Introduction
Streptococcus suis serotype 2 (S. suis
2) is an important zoonotic pathogen that plays a vital role in
the transmission of Streptococcus disease in swine; thus,
the bacterium poses a potential threat to susceptible swine
populations and humans involved in the raising and processing these
animals (1). S. suis 2 is
transmitted via open wounds and may be isolated from animal waste
and decaying corpses. However, not all types of S. suis 2
induce disease, and virulence varies among different strains. S.
suis 2 strains may be divided into three types, namely
virulent, attenuated and avirulent (2,3).
Virulent strains result in evident clinical symptoms, such as acute
septicemia, meningitis, arthritis and endocarditis, and may lead to
mortality. Virulent S. suis 2 infections in humans are
associated with a very high mortality rate (1,4).
Carbon catabolite repression (CCR) is involved in
the microbial metabolic process. CCR is a physiological phenomenon
in which microorganisms use quick-impact carbon sources, such as
glucose among mixed carbon sources, in the process of fermentation.
Catabolite control protein A (CcpA) is a key regulator of
pleiotropic functions in the process of CCR (5). Furthermore, CcpA is involved in
numerous physiological processes, including the regulation of
central carbon and nitrogen metabolism, biofilm formation and the
expression of virulent genes. CcpA-mediated CCR exists in a number
of low-GC Gram-positive bacteria, where it is used to regulate
crucial genes in several metabolic pathways through specific
functional domains. The CcpA-mediated CCR process exerts a
regulatory function by altering CcpA expression. The specific
regulation of CCR may emerge as a notable area of study in the near
future (6).
In addition, CcpA is a key regulatory protein of
carbon metabolism. Low-GC Gram-positive bacteria adapt to
environmental changes by modulating CcpA activity (7). CcpA is a member of the LacI/GalR family
of transcriptional regulators. CcpA is not affected by CCR,
although the protein regulates multiple metabolic pathways in
numerous Gram-positive bacteria (8).
Previous studies (9) have identified
a number of important virulence-associated factors associated with
the growth phase in Streptococci, including arcABC, sly,
ofs, sao and cps2A. The arcABC operon is controlled by CCR, and
virulence-associated factors are indirectly regulated by factors in
carbon metabolism, such as sucrose and glucose metabolism. In
addition, CcpA has been observed to affect virulence through its
role as a key regulatory factor of carbon metabolism in a variety
of bacteria, including Staphylococcus aureus (10), Streptococcus pneumoniae
(11), Clostridium
perfringens (12) and S. suis
2 (9).
As a key transcription regulatory factor, CcpA is
able to effectively regulate the metabolism of products, such as
glucose, in various Gram-positive bacteria. However, the CcpA gene
is well-conserved and its CCR function is stable in numerous low-GC
Gram-positive bacteria (13). CcpA
exhibits similar regulatory functions in the metabolism of
Bacillus cereus (14),
Staphylococcus xylosus (15),
Lactococcus lactis (16),
S. pneumoniae (11),
Streptococcus mutans (17)
and Listeria monocytogenes (18). Therefore, further research into the
metabolic mechanisms of bacteria may aid understanding of metabolic
processes.
With the increasing research in this field,
including S. suis 2 DNA sequences obtained from outbreaks in
China (19), the present study
investigated the role of CcpA in functional regulation by analyzing
gene expression profiles. In addition, the effect of CcpA on carbon
metabolism was investigated using bioinformatics analysis. The
current study presents data at a gene expression level to
facilitate further study into the role of CcpA in the metabolic
regulation of S. suis 2 and the interaction among various
genes.
Materials and methods
Strains
S. suis 2 and CcpA mutant strains (20) were cultivated to a logarithmic phase
in Todd-Hewitt broth medium (Oxoid Ltd., Basingstoke, UK) at 37°C
in an atmosphere of 5% CO2. The strains were stored in
our laboratory at the Institute of Military Veterinary (Changchun,
China) until required for further use.
Extraction and pretreatment
Total S. suis 2 RNA was extracted using a
Bacteria total RNA Isolation Kit (Shanghai Sangon Biological
Engineering Technology & Services Co., Ltd., Shanghai, China).
Total RNA was examined and quantified using an Agilent 2100
Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). The
total RNA was used for library construction if the RNA integrity
number of 1 µg of total RNA was >7. Paired-end index libraries
were constructed using the NEBNext® Ultra™RNA Library Prep kit
(Illumina®; New England BioLabs, Inc., Ipswich, MA, USA). Large
ribosomal RNA among the total DNA was removed using a RiboMinus™
Transcriptome Isolation kit (Invitrogen Life Technologies,
Carlsbad, CA, USA).
cDNA library construction
First and second chains of cDNA were synthesized
using ProtoScript II Reverse Transcriptase and Second Strand
Synthesis Enzyme Mix (New England BioLabs, Inc.), respectively.
Double-chain cDNA was purified using the AxyPrep Mag Polymerase
Chain Reaction (PCR) Clean-up kit (Axygen; Corning, Inc., Corning,
NY, USA), and End Prep Enzyme Mix (New England BioLabs, Inc.) was
used to repair and join the DNA. DNA fragments (<400 bp) with
inserts of ~250 bp were selected using the AxyPrep Mag PCR Clean-up
kit, and the PCR products were purified using the AxyPrep Mag PCR
Clean-up kit (Axygen; Corning, Inc.). The quality of the library
preparations was assessed using the Bioanalyzer 2000 system
(Agilent Technologies, Inc.). In addition, the quantity and quality
of the PCR products were verified by quantum computing and
quantitative PCR (Applied Biosystems Life Technologies, Foster
City, CA, USA). Subsequent to confirmation that the library
preparations were qualified, sequencing was performed using a HiSeq
2500 Sequencing System (Illumina, Inc., San Diego, CA, USA). The
sequencing strategy was SE50 with a length of 1×50 bp (1×50
single-end). Every library consisted of 10 million reads (0.5 Gb)
sequencing products or so with a total length of >1.0 Gb. Data
analyses were performed using HiSeq Control software and
GAPipeline-1.6 with a HiSeq instrument (Illumina, Inc.). The
Student's t-test was adopted for statistical analyses, where
P<0.05 was considered to indicate a statistically significant
difference.
Data analyses
After sequencing, raw data of each sample were
processed by base calling and quality analysis. Certain reads were
filtered in order to clean the data. Data filtering was performed
using the Next Generation Sequencing Quality Control Toolkit
version 2.3 (http://59.163.192.90:8080/ngsqctoolkit/). Low-quality
and joint sequences were filtered with a QualScore cut-off value of
30. The S. suis 2 sequence was compared with that of S.
suis 05AYH33 (GenBank accession no. CP000407.1) using
Burrows-Wheeler Aligner version 0.7.5a-r405 (http://sourceforge.net/projects/bio-bwa/files/).
Gene quantification was performed using the reads per kilobase per
million mapped reads method. Differences in gene expression
profiles were analyzed using EdgeR software, version 2.13
(http://bioconductor.org/news/bioc_2_13_release/) with
the EdgeR algorithm. More than one difference in gene expression
profiles with a false discovery rate of ≤0.05 was considered
significant.
Enrichment of Gene Ontology (GO) terms was analyzed
using the GO::TermFinder package (http://search.cpan.org/dist/GO-TermFinder/), and a Web
Gene Ontology Annotation Plot (WEGO) image was produced at
http://wego.genomics.org.cn/cgi-bin/wego/index.pl.
Enrichment analysis was performed using the Kyoto Encyclopedia of
Genes and Genomes (KEGG) with the KEGG Orthology (KO) Based
Annotation System version 2.0 super geometry algorithm (www.genome.jp/kegg/ko.html). All analyses of gene
expression profiles and bioinformatics were outsourced to GENEWIZ,
Inc. (South Plainfield, NJ, USA).
Metabolic pathway analyses
In order to further elucidate the role of CcpA in
the metabolic regulation of S. suis 2, the effects of CcpA
on regulation mechanisms were investigated using gene expression
profiles based on previous metabonomic analyses (20). In addition, KEGG network data were
used to establish a network diagram of interactions among
metabolites, genes and proteins. Furthermore, the function of
mutants in the interaction of metabolites, genes and proteins in
the carbon metabolism pathway were analyzed.
Results
Analysis of gene expression
profiles
Analysis of gene expression profiles using sequence
analysis identification revealed 549 gene differences between S.
suis 2 and CcpA mutants (Fig.
1). Differential transcripts selected according to the standard
of a significant difference in expression levels identified 299
genes (Fig. 2).
GO information and the functional category of each
gene were obtained by GO function analyses of GO using the blast2GO
algorithm (http://www.blast2go.com), which
facilitated the elucidation of the distribution characteristics of
gene function of species or samples at the macro level. Significant
enrichment analysis of GO function enriched three GOs (P≤0.05). A
total of 1,782 reference genes were annotated in the GO database,
while 235 differential genes were annotated. Enrichment results of
the differential genes at three levels of GO were obtained using a
hypergeometric algorithm, while enrichment results of the genes
with different functions were obtained by WEGO analyses (Fig. 3).
Enrichment analysis of differentially
expressed gene pathways
Target protein sequences of S. suis 2 were
compared with Streptococcus sequences in KEGG GENES using
the KEGG Automatic Annotation Server (http://www.genome.jp/kegg/kaas/). Subsequently, the KO
number of an identical or similar protein was annotated to the KEGG
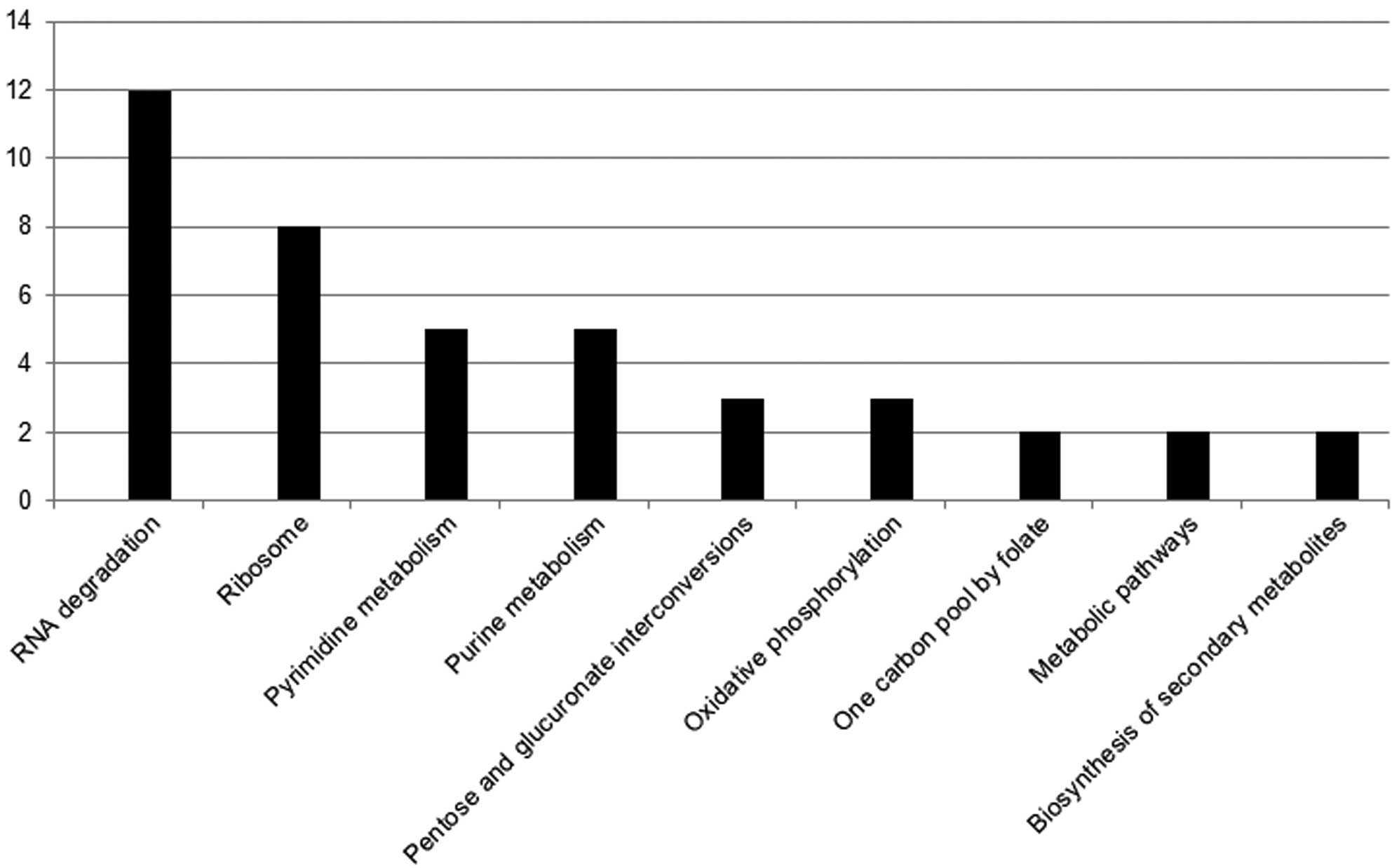
pathway. Pathway analysis of differential gene expression revealed
marked values among the pathways of oxidative phosphorylation,
metabolic pathways, one carbon pool by folate, pentose and
glucuronate interconversions, and the biosynthesis of secondary
metabolites.
Analysis of carbon metabolic
regulation
Based on previous studies of associated gene
metabonomics, the results demonstrated that CcpA affected carbon
metabolism to a certain extent by connecting differential
metabolites, genes and proteins (20). According to the network database and
the results of the present study, succinic, aspartic and citric
acid concentrations decreased and affected the Streptococcus
genome under the influence of the CcpA gene, which was verified by
previous research from metabolite experiments (20). Furthermore, downregulation of
ssu05_1508, ssu05_1065, ssu05_1164, ssu05_1640 and ssu05_2154, and
upregulation of ssu05_1885 and ssu05_1839 was observed (Fig. 4). These observations indicated that
the carbon metabolism of S. suis 2 was partially regulated
by CcpA. Metabolites may alter with modifications in the
corresponding genes. Network analysis may facilitate a more
comprehensive understanding of the metabolic mechanisms of CcpA in
S. suis 2 and provide a foundation for future studies.
Discussion
Streptococci infections of swine herds are
common worldwide and lead to significant financial losses for the
swine industry. S. suis 2 is a hazardous zoonotic pathogen
that may potentially result in fatal infections in humans.
Previously, a massive outbreak of S. suis 2 occurred in
Jiangsu and Sichuan provinces, China, resulting in severe swine
herd losses (21,22).
CcpA widely participates in the regulation of carbon
and nitrogen metabolism in bacteria (17). In addition, CcpA is involved in the
specific physiological processes of certain microorganisms, such as
the production of spores and solvents, and the expression of
virulence genes (23). In S.
aureus, CcpA directly regulates and activates the expression of
several virulence genes (10,24).
Furthermore, in a number of bacteria, deletion of CcpA leads to
reduced glycolytic enzyme activity, such as enolase, particularly
in infected hosts. Due to specific metabolic associations between
particular enzymes and pathogenic virulence, the toxicity of
strains without CcpA is notably reduced compared with wild-type
parental strains (25). CcpA in
Streptococcus pyogenes directly activates the expression of
virulence genes (24,26). As a result of the pleiotropic
regulatory function of CcpA, numerous metabolic processes are
interrupted by knockout of CcpA. Therefore, the association between
CcpA structure and function requires further study. Furthermore,
mutation analysis may be key to further clarifying the functional
domains and active sites of CcpA.
As CcpA is able to regulate genes involved in key
metabolic pathways through specific functional domains, the
clarification of its regulatory function by transformation of
functional domains and complicated pleiotropy into simple
specificity requires further investigation. Previous studies
investigating S. suis 2 metabonomics indicated that CcpA
serves a key function in the regulation of carbon metabolism in
amino and nucleic acids and lipids in S. suis 2 (6). Furthermore, pathway analysis indicated
that CcpA exerts an indirect influence on the regulation of
succinic, nucleic and aspartic acid. In this type of metabolic
regulation, the metabolic products are associated with glucose and
pyruvic acid, amongst other molecules. As glucose is a readily
available carbon source, glucose-induced CcpA-mediated CCR exerts a
wide influence on central metabolic pathways (11,17).
Thus, CcpA activation and the suppression of certain genes may
alter the concentrations of specific metabolites and intermediates.
In the present study, reductions in the concentrations of succinic,
aspartic and citric acid were also shown to indirectly alter the
availability of glucose and similar carbon metabolites, which
further affected metabolic regulation and functional changes in
S. suis 2. For example, studies on Bacillus subtilis
have suggested that CCR relies primarily on the mediation of CcpA
(27). In response to environmental
glucose, CcpA has been shown to affect catabolite responsive
element boxes of target genes via the combined activity of
trans-effect factors and cofactors, which subsequently inhibits or
activates the transcription of target genes (28). Glucose serves a crucial function in
the regulation of numerous central metabolic pathways. The pathway
in which CcpA activates or inhibits central metabolism may
influence expression levels of particular genes and result in
alterations in the secretion of metabolic products. As a signaling
molecule, metabolic products indirectly regulate the expression of
various genes. Thus, glucose-dependent modifications of central
metabolic pathways that alter the distribution of metabolites vary
with changing glucose content by altering the transcriptional level
of associated genes (29).
In the present study, the Illumina-based analysis of
the genome expression of CcpA in S. suis 2 has provided a
basis for the further study of the mechanism underlying CcpA
activity. In particular, large-scale gene expression analyses offer
a suitable analytical method for the study of metabolic and
pathogenic mechanisms in bacteria. Gene expression profile analyses
were employed to identify genes with varying expression levels
involved in metabolic and pathogenic processes of bacteria.
Therefore, study into the mechanisms underlying CcpA expression and
regulation of possible target genes in S. suis 2 by gene
expression profile analysis provides a theoretical basis on which
to investigate the role of similar genes in pathogenic mechanisms.
Compared with previous studies of CcpA gene transcriptomics in
B. subtilis, CcpA was demonstrated to directly or indirectly
regulate the expression of several genes and operons, at least to a
certain extent (30,31). Similarly, CcpA plays an important
role in the regulation of carbon metabolism and virulence
regulation in S. suis 2, which has been investigated
previously using other methods (6,32). In
particular, the study of bacteria in previous studies of functional
genomics have been crucial in describing the specific gene
expression alterations of a species and have generated large
quantities of information concerning the gene expression of cells
or tissues under specific conditions. This method is particularly
useful in being able to rapidly detect the specific gene
organization of a species in a particular state of the expression.
However, the results of the present study may facilitate future
studies, using the techniques described.
Acknowledgements
This study was supported by a grant from the
National Natural Science Foundation of China (no. 31101790/C1803).
Analyses of gene expression profiles and bioinformatics were
performed by GENEWIZ, Inc. (Beijing, China). Metabolomic pathway
analyses were performed by Shanghai Sensichip Infotech Co. Ltd.
(Shanghai, China).
References
|
1
|
Staats JJ, Feder I, Okwumabua O and
Chengappa MM: Streptococcus suis: Past and present. Vet Res Commun.
21:381–407. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Busque P, Higgins R, Caya F and Quessy S:
Immunization of pigs against Streptococcus suis serotype 2
infection using a live avirulent strain. Can J Vet Res. 61:275–279.
1997.PubMed/NCBI
|
|
3
|
Vecht U, Stockhofe-Zurwieden N, Tetenburg
BJ, Wisselink HJ and Smith HE: Virulence of Streptococcus suis type
2 for mice and pigs appeared host-specific. Vet Microbiol.
58:53–60. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gottschalk M, Xu J, Calzas C and Segura M:
Streptococcus suis: A new emerging or an old neglected zoonotic
pathogen? Future Microbiol. 5:371–391. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sanchez S and Demain AL: Metabolic
regulation and overproduction of primary metabolites. Microb
Biotechnol. 1:283–319. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Willenborg J, de Greeff A, Jarek M,
Valentin-Weigand P and Goethe R: The CcpA regulon of Streptococcus
suis reveals novel insights into the regulation of the
streptococcal central carbon metabolism by binding of CcpA to two
distinct binding motifs. Mol Microbiol. 92:61–83. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kaufman GE and Yother J: CcpA-dependent
and -independent control of beta-galactosidase expression in
Streptococcus pneumoniae occurs via regulation of an upstream
phosphotransferase system-encoding operon. J Bacteriol.
189:5183–5192. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sumby P, Barbian KD, Gardner DJ, et al:
Extracellular deoxyribonuclease made by group A Streptococcus
assists pathogenesis by enhancing evasion of the innate immune
response. Proc Natl Acad Sci USA. 102:1679–1684. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Willenborg J, Fulde M, de Greeff A, et al:
Role of glucose and CcpA in capsule expression and virulence of
Streptococcus suis. Microbiology. 157:1823–1833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Seidl K, Stucki M, Ruegg M, et al:
Staphylococcus aureus CcpA affects virulence determinant production
and antibiotic resistance. Antimicrob Agents Chemother.
50:1183–1194. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Iyer R, Baliga NS and Camilli A:
Catabolite control protein A (CcpA) contributes to virulence and
regulation of sugar metabolism in Streptococcus pneumoniae. J
Bacteriol. 187:8340–8349. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Varga J, Stirewalt VL and Melville SB: The
CcpA protein is necessary for efficient sporulation and enterotoxin
gene (cpe) regulation in Clostridium perfringens. J Bacteriol.
186:5221–5229. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Warner JB and Lolkema JS: CcpA-dependent
carbon catabolite repression in bacteria. Microbiol Mol Biol Rev.
67:475–490. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
van der Voort M, Kuipers OP, Buist G, de
Vos WM and Abee T: Assessment of CcpA-mediated catabolite control
of gene expression in Bacillus cereus ATCC 14579. BMC Microbiol.
8:622008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jankovic I, Egeter O and Brückner R:
Analysis of catabolite control protein A-dependent repression in
Staphylococcus xylosus by a genomic reporter gene system. J
Bacteriol. 183:580–586. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zomer AL, Buist G, Larsen R, Kok J and
Kuipers OP: Time-resolved determination of the CcpA regulon of
Lactococcus lactis subsp. cremoris MG1363. J Bacteriol.
189:1366–1381. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abranches J, Nascimento MM, Zeng L, et al:
CcpA regulates central metabolism and virulence gene expression in
Streptococcus mutans. J Bacteriol. 190:2340–2349. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Behari J and Youngman P: A homolog of CcpA
mediates catabolite control in Listeria monocytogenes but not
carbon source regulation of virulence genes. J Bacteriol.
180:6316–6324. 1998.PubMed/NCBI
|
|
19
|
Chen C, Tang J, Dong W, et al: A glimpse
of streptococcal toxic shock syndrome from comparative genomics of
S. suis 2 Chinese isolates. PLoS One. 2:e3152007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lang X, Wan Z, Bu Z and Wang X and Wang X,
Zhu L, Wan J, Sun Y and Wang X: Catabolite control protein A is an
important regulator of metabolism in Streptococcus suis type 2.
Biomed Rep. 2:709–712. 2014.PubMed/NCBI
|
|
21
|
Tang J, Wang C, Feng Y, et al:
Streptococcal toxic shock syndrome caused by Streptococcus suis
serotype 2. PLoS Med. 3:e1512006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sriskandan S and Slater JD: Invasive
disease and toxic shock due to zoonotic Streptococcus suis: An
emerging infection in the East? PLoS Med. 3:e1872006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ren C, Gu Y, Wu Y, Zhang W, Yang C, Yang S
and Jiang W: Pleiotropic functions of catabolite control protein
CcpA in Butanol-producing Clostridium acetobutylicum. BMC Genomics.
13:3492012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Poncet S, Milohanic E, Mazé A, et al:
Correlations between carbon metabolism and virulence in bacteria.
Contrib Microbiol. 16:88–102. 2009.PubMed/NCBI
|
|
25
|
Wang Y, Dang Y, Wang X, et al: Comparative
proteomic analyses of Streptococcus suis serotype 2 cell
wall-associated proteins. Curr Microbiol. 62:578–588. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Deutscher J, Herro R, Bourand A, Mijakovic
I and Poncet S: P-Ser-HPr - A link between carbon metabolism and
the virulence of some pathogenic bacteria. Biochim Biophys Acta.
1754:118–125. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Seidel G, Diel M, Fuchsbauer N and Hillen
W: Quantitative interdependence of coeffectors, CcpA and cre in
carbon catabolite regulation of Bacillus subtilis. FEBS J.
272:2566–2577. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Blencke HM, Homuth G, Ludwig H, et al:
Transcriptional profiling of gene expression in response to glucose
in Bacillus subtilis: Regulation of the central metabolic pathways.
Metab Eng. 5:133–149. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kraus A, Hueck C, Gärtner D and Hillen W:
Catabolite repression of the Bacillus subtilis xyl operon involves
a cis element functional in the context of an unrelated sequence
and glucose exerts additional xylR-dependent repression. J
Bacteriol. 176:1738–1745. 1994.PubMed/NCBI
|
|
30
|
Yoshida K, Kobayashi K, Miwa Y, et al:
Combined transcriptome and proteome analysis as a powerful approach
to study genes under glucose repression in Bacillus subtilis.
Nucleic Acids Res. 29:683–692. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lulko AT, Buist G, Kok J and Kuipers OP:
Transcriptome analysis of temporal regulation of carbon metabolism
by CcpA in Bacillus subtilis reveals additional target genes. J Mol
Microbiol Biotechnol. 12:82–95. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tang Y, Wu W, Zhang X, Lu Z, Chen J and
Fang W: Catabolite control protein A of Streptococcus suis type 2
contributes to sugar metabolism and virulence. J Microbiol.
50:994–1002. 2012. View Article : Google Scholar : PubMed/NCBI
|