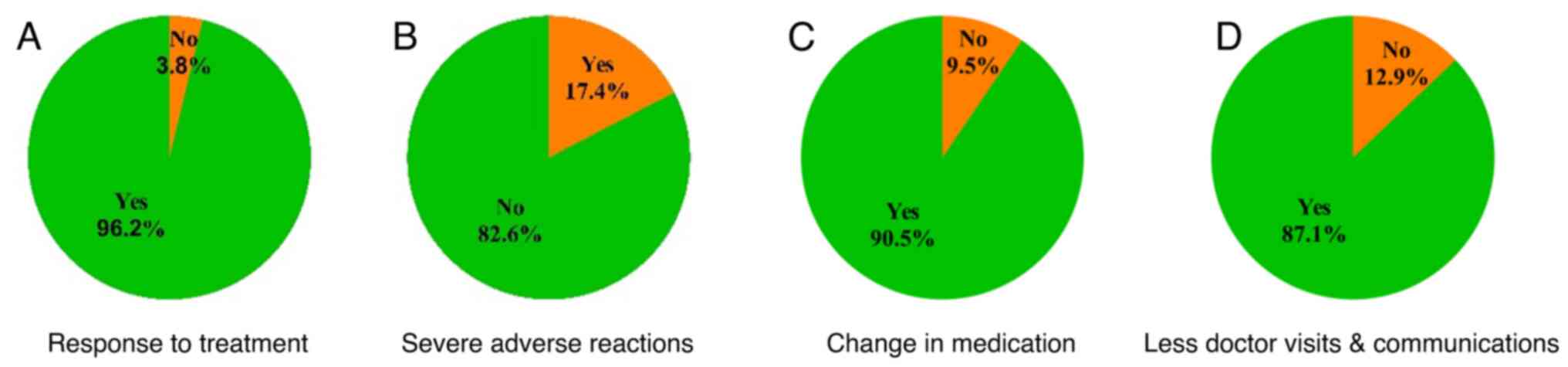
In patients diagnosed with MDD that followed
personalized drug and dosage selection by their doctor through PGx
guidance, significant improvements in symptoms were noted by 96.2%
(P<0.01) of individuals (Fig.
3A and Table I). Furthermore,
82.6% (P<0.01) did not report severe adverse events (Fig. 3B and Table I). In 90.5% (P<0.001) of
patients, treatment modifications were made, including dosage
adjustments or selection of an alternative medication (Fig. 3C and Table I). Finally, 87.1% (P<0.01) of
patients reported a reduction in visits and communications with
their doctors following pharmacogenetically guided treatment
(Fig. 3D and Table I).
 | Table IOutcomes in patients suffering from
major depressive disorder (n=132) following pharmacogenetic-guided
treatment. |
Table I
Outcomes in patients suffering from
major depressive disorder (n=132) following pharmacogenetic-guided
treatment.
| | Response to
treatment | Severe adverse
reactions | Change in
medication | Fewer doctor visits
and communications |
|---|
| Yes, n (%) | 127 (96.2) | 23 (17.4) | 119 (90.5) | 115 (87.1) |
| No, n (%) | 5 (3.8) | 109 (82.6) | 13 (9.5) | 17 (12.9) |
| P-value | <0.01 | <0.01 | <0.01 | <0.01 |
Discussion
Depression, a prevalent and debilitating mental
health disorder, affects millions of individuals worldwide,
presenting a significant challenge for clinicians striving to find
effective treatments for their patients (2). To date, therapy is based on a
trial-and-error approach, leading to delayed relief, increased
healthcare costs, and patient hardship (6). However, recent advancements in
PGx-guided therapy offer a promising avenue for personalized
treatment, leveraging genetics to improve medication selection and
dosage adjustments.
The efficacy and tolerability of antidepressants
vary widely among individuals, partly due to genetic differences
influencing drug metabolism and other molecular mechanisms. PGx
studies have identified several key genes implicated in
antidepressant response, including those encoding cytochrome P450
enzymes (1,14). By analyzing genetic variants
relevant to drug metabolism and pharmacodynamics, clinicians can
tailor treatment strategies to match genetic profiles of patients,
potentially improving outcomes and minimizing adverse effects
(5).
A growing body of evidence supports the utility of
PGx testing in guiding antidepressant therapy (15-19).
Studies have demonstrated that individuals receiving PGx-guided
treatment experience higher response rates, a more rapid
improvement of symptoms and fewer adverse events compared to those
managed conventionally (8,9). A recent umbrella review and updated
meta-analysis aimed to evaluate the effectiveness and safety of PGx
in guiding antidepressant prescribing for patients with depression
(20). Patients receiving
PGx-guided therapy had a significantly higher likelihood of
achieving remission, with rates ranging from 41 to 78%. The
response rate for these patients was also improved, ranging from 20
to 49%, compared to those receiving TAU (20). Although the present study lacks a
control group, the findings with regards to symptom improvement,
reported by 96.2% of patients, are in the same direction. Moreover,
while there is limited available evidence on the safety of
PGx-guided treatment (20), the
present study demonstrawted that 82.6% of patients did not report
any side-effects following PGx-guided medication, indicating that
PGx guidance may lead to better safety and tolerability.
Furthermore, integrating PGx testing into routine clinical practice
has shown feasibility and cost-effectiveness, with some healthcare
systems adopting it as standard care (21-23).
The real-world data presented herein suggest that PGx-guided
therapy enhances treatment adherence, reduces healthcare
utilization, and enhances patient satisfaction, highlighting its
potential to revolutionize depression management.
Despite its promise, several challenges remain in
the widespread implementation of PGx-guided therapy for depression.
These include issues related to test accessibility, interpretation
of results, clinician education and reimbursement. Additionally,
further research is required to elucidate optimal testing
protocols, refine bioinformatic algorithms and assess long-term
health outcomes. Collaborative efforts among researchers,
clinicians, policymakers and industry stakeholders are crucial to
address these challenges and maximize the clinical utility of
pharmacogenetics.
PGx-guided therapy represents a groundbreaking
approach to individualizing depression treatment, offering the
potential to enhance efficacy, safety, and patient satisfaction.
While challenges persist, accumulating evidence supports its
integration into routine clinical practice, heralding a new era of
precision medicine in mental health care. By leveraging genetic
insights, clinicians can empower patients with personalized
treatment plans, ultimately improving outcomes and quality of life
for those living with depression.
Acknowledgements
Not applicable.
Funding
Funding: The present study was privately funded by iDNA
Laboratories.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
NP conceptualized the study. NP, AS and TF analyzed
the data. EN and ES performed laboratory analysis, DNA isolation
and genotyping. NP and AS wrote the manuscript. All authors confirm
the authenticity of all the raw data. All authors reviewed and
edited the manuscript, and all authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study used pre-existing anonymized data
that does not allow for the identification of individuals. The
present study did not involve any invasive procedures or direct
interaction with patients. The present study only utilized
pre-existing anonymized genetic data. These factors place the
research outside the scope of requiring formal ethical review, as
it poses no foreseeable risks to individuals. Nevertheless, it was
ensured that the study adhered to the highest ethical standards
available and was carried out in accordance with the Declaration of
Helsinki. All participants were able to provided informed consent,
allowing for the use of their anonymized genetic data for research
and statistical purposes. This also applies to the questionnaire
survey for which all participants were capable of and provided
informed consent, allowing for the use of their answers for
research and statistical purposes. Moreover, all participants had
the right to withdraw their data at any time, ensuring their
autonomy and control over their information.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Use of artificial intelligence tools
During the preparation of this work, AI tools were
used to improve the readability and language of the manuscript or
to generate images, and subsequently, the authors revised and
edited the content produced by the AI tools as necessary, taking
full responsibility for the ultimate content of the present
manuscript.
References
|
1
|
van Westrhenen R, van Schaik RHN, van
Gelder T, Birkenhager TK, Bakker PR, Houwink EJF, Bet PM,
Hoogendijk WJG and van Weelden-Hulshof MJM: Policy and practice
review: A first guideline on the use of pharmacogenetics in
clinical psychiatric practice. Front Pharmacol.
12(640032)2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Moncrieff J and Kirsch I: Efficacy of
antidepressants in adults. BMJ. 331:155–157. 2005.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Mitchell AJ: Depressed patients and
treatment adherence. Lancet. 367:2041–2043. 2006.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sawada N, Uchida H, Suzuki T, Watanabe K,
Kikuchi T, Handa T and Kashima H: Persistence and compliance to
antidepressant treatment in patients with depression: A chart
review. BMC Psychiatry. 9(38)2009.PubMed/NCBI View Article : Google Scholar
|
|
5
|
van Westrhenen R, Aitchison KJ,
Ingelman-Sundberg M and Jukić MM: Pharmacogenomics of
antidepressant and antipsychotic treatment: How far have we got and
where are we going? Front Psychiatry. 11(94)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Altar CA, Carhart J, Allen JD, Hall-Flavin
D, Winner J and Dechairo B: Clinical utility of combinatorial
Pharmacogenomics-guided antidepressant therapy: Evidence from three
clinical studies. Mol Neuropsychiatry. 1:145–155. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhou Y, Ingelman-Sundberg M and Lauschke
VM: Worldwide distribution of cytochrome P450 alleles: A
Meta-analysis of Population-scale sequencing projects. Clin
Pharmacol Ther. 102:688–700. 2017.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Bunka M, Wong G, Kim D, Edwards L, Austin
J, Doyle-Waters MM, Gaedigk A and Bryan S: Evaluating treatment
outcomes in pharmacogenomic-guided care for major depression: A
rapid review and meta-analysis. Psychiatry Res.
321(115102)2023.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Skryabin V, Rozochkin I, Zastrozhin M,
Lauschke V, Franck J, Bryun E and Sychev D: Meta-analysis of
pharmacogenetic clinical decision support systems for the treatment
of major depressive disorder. Pharmacogenomics J. 23:45–49.
2023.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Binder EB, Salyakina D, Lichtner P,
Wochnik GM, Ising M, Pütz B, Papiol S, Seaman S, Lucae S, Kohli MA,
et al: Polymorphisms in FKBP5 are associated with increased
recurrence of depressive episodes and rapid response to
antidepressant treatment. Nat Genet. 36:1319–1325. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Zou YF, Wang F, Feng XL, Li WF, Tao JH,
Pan FM, Huang F and Su H: Meta-analysis of FKBP5 gene polymorphisms
association with treatment response in patients with mood
disorders. Neurosci Lett. 484:56–61. 2010.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Bothos E, Ntoumou E, Kelaidoni K, Roukas
D, Drakoulis N, Papasavva M, Karakostis FA, Moulos P and Karakostis
K: Clinical pharmacogenomics in action: Design, assessment and
implementation of a novel pharmacogenetic panel supporting drug
selection for diseases of the central nervous system (CNS). J
Transl Med. 19(151)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Roukas D, Bothos E, Ntoumou E, Kelaidoni
K, Drakoulis N, Papasavva M, Karakostis FA, Moulos P and Karakostis
K: P.0221 A rapid, concise and robust companion diagnostic pgx test
supporting drug selection for diseases of the central nervous
system. Eur Neuropsychopharmacol. 53 (Suppl 1):S159–S160. 2021.
|
|
14
|
Beunk L, Nijenhuis M, Soree B, de
Boer-Veger NJ, Buunk AM, Guchelaar HJ, Houwink EJF, Risselada A,
Rongen GAPJM, van Schaik RHN, et al: Dutch pharmacogenetics Working
Group (DPWG) guideline for the gene-drug interaction between
CYP2D6, CYP3A4 and CYP1A2 and antipsychotics. Eur J Hum Genet.
32:278–285. 2024.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Hall-Flavin DK, Winner JG, Allen JD,
Carhart JM, Proctor B, Snyder KA, Drews MS, Eisterhold LL, Geske J
and Mrazek DA: Utility of integrated pharmacogenomic testing to
support the treatment of major depressive disorder in a psychiatric
outpatient setting. Pharmacogenet Genomics. 23:535–548.
2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Singh AB: Improved antidepressant
remission in major depression via a pharmacokinetic pathway
polygene pharmacogenetic report. Clin Psychopharmacol Neurosci.
13:150–156. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Pérez V, Salavert A, Espadaler J, Tuson M,
Saiz-Ruiz J, Sáez-Navarro C, Bobes J, Baca-García E, Vieta E,
Olivares JM, et al: Efficacy of prospective pharmacogenetic testing
in the treatment of major depressive disorder: Results of a
randomized, double-blind clinical trial. BMC Psychiatry.
17(250)2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Bradley P, Shiekh M, Mehra V, Vrbicky K,
Layle S, Olson MC, Maciel A, Cullors A, Garces JA and Lukowiak AA:
Improved efficacy with targeted pharmacogenetic-guided treatment of
patients with depression and anxiety: A randomized clinical trial
demonstrating clinical utility. J Psychiatr Res. 96:100–107.
2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Greden JF, Parikh SV, Rothschild AJ, Thase
ME, Dunlop BW, DeBattista C, Conway CR, Forester BP, Mondimore FM,
Shelton RC, et al: Impact of pharmacogenomics on clinical outcomes
in major depressive disorder in the GUIDED trial: A large, patient-
and rater-blinded, randomized, controlled study. J Psychiatr Res.
111:59–67. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Tesfamicael KG, Zhao L,
Fernández-Rodríguez R, Adelson DL, Musker M, Polasek TM and Lewis
MD: Efficacy and safety of pharmacogenomic-guided antidepressant
prescribing in patients with depression: An umbrella review and
updated meta-analysis. Front Psychiatry. 15(1276410)2024.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chatziandreou E and Panagiotou N: EE77
Cost-effectiveness analysis of Pharmacogenetic-guided treatment in
drug resistant depression. Value in Health. 25 (Suppl
1)(S68)2022.
|
|
22
|
Morris SA, Alsaidi AT, Verbyla A, Cruz A,
Macfarlane C, Bauer J and Patel JN: Cost effectiveness of
pharmacogenetic testing for drugs with clinical pharmacogenetics
implementation consortium (CPIC) guidelines: A systematic review.
Clin Pharmacol Ther. 112:1318–1328. 2022.PubMed/NCBI View
Article : Google Scholar
|
|
23
|
Carrascal-Laso L, Franco-Martín M,
Marcos-Vadillo E, Ramos-Gallego I, García-Berrocal B, Mayor-Toranzo
E, Sánchez-Iglesias S, Lorenzo C, Sevillano-Jiménez A,
Sánchez-Martín A, et al: Economic impact of the application of a
precision medicine model (5SPM) on psychotic patients.
Pharmgenomics Pers Med. 14:1015–1025. 2021.PubMed/NCBI View Article : Google Scholar
|