Introduction
MicroRNAs (miRNAs) are an abundant class of short
regulatory non-coding RNAs (ncRNAs; length, 20–24 nucleotides) that
act as post-transcriptional repressors by binding the
3′-untraslated regions (UTRs) of target messenger RNAs (mRNAs). The
regulatory activity of miRNAs is exerted at the
post-transcriptional level and it is estimated that they act on up
to one-third of the protein coding genes. miRNAs have been
implicated in various biological and pathological processes, and
have emerged as important contributors to cell homeostasis
(1).
Cells actively secrete miRNAs, and it is possible to
detect those small ncRNAs in all biological fluids. As a
consequence, circulating miRNAs have great potential as biomarkers,
which has already been reported for various types of human disease,
such as cancer (2), metabolic
disorders (3) and cardiac conditions
(4). In systemic infections, the
presence of a pathogenic agent often induces a significant change
in the profile of circulating miRNAs that facilitates their use as
biomarkers of disease establishment and progression. In certain
cases, such as viral infections, these changes in circulating
miRNAs are associated with the targeted cell, as demonstrated in
the acute and chronic hepatitis C virus infection (5,6). Parasitic
flatworms, such as Schistosoma japonicum induce a
differential expression of circulating miRNAs (7). Furthermore, previous studies have
suggested the involvement of circulating miRNAs in response to
bacterial infections, including active pulmonary tuberculosis
(8). However, the potential use of
circulating miRNAs as biomarkers in fungal infections has been
investigated comparatively less. In this context, in response to
infection with Candida albicans, miR-455, miR-125a, miR-146
and miR-155 were upregulated in mouse macrophages and miR-204 and
miR-211 were downregulated in renal mice tissues (9,10). In
monocytes and dendritic cells infected with Aspergillus
fumigatus, miR-132 and miR-155 were identified to be
differentially expressed (11).
The incidence of fungal infections has increased in
recent years and is associated with significant morbidity and
mortality (12).
Paracoccidioidomycosis (PCM) is a human systemic mycosis that is
considered to be clinically important, due to the increase of
frequency, the severity of their clinical manifestations and the
associated mortality rates. PCM is caused by dimorphic fungus of
the Paracoccidioides species (Paracoccidioides
brasiliensis and Paracoccidioides lutzii) and is endemic
in Central and South America, with a predominance of clinical
disease in adult men and labourers from rural areas (13). In patients with PCM, the fungus is
initially observed in the lungs; however, the yeast spreads to
other organs (14).
Considering the central role of miRNAs in infectious
diseases and that little is known about the molecular mechanisms
underlying PCM, the aim of the present study was to verify the
presence of differentially expressed circulating miRNAs in patients
with PCM, which could serve as novel biomarkers in the diagnosis of
this fungal disease.
Materials and methods
Study subjects
Four male patients diagnosed with chronic PCM, and
four male healthy control subjects free of the PCM infection and
free of any clinical symptoms of any infectious disease were
recruited from the School of Pharmaceutical Sciences, São Paulo
State University (UNESP; Araraquara, Brazil) and included in the
current study. Venous blood (1 ml) was collected from each
participant and samples were centrifuged at 1,800 × g for 10 min at
4°C. The supernatant serum was quickly removed, aliquoted, and
frozen at −80°C until the experiments were performed. The present
study was approved by the local ethics committee of the School of
Pharmaceutical Sciences, São Paulo State University (UNESP)
(approval no. 33397114.5.0000.5426) and was performed in accordance
with the Declaration of Helsinki. Written informed consent was
obtained prior to blood sample collection.
Extraction of RNA
The extraction of RNA, including miRNAs, from 200 µl
serum was performed using miRCURY™ RNA Isolation kit - Biofluids
(Exiqon A/S, Vedbaek, Denmark), according to the manufacturer's
instructions.
Synthesis of cDNA and analysis by
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
cDNA was synthesized from RNA using a Universal cDNA
synthesis kit (Exiqon A/S) according to the manufacturer's
instructions. For miRNA screening, a global RT-qPCR low-density
panel (microRNA Ready-to-Use PCR, Human panel I+II, V.2 M/R; Exiqon
A/S) was used, which contains locked nucleic acid specific primers
for 752 unique human miRNAs. The reaction mixture included 40 µl
cDNA, 40 ml SYBR-Green PCR Master Mix (Exiqon A/S) and 40 ml
RNase-free water. Then, 10 µl of the reaction mixture was added to
each well of the panel. Amplification was performed by RT-qPCR
(ViiA™ 7 Real-Time PCR System; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) followed by the determination of the melting
curve according the following conditions: Denaturation at 95°C for
10 min, followed by 45 cycles of 95°C for 10 sec and 60°C for 60
sec. The experiment was performed in triplicate. The raw Cq values
were normalized using the global normalization method in each
sample, and the expression levels of all miRNAs were calculated
according to the ΔΔCq method (15), as
implemented in DataAssist v2 software (Applied Biosystems; Thermo
Fisher Scientific, Inc.).
Statistical analysis
The statistics toolbox of the DataAssist v2 software
(Applied Biosystems; Thermo Fisher Scientific, Inc.) was used and
P<0.05 was considered to indicate a statistically significant
difference. A volcano plot was constructed for the analyzed miRNAs
and miRBase was used to identify them (16). For bioinformatics analysis,
differentially expressed miRNAs were selected and submitted to
in silico pathways analysis for Gene Ontology (GO)
annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG),
using the bioinformatics tool, Database for Annotation,
Visualization and Integrated Discovery (DAVID).
Results
Circulating miRNA levels in the
PCM
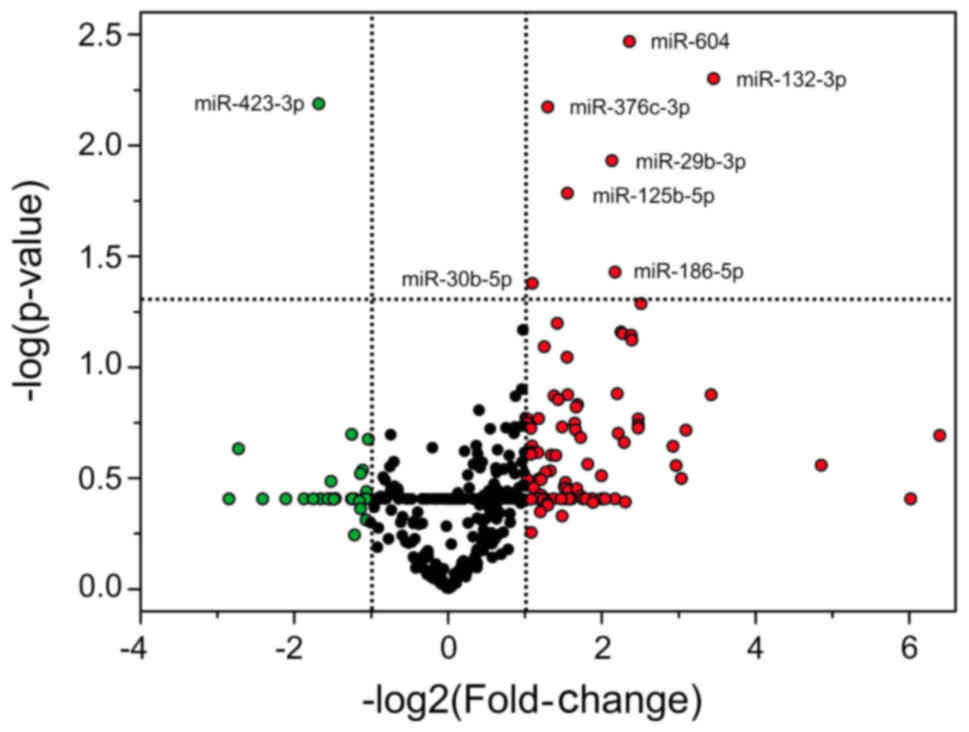
The differentially expressed miRNAs in PCM patients
are demonstrated by a non-symmetrical distribution in a volcano
plot (Fig. 1). The x-axis represents
the log2 ratio of the expression difference of miRNA
between the groups and the y-axis demonstrates the log of the
P-values. From the 752 miRNAs analyzed, seven were overexpressed
(miR-132-3p, miR-604, miR-186-5p, miR-29b-3p, miR-125b-5p,
miR-376c-3p and miR-30b-5p) and one was underexpressed (miR-423-3p)
in the serum of patients with PCM when compared with the healthy
control subjects (Table I). The data
were regarded as significantly different at P<0.05 and with a
fold-change ≥2. Furthermore, the expression profile of the
investigated microRNAs was similar among the subjects of each
group.
 | Table I.Differentially expressed miRNAs in
patients with paracoccidioidomycosis relative to healthy control
subjects. |
Table I.
Differentially expressed miRNAs in
patients with paracoccidioidomycosis relative to healthy control
subjects.
| miRNA | Fold-change | P-value |
|---|
| Upregulated |
|
|
|
hsa-miR-132-3p | 10.97 | 0.0050 |
|
hsa-miR-604 | 5.13 | 0.0034 |
|
hsa-miR-186-5p | 4.51 | 0.0372 |
|
hsa-miR-29b-3p | 4.38 | 0.0117 |
|
hsa-miR-125b-5p | 2.93 | 0.0164 |
|
hsa-miR-376c-3p | 2.45 | 0.0067 |
|
hsa-miR-30b-5p | 2.14 | 0.0418 |
| Downregulated |
|
|
|
hsa-miR-423-3p | 0.31 | 0.0065 |
Pathway analysis of miRNA-controlled
pathways
Based on the prediction of pathways, using the gene
annotation tool DAVID (Table II), the
differentially expressed miRNAs in PCM participate in the processes
of apoptosis, immune response, adhesion and the infection processes
of fungus to host cells. The biological processes identified may
regulate genes that are included in the mitogen-activated protein
kinases cascade, inflammatory response, activation of leukocytes, T
and B cells, regulation of cytokine production, regulation of
adhesion to the extracellular matrix (ECM) and endocytosis.
 | Table II.Signaling pathways regulated by eight
microRNAs identified in patients with paracoccidioidomycosis. |
Table II.
Signaling pathways regulated by eight
microRNAs identified in patients with paracoccidioidomycosis.
| Signaling
pathway | P-value |
|---|
| Regulation of
apoptosis | 1.8E-28 |
| Negative regulation
of transcription | 3.2E-16 |
| Metabolism
regulation of cellular proteins | 2.3E-18 |
| Regulation of DNA
binding | 1.7E-18 |
| Regulation of the
cascade of protein kinases | 3.8E-12 |
| Regulation of cell
migration | 8.9E-15 |
| Inflammatory
response | 2.3E-8 |
| Mitogen activated
protein kinases cascade | 2.4E-5 |
| Activation of
leukocytes | 1.1E-9 |
| Activation of T
cells | 4.5E-6 |
| Activation of B
cells | 2.7E-3 |
| Regulation of
cytokine production | 8.5E-10 |
| Regulation of
adhesion to the extracellular matrix | 8.2E-7 |
| Endocytosis | 2.7E-5 |
| Binding to the
tumor necrosis factor receptor superfamily | 3.6E-6 |
Discussion
Previous studies have shown the involvement of
circulating miRNAs in certain infectious and non-infectious
diseases (2,8,17,18). miRNAs may be used for diagnosis, as
well as for monitoring disease progression and treatment efficacy
(19). This is a preliminary study
that compared the miRNA profile in the serum of patients diagnosed
with chronic PCM with healthy control subjects. Eight miRNAs
(miR-132-3p, miR-604, miR-186-5p, miR-29b-3p, miR-125b-5p,
miR-376c-3p, miR-30b-5p and miR-423-3p) were identified to be
differentially expressed in serum samples obtained from patients
with PCM.
The identification of miRNAs differentially
expressed in PCM in the present study is considered to be
important. It facilitates the understanding of the regulatory
mechanisms involved in host-parasite interactions and the
infectious process. The in silico analysis demonstrated that
apoptosis, immune response and adhesion of fungus to host cells are
important signaling pathways that are regulated by eight
differentially expressed miRNAs in PCM. Following inhalation by the
host, Paracoccidioides spp. adhere to ECM components, such
as collagen, fibronectin and laminin. This process avoids
elimination of the fungi by lung ciliary cells and contributes to
establishment of the infection (20).
Apoptosis is an important mechanism, as described in certain
infections, including PCM, and has been implicated in
immunosuppressive events, which works in favor of the parasite to
limit the exaggerated inflammatory response. The process of
apoptosis has been described in PCM patients and in a PCM murine
model (21,22). P. brasiliensis induces apoptosis
in macrophages via caspases (23,24). In
addition, the participation of macrophages, cytokines, interleukins
(ILs) and tumor necrosis factor (TNF) is observed in PCM (20,25).
In addition, the majority of miRNAs identified in
the present study have been previously described in the processes
of apoptosis and immune response to other infections. miR-132 and
miR-186, for example, have been described to be associated with the
promotion of apoptosis, which activates the signaling pathway via
caspases (26). Furthermore, the
expression of miR-132 was induced in cells of the immune system by
the A. fumigatus and Mycobacterium tuberculosis
infection (11,27). Members of the miR-29 family may act in
the Wnt signaling pathway, which regulates the activation of
macrophages in the inflammatory process (28,29).
Furthermore, miR-29 was identified to be overexpressed in the serum
and sputum of patients with active pulmonary tuberculosis (8). Comparable with miR-29, miR-376c was
recently reported to be differentially expressed in the serum of
patients with active tuberculosis (30). In addition, miR-125 may be involved in
the immune response to infections. It has been shown that M.
tuberculosis induces the expression of miR-125b, which binds to
the 3′-UTR of the mRNA of TNF in human macrophages and destabilizes
the transcript. Blocking biosynthesis of the TNF is an important
process in response to bacterial infection (31). miR-30 may also be associated with the
immune response process, as it suppresses the polypeptide
N-acetylgalactosaminyltransferase 7 gene, which leads to an
increase of IL-10 (32). By contrast,
two miRNAs (has-miR-604 and miR-423-3p) were described, but were
not associated with infectious disease (33,34).
miRNAs detected in the serum of patients with PCM
may serve as biomarkers. The diagnosis of PCM is based on clinical
and laboratory findings. Typically, the direct microscopic
examination of lesions or tissue samples that are collected is made
with 10% potassium hydroxide, 4% sodium hydroxide or calcofluor
white to observe the fungi. The culture to observe the thermal
dimorphism of Paracoccidioides spp. is necessary for a
definitive diagnosis; however, the fungus grows slowly. Serologic
tests facilitate with diagnosis, however, certain tests for PCM are
not well standardized, which may result in cross-reactions with
other infections (35). Thus, the
production of antigenic preparations from whole yeast cells and
culture filtrate is important to ensure the sensitivity and
specificity of the tests. Gp43 has served as the main antigen in
the diagnosis of PCM; however, it was demonstrated that high
percentages of false negative results for PCM were caused by P.
lutzii (36). Thus, novel
biomarkers along with existing techniques may contribute to
improved diagnosis of PCM. In this context, miRNAs have been
presented as ‘fingerprints’ of various pathological conditions
(37). These molecules are
advantageous due to their stability in body fluids, as they are
normally packaged into exosome-like microparticles that protect
them against endogenous RNase activity. Circulating miRNAs have
been shown to be stable during repeated freezing and thawing, as
well as in acidic and alkaline environments (38). Furthermore, detection techniques,
including PCR and microarray, are sensitive and rapid (1,39).
In conclusion, eight differentially expressed miRNAs
were identified in serum samples from patients with PCM. These
miRNAs are associated with apoptosis, immune responses and adhesion
processes. Although the small sample size is a limitation, the
present study provided, to the best of our knowledge, the first
description of circulating miRNAs as potential biomarkers for the
diagnosis of PCM. However, further studies are required to extend
the identification of miRNAs for a larger number of patients with
PCM and to clarify the role of theses miRNAs in this mycosis.
Acknowledgements
The present study was supported by grants from the
Coordenação de Aperfeiçoamento de Pessoal de Nível
Superior/Fundação para a Ciência e a Tecnologia (grant no. 345/13),
the Conselho Nacional de Pesquisa e Desenvolvimento the Fundação de
Amparo à Pesquisa do Estado de São Paulo (grant no. 2014/10446-9)
and Programa de Apoio ao Desenvolvimento Científico da Faculdade de
Ciências Farmacêuticas da UNESP.
References
|
1
|
Hu G, Drescher KM and Chen XM: Exosomal
miRNAs: Biological properties and therapeutic potential. Front
Genet. 3:562012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu X, Liu L, Xu Q, Wu P, Zuo X and Ji A:
MicroRNA as a novel drug target for cancer therapy. Expert Opin
Biol Ther. 12:573–580. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Peng Y, Yu S, Li H, Xiang H, Peng J and
Jiang S: MicroRNAs: Emerging roles in adipogenesis and obesity.
Cell Signal. 26:1888–1896. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Goretti E, Zangrando J, Wagner DR and
Devaux Y: Unity is strength - a panel of 4 microRNAs decreases
cardiomyocyte hypertrophy. Int J Cardiol. 182:62–64. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
El-Diwany R, Wasilewski LN, Witwer KW,
Bailey JR, Page K, Ray SC, Cox AL, Thomas DL and Balagopal A: Acute
Hepatitis C virus infection induces consistent changes in
circulating microRNAs that are associated with nonlytic hepatocyte
release. J Virol. 89:9454–9464. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shrivastava S, Petrone J, Steele R, Lauer
GM, Di Bisceglie AM and Ray RB: Up-regulation of circulating
miR-20a is correlated with hepatitis C virus-mediated liver disease
progression. Hepatology. 58:863–871. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cai P, Gobert GN, You H, Duke M and
McManus DP: Circulating miRNAs: Potential novel biomarkers for
hepatopathology progression and diagnosis of schistosomiasis
japonica in two murine models. PLoS Negl Trop Dis. 9:e00039652015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fu Y, Yi Z, Wu X, Li J and Xu F:
Circulating microRNAs in patients with active pulmonary
tuberculosis. J Clin Microbiol. 49:4246–4251. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Monk CE, Hutvagner G and Arthur JS:
Regulation of miRNA transcription in macrophages in response to
Candida albicans. PLoS One. 5:e136692010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li XY, Zhang K, Jiang ZY and Cai LH:
miR-204/miR-211 downregulation contributes to candidemia-induced
kidney injuries via derepression of Hmx1 expression. Life Sci.
102:139–144. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Das Gupta M, Fliesser M, Springer J,
Breitschopf T, Schlossnagel H, Schmitt AL, Kurzai O, Hünniger K,
Einsele H and Löffler J: Aspergillus fumigatus induces microRNA-132
in human monocytes and dendritic cells. Int J Med Microbiol.
304:592–596. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pfaller MA and Diekema DJ: Epidemiology of
invasive mycoses in North America. Crit Rev Microbiol. 36:1–53.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shikanai-Yasuda MA, Filho FeQ Telles,
Mendes RP, Colombo AL and Moretti ML: Guidelines in
paracoccidioidomycosis. Rev Soc Bras Med Trop. 39:297–310. 2006.(In
Portuguese). View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Marques SA: Paracoccidioidomycosis. Clin
Dermatol. 30:610–615. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:D140–D144. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reid G, Kirschner MB and van Zandwijk N:
Circulating microRNAs: Association with disease and potential use
as biomarkers. Crit Rev Oncol Hematol. 80:193–208. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen Y, Li L, Zhou Z, Wang N, Zhang CY and
Zen K: A pilot study of serum microRNA signatures as a novel
biomarker for occult hepatitis B virus infection. Med Microbiol
Immunol (Berl). 201:389–395. 2012. View Article : Google Scholar
|
|
19
|
Weiland M, Gao XH, Zhou L and Mi QS: Small
RNAs have a large impact: Circulating microRNAs as biomarkers for
human diseases. RNA Biol. 9:850–859. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
de Oliveira HC, Assato PA, Marcos CM,
Scorzoni L, de Paula E, Silva AC, Da Silva JF, Singulani JL,
Alarcon KM, Fusco-Almeida AM and Mendes-Giannini MJ:
Paracoccidioides-host Interaction: An overview on recent advances
in the Paracoccidioidomycosis. Front Microbiol. 6:13192015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cacere CR, Romano CC, Giannini MJ Mendes,
Duarte AJ and Benard G: The role of apoptosis in the
antigen-specific T cell hyporesponsiveness of
paracoccidioidomycosis patients. Clin Immunol. 105:215–222. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Verícimo MA, França KM, Arnholdt AC and
Kipnis TL: Increased apoptosis during the early phase of
experimental paracoccidioidomycosis as a phenotypic marker of
resistance. Microbes Infect. 8:2811–2820. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Silva SS, Tavares AH, Passos-Silva DG,
Fachin AL, Teixeira SM, Soares CM, Carvalho MJ, Bocca AL,
Silva-Pereira I, Passos GA and Felipe MS: Transcriptional response
of murine macrophages upon infection with opsonized
Paracoccidioides brasiliensis yeast cells. Microbes Infect.
10:12–20. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Del Vecchio A, Silva JF, Silva JL,
Andreotti PF, Soares CP, Benard G and Giannini MJ: Induction of
apoptosis in A549 pulmonary cells by two Paracoccidioides
brasiliensis samples. Mem Inst Oswaldo Cruz. 104:749–754. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fortes MR, Miot HA, Kurokawa CS, Marques
ME and Marques SA: Immunology of paracoccidioidomycosis. An Bras
Dermatol. 86:516–524. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang B, Lu L and Zhang X, Ye W, Wu J, Xi
Q and Zhang X: hsa-miR-132 regulates apoptosis in non-small cell
lung cancer independent of acetylcholinesterase. J Mol Neurosci.
53:335–344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ni B, Rajaram MV, Lafuse WP, Landes MB and
Schlesinger LS: Mycobacterium tuberculosis decreases human
macrophage IFN-γ responsiveness through miR-132 and miR-26a. J
Immunol. 193:4537–4547. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kapinas K, Kessler C, Ricks T, Gronowicz G
and Delany AM: miR-29 modulates Wnt signaling in human osteoblasts
through a positive feedback loop. J Biol Chem. 285:25221–25231.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pereira CP, Bachli EB and Schoedon G: The
wnt pathway: A macrophage effector molecule that triggers
inflammation. Curr Atheroscler Rep. 11:236–242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang H, Sun Z, Wei W, Liu Z, Fleming J,
Zhang S, Lin N, Wang M, Chen M, Xu Y, et al: Identification of
serum microRNA biomarkers for tuberculosis using RNA-seq. PLoS One.
9:e889092014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rajaram MV, Ni B, Morris JD, Brooks MN,
Carlson TK, Bakthavachalu B, Schoenberg DR, Torrelles JB and
Schlesinger LS: Mycobacterium tuberculosis lipomannan blocks TNF
biosynthesis by regulating macrophage MAPK-activated protein kinase
2 (MK2) and microRNA miR-125b. Proc Natl Acad Sci USA.
108:17408–17413. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gaziel-Sovran A, Segura MF, Di Micco R,
Collins MK, Hanniford D, Vega-Saenz de Miera E, Rakus JF, Dankert
JF, Shang S, Kerbel RS, et al: miR-30b/30d regulation of GalNAc
transferases enhances invasion and immunosuppression during
metastasis. Cancer Cell. 20:104–118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sabina S, Pulignani S, Rizzo M, Cresci M,
Vecoli C, Foffa I, Ait-Ali L, Pitto L and Andreassi MG: Germline
hereditary, somatic mutations and microRNAs targeting-SNPs in
congenital heart defects. J Mol Cell Cardiol. 60:84–89. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Guan G, Zhang D, Zheng Y, Wen L, Yu D, Lu
Y and Zhao Y: microRNA-423-3p promotes tumor progression via
modulation of AdipoR2 in laryngeal carcinoma. Int J Clin Exp
Pathol. 7:5683–5691. 2014.PubMed/NCBI
|
|
35
|
Caldini CP, Xander P, Kioshima ÉS, Bachi
AL, de Camargo ZP, Mariano M and Lopes JD: Synthetic peptides mimic
gp75 from Paracoccidioides brasiliensis in the diagnosis of
paracoccidioidomycosis. Mycopathologia. 174:1–10. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Batista J Jr, de Camargo ZP, Fernandes GF,
Vicentini AP, Fontes CJ and Hahn RC: Is the geographical origin of
a Paracoccidioides brasiliensis isolate important for antigen
production for regional diagnosis of paracoccidioidomycosis?
Mycoses. 53:176–180. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Polakovičová M, Musil P, Laczo E, Hamar D
and Kyselovič J: Circulating microRNAs as potential biomarkers of
exercise response. Int J Mol Sci. 17:E15532016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim T and Reitmair A: Non-Coding RNAs:
Functional aspects and diagnostic utility in oncology. Int J Mol
Sci. 14:4934–4968. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Madhavan D, Cuk K, Burwinkel B and Yang R:
Cancer diagnosis and prognosis decoded by blood-based circulating
microRNA signatures. Front Genet. 4:1162013. View Article : Google Scholar : PubMed/NCBI
|