Introduction
Nearly 1% of the world population is affected by
schizophrenia (SCZ), which is a severe neuropsychiatric disorder
that is characterized by cognitive dysfunction (1–3). The
etiology of SCZ pathogenesis is complex and lacks effective
therapeutic program. The role of genetics in SCZ has been widely
explored. Studies on twins and families have indicated that genetic
factors play a key role in the onset of SCZ. The concordance rate
of monozygotic twins is ~50%, which is higher than fraternal twins
and other siblings (4). In a family
without SCZ patients, the morbidity of first relatives is lower
than the lineal consanguinity of SCZ patients (5). Estimated heritability of SCZ ranges
from 65 to 80% (6,7). Recently, genome-wide association
studies (GWAS) has been used to identify molecular and genetic
mechanisms underlying SCZ (8) and
several hundred loci that are related to SCZ risk have been
identified (9–20). However, analyses of GWAS results are
not robust and account for only a small fraction of critical
genes.
Although many genes are associated with SCZ
susceptibility, the mechanistic details are yet to be elucidated.
The in silico methods are the best tools available to
functionally annotate genetic factors that contribute to SCZ. Among
these, the pathway-based method is the most reliable way to
functionally analyze GWAS data. This method assumes that genes
belonging to the same pathway are most likely to confer the risk
for a complex disease and is used to identify the enriched
functional categories (21,22). For example, Wang et al used
the pathway-based approach to identify 102 genes in sterol
transport and metabolism that were associated with high-density
lipoprotein cholesterol levels (23). The pathway-based analysis provides
biological insights that are not readily available from a single
marker analysis and is more effective in replicating different
independent studies. Therefore, in this study, we performed Gene
Set Enrichment Analysis (GSEA) of two independent SCZ-related GWAS
to identify the pathways and genetic factors that contribute to
SCZ. We further validated our findings in a two-step case-control
study involving Shandong migrants from northeastern China.
Materials and methods
Ethics statement
All procedures in this study were conducted as
approved by the Ethics Committee of Qiqihar Medical University
(Qiqihar, China) and in accordance with the Helsinki Declaration.
All participants or their guardians gave written informed consent
prior to inclusion.
Study population
The northeastern part of China is a border area with
frequent population migration, especially from the Shandong
province. In this study, we included two independent cohorts of
migrants that self-reported as originating from Shandong. The
initial discovery cohort consisted of 600 unrelated Han SCZ
patients (286 females and 314 males; mean age, 37.3 years) and 600
control subjects (278 females and 322 males; mean age, 38.2 years).
For validation, we included an independent sample set consisting of
1,731 SCZ patients (829 females and 902 males; mean age, 34.6
years) and 1,849 controls (936 females and 913 males; mean age,
35.0 years). The patients were recruited from the Second Affiliated
Hospital of Qiqihar Medical University, Harbin First Specific
Hospital, Mudanjiang Mental Hospital, Wealth Mental Hospital and
Beian Mental Hospital in Heilongjiang province. The patients were
clinically diagnosed according to the Chinese Classification of
Mental Disorders version 3 (CCMD-3) and the Diagnostic and
Statistical Manual of Mental Disorders-IV (DSM-IV). All control
individuals were enrolled from the Third Affiliated Hospital of
Qiqihar Medical University and matched for age and sex to patients.
Two experienced psychiatrists interviewed the healthy controls
individually using the Structured Clinical Interview for DSM-IV-TR
Axis I Disorders-Nonpatient Edition (SCID-NP), and controls were
clinically determined to be free of psychiatric disorders and
individual or family history of mental illness.
Patient sample preparation and
genotyping
Genomic DNA was isolated from peripheral white blood
cells using a commercial Axygen Blood Genomic DNA Miniprep kit
(Axygen Biosciences, Hangzhou, China). The DNA samples were
verified by 0.8% agarose gel electrophoresis and only those
preparations with no signs of degradation were used to construct
DNA pools. The DNA concentrations were quantified in a NanoDrop™
2000 spectrophotometer (Thermo Fisher Scientific, Inc., Waltham,
MA, USA) and each sample was diluted to 50 ng/µl. The DNA pooling
was performed thrice.
For allelotyping, six DNA pools from the discovery
sample set were individually genotyped on Affymetrix GeneChip Human
Mapping 6.0 chips (Affymetrix: Thermo Fisher Scientific, Inc.,
Santa Clara, CA, USA). The processing of the genomic DNA and the
array hybridization were performed according to the manufacturer's
instructions. The hybridization intensities of two probes for each
allele were derived from the raw scanning files.
For the validation of the candidate loci, DNA
samples from the validation panel consisting of 1,731 SCZ patients
and 1,849 controls were genotyped using ABI 7900HT Real-Time PCR
system (Applied Biosystems: Thermo Fisher Scientific, Inc., Foster
City, CA, USA).
Statistical analysis
The age and sex distributions between SCZ and
control subjects in the two stages were tested by Student's t-test
and Chi-square test, respectively. P<0.05 was considered
statistically significant. The relative allele signal (RAS) score
for each SNP was estimated as the ratio of signal intensity from
each allele as analyzed by the R package SNP MaP. The differences
in allele frequency between SCZ and controls were evaluated by the
Z combined test in the pooled dataset. Association analysis was
performed with PLINK v1.07 software (http://pngu.mgh.harvard.edu/~purcell/plink/) using
additive, recessive and dominant models in the validation stage. We
assumed that the prevalence of SCZ is 1% and used GAS Power
Calculator to perform the power calculation. The sample size used
in the validation stage could detect a risk allele with 73% power
at a false-positive rate of 5%.
Integrative analysis of published GWAS
data
The P-values for association between the two
SCZ-related GWAS were derived from dbGaP database (http://www.ncbi.nlm.nih.gov/gap/) and downloaded
for integrative analysis. The first study (dbGaP accession no.
pha002857) consisted of 1,351 SCZ patients and 1,378 control
subjects of European ancestry, whereas the second study (dbGaP
accession no. pha002859) included 1,195 SCZ patients and 954
control subjects of American ancestry. Raw data of both studies
were generated from Affymetrix GeneChip Human Mapping 6.0 chips.
The detailed information of the two studies is shown in Table I. Moreover, SCZ meta-analysis data
were obtained from the Psychiatric Genomics Consortium (https://www.med.unc.edu/pgc).
 | Table I.Basic characteristics of European and
American SCZ GWAS datasets. |
Table I.
Basic characteristics of European and
American SCZ GWAS datasets.
|
|
|
| Genes |
|---|
|
|
|
|
|
|---|
| Study datasets | Study
population | Sample size
(case/control) | SNPs passing
QC | With SNPs |
P<10−5 |
P<10−2 |
|---|
| pha002857 | European | 1,351/1,378 | 700,487 | 44,462 | 12 | 4,703 |
| pha002859 | American | 1,195/954 | 809,854 | 44,840 | 4 | 5,863 |
| Overlap |
| – | 537,598 | 44,410 | 0 | 1,098 |
Gene-SNP mapping
We downloaded the gene annotation file from the UCSC
human genome annotation database (http://hgdownload.cse.ucsc.edu). The physical position
of SNPs was obtained from the Affymetrix chip annotation file. The
positional information of the genes and the SNPs was based on the
human genome assembly GRCh37 (Feb. 2009). Based on the positional
information, the SNPs identified in all three studies were mapped
to the gene region and 20 kb upstream and downstream of gene
boundaries to cover the coding sequence and most of the regulatory
features with the PLINK v1.07 software (http://zzz.bwh.harvard.edu/plink/index.shtml).
The SNP set P-value for a gene was calculated from the minimum
P-value of the gene.
GSEA and pathway enrichment
analysis
We performed functional analysis for all candidate
genes using the GSEA method. The number of minimum candidate genes
included in a pathway was set to 2 and it was considered to be
significant with adjusted P<0.01. To test the repeatability of
both GWAS data, the enrichment of one dataset was analyzed with the
other dataset as the background. The pathway enrichment analysis
was performed using the GEne SeT AnaLysis Toolkit (WebGestalt)
(24). The pathway files were
compiled from the KEGG pathway (http://www.genome.jp/kegg/pathway.html). P-value was
calculated based on the hypergeometric distribution.
Results
Integrative analysis of the European
and American SCZ GWAS datasets
We mapped SNPs to their corresponding genes to
identify genetic factors related to SCZ in the European and
American GWAS datasets. We used P<0.01 as the cutoff and
identified 4,703 candidate genes in the European samples and 5,863
candidate genes in the American samples. Then, we integrated the
two candidate gene lists and identified 1,098 common genes
(~11.59%, 1,098/9,468), which was 1.79 times of the expected value
of each individual pair-wise comparison (614 genes,
χ2=471.4, df=1, P<10−4). This implied
considerable replication between the two GWAS datasets for
individual genes (Table I).
Moreover, we performed GSEA to confirm the
replication between the two studies. We compared 4,703 candidate
genes from the European study against the entire gene set from the
American study, which were ranked according to their P-values. The
candidate genes of the European study were enriched at the top of
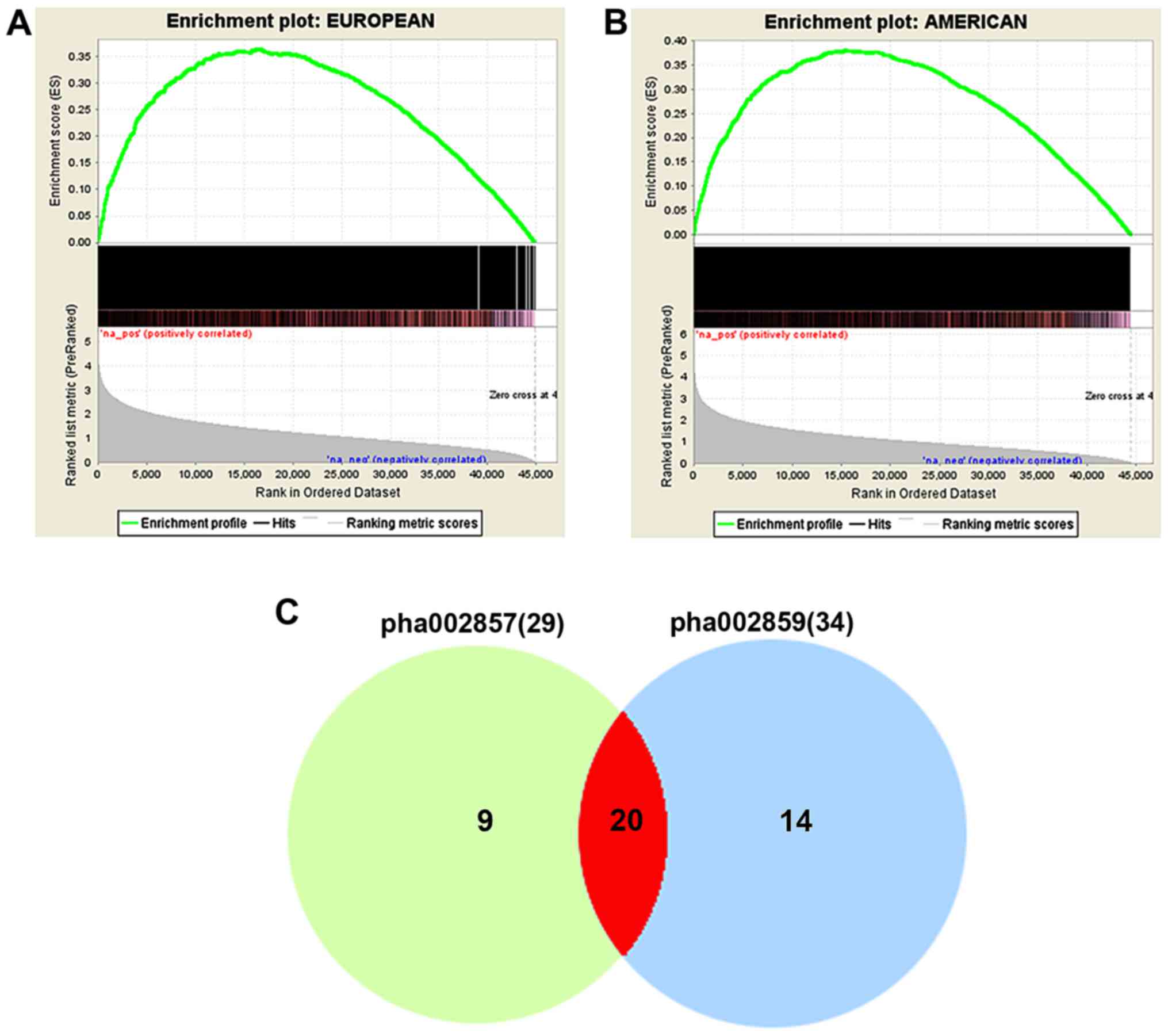
the gene list from the American study (Fig. 1A, ES=0.36390138, NES=1.7276831,
nominal P<10−3). The GSEA of 5,863 candidate genes
from the American study also showed analogous enrichment at the top
of the European gene list (Fig. 1B,
ES=0.381695544, NES=1.6385169, nominal P<10−3). These
results suggested that the two SCZ GWAS datasets were highly
overlapping and contained SCZ-related genes with similar biological
function.
Analysis of SCZ-related pathways
For better insight into the molecular mechanism of
SCZ, we performed KEGG pathway enrichment analysis for candidate
genes identified by both studies with the WebGestalt software
(24). We identified 29 enriched
KEGG pathways for the European study and 34 for the American study
(adjusted P<0.01). As shown in Fig.
1C, 20 pathways (~83.33%, 20/34) were shared by the two
studies.
Next, we compared the 20 common pathways to identify
susceptibility genes in SCZ meta-analysis data from the Psychiatric
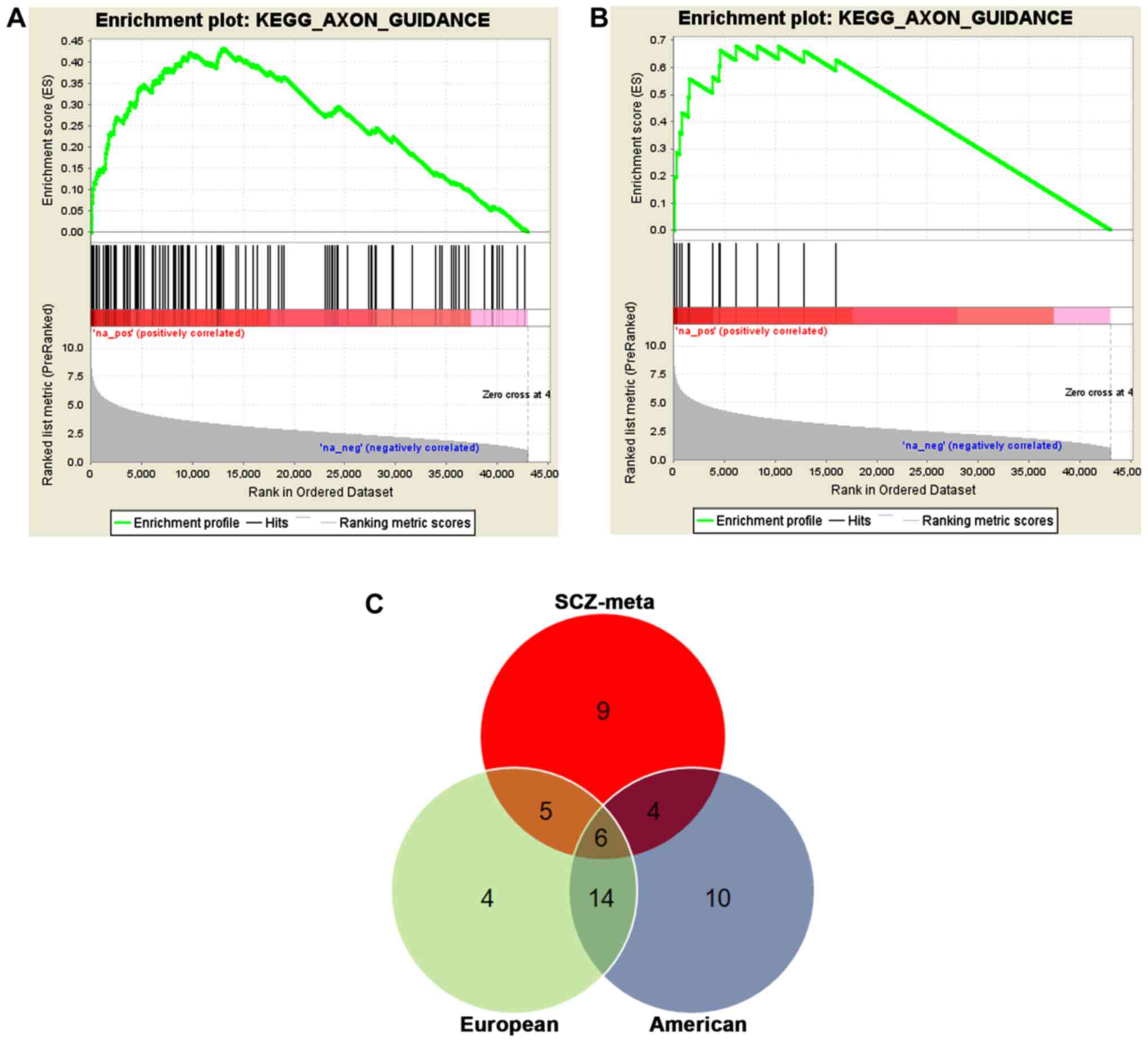
Genomics Consortium. As shown in Fig.
2C, we validated six pathways, namely, axon guidance, vascular
smooth muscle contraction, calcium signaling pathway, long term
potentiation, MAPK signaling pathway and arrhythmogenic right
ventricular cardiomyopathy (ARVC). In both studies, axon guidance,
long term potentiation and ARVC were highly significant
(P<10−3). In particular, the axon guidance represents
a key stage in the formation of neuronal network and the pathway
deserves our further study. Therefore, we focused on the axon
guidance pathway in this study. The European, American and SCZ
meta-analysis identified 23, 39 and 109 genes related to axon
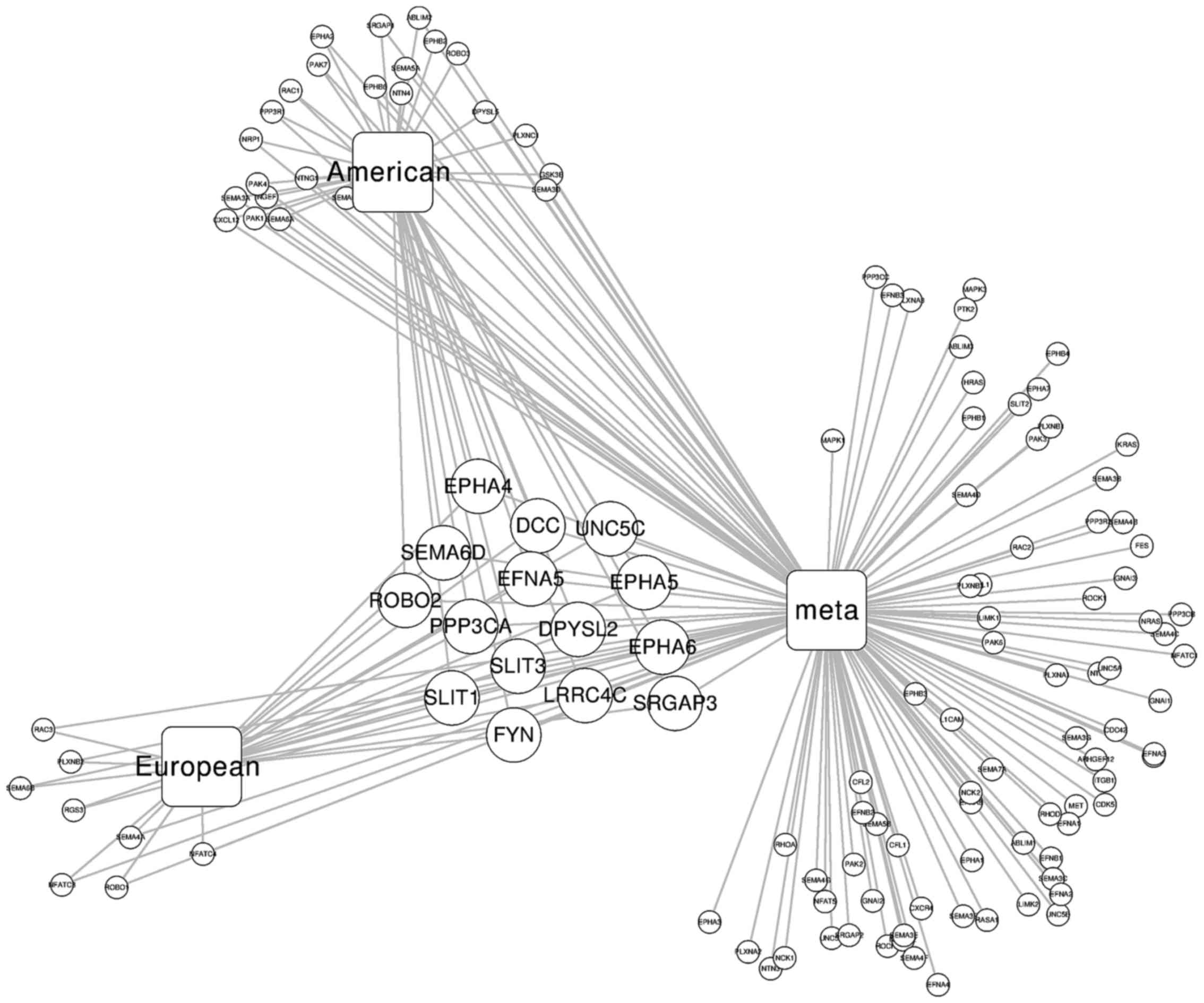
guidance pathway, respectively. Integrating the three datasets, we
identified 15 axon guidance pathway genes that were shared among
the three studies (Table II,
Fig. 3).
 | Table II.The 15 common axon guidance
genes. |
Table II.
The 15 common axon guidance
genes.
|
| -log10
(P-value) |
|---|
|
|
|
|---|
| Gene | European | American | SCZ
meta-analysis |
|---|
| DCC | 2.8091083 | 3.7437634 | 7.001612 |
| DPYSL2 | 2.352617 | 2.5673513 | 3.7548857 |
| EFNA5 | 3.3699791 | 2.5774102 | 4.387744 |
| EPHA4 | 2.031097 | 2.1434524 | 3.0377238 |
| EPHA5 | 2.103915 | 2.219971 | 5.9553895 |
| EPHA6 | 2.111259 | 2.9742846 | 4.5623975 |
| FYN | 2.79588 | 2.2160964 | 5.4071918 |
| LRRC4C | 2.3809066 | 2.3208447 | 7.528907 |
| PPP3CA | 2.2432117 | 2.5166981 | 3.2800066 |
| ROBO2 | 2.7068586 | 3.1870866 | 7.847146 |
| SEMA6D | 2.2464936 | 2.6424463 | 5.4734087 |
| SLIT1 | 3.4405723 | 2.8052082 | 3.5275486 |
| SLIT3 | 2.6044989 | 2.230475 | 6.288213 |
| SRGAP3 | 3.5915904 | 2.2222834 | 4.070334 |
| UNC5C | 3.3293831 | 2.4387794 | 4.3944135 |
Validation of SCZ susceptibility genes
in clinical samples
Next, we performed pooled GWAS on the clinical
samples to verify the SCZ susceptibility genes involved in axon
guidance in the Han population. GSEA pathway analysis identified
the 15 susceptibility genes in the pooled GWAS data (Fig. 2A). The 15 SCZ susceptibility genes
were deleted in colorectal cancer (DCC) (P=9.96e-08),
dihydropyrimidinase-like 2 (DPYSL2) (P=1.75e-04), ephrin-A5
(EFNA5) (P=4.09e-05), EPH receptor A4 (EPHA4)
(P=0.00092), EPH receptor A5 (EPHA5) (P=1.11e-06), EPH
receptor A6 (EPHA6) (P=2.74e-05), FYN proto-oncogene, Src
family tyrosine kinase (FYN) (P=3.92e-06), leucine rich
repeat containing 4C (LRRC4C) (P=2.96e-08), protein
phosphatase 3 catalytic subunit α (PPP3CA) (P=0.00052),
roundabout guidance receptor 2 (ROBO2) (P=1.42e-08),
semaphorin 6D (SEMA6D) (P=3.37e-06), slit guidance ligand 1
(SLIT1) (P=0.0003), slit guidance ligand 3 (SLIT3)
(P=5.15e-07), SLIT-ROBO Rho GTPase activating protein 3
(SRGAP3) (P=8.51e-05) and unc-5 netrin receptor C
(UNC5C) (P=4.04e-05). The pathway enrichment of these 15
axon guidance genes was significant with P<10−3
(Fig. 2B). DCC, LRRC4C and
ROBO2 were the top genes in the list with
P<10−7.
We further investigated these 15 genes in the pooled
GWAS and SCZ meta-analysis datasets, and identified 7,237 common
loci related to the 15 genes. After Bonferroni correction, we
obtained 14 significant loci in four genes, DCC, FYN, ROBO2
and SEMA6D (Table III).
Among the 14 candidate loci, two SNPs, rs9944880 in DCC gene
and rs7630091 in ROBO2 gene were significant in both the
meta-analysis and pooled GWAS datasets (P<10−4).
 | Table III.The SCZ-associated loci after
Bonferroni correction. |
Table III.
The SCZ-associated loci after
Bonferroni correction.
| Gene | Chromosome | SNP | Meta P-value | Pooled P-value |
|---|
| DCC | 18 | rs9944880 | 4.19E-06 | 5.55109E-05 |
| FYN | 6 | rs7764009 | 1.86E-04 | 0.375738765 |
| ROBO2 | 3 | rs7636341 | 1.42E-05 | 0.447750198 |
| ROBO2 | 3 | rs1516457 | 1.63E-05 | 0.163967215 |
| ROBO2 | 3 | rs11128505 | 1.98E-05 | 0.013580237 |
| ROBO2 | 3 | rs7633954 | 2.65E-05 | 0.526997858 |
| ROBO2 | 3 | rs7630091 | 4.14E-05 | 3.95512E-05 |
| ROBO2 | 3 | rs3849481 | 5.69E-05 | 0.086994752 |
| ROBO2 | 3 | rs6802433 | 8.72E-05 | 0.657963545 |
| SEMA6D | 15 | rs1559677 | 1.02E-05 | 0.00947373 |
| SEMA6D | 15 | rs281299 | 1.24E-05 | 0.019215293 |
| SEMA6D | 15 | rs1353580 | 1.24E-05 | 0.006132857 |
| SEMA6D | 15 | rs13313462 | 1.44E-05 | 0.163967215 |
| SEMA6D | 15 | rs1948572 | 1.51E-05 | 0.899343456 |
We validated rs9944880 and rs7630091 in an
independent pooled dataset that included 1,731 SCZ patients and
1,849 health controls. The rs9944880 located in DCC gene was
associated with SCZ patients (Table
IV). The T allele was associated with SCZ risk under additive
model (P=0.004, OR=1.19, 95% CI=1.06–1.33). Individuals with
rs9944880TT genotype were more likely to contract SCZ under
recessive model (P=0.0001, OR=1.67, 95% CI=1.29–2.17 for TT vs.
C+CT). No significant association was found in dominant model
(P=0.218, OR=1.10, 95% CI=0.95–1.26). However, the SNP rs7630091 in
ROBO2 was not associated with SCZ risk in our validation
cohort under the three models.
 | Table IV.Verification of gene loci in SCZ
patients from Shandong migrants. |
Table IV.
Verification of gene loci in SCZ
patients from Shandong migrants.
|
|
| Genotype count | Allele
frequency |
|
|
|
|---|
|
|
|
|
|
|
|
|
|---|
| SNP | Group | AA | AB | BB | A | B |
Padd |
Pdom |
Prec |
|---|
| rs9944880 |
| CC | CT | TT | C | T | 0.004 | 0.218 | 0.0001 |
|
| SCZ | 1,167 | 410 | 154 | 0.793 | 0.207 |
|
|
|
|
| Controls | 1,283 | 464 | 102 | 0.819 | 0.181 |
|
|
|
| rs7630091 |
| GG | AG | AA | G | A | 0.680 | 0.693 | 0.134 |
|
| SCZ | 913 | 602 | 216 | 0.701 | 0.299 |
|
|
|
|
| Controls | 962 | 687 | 200 | 0.706 | 0.294 |
|
|
|
Discussion
SCZ is a complex polygenic disease, which is
associated with many genes and non-coding loci (25). Traditional GWAS analysis has mainly
focused on the most significant SNPs that are associated with the
pathogenesis of SCZ (26).
Therefore, the less significant genes have not yet been analyzed.
Therefore, we developed a comprehensive approach by combining a
sub-significance strategy with pathway analysis. When we used
stringent common genome-wide significance level of
P<10−5, only 12 and 4 candidate genes were identified
in the European and American studies, respectively. Moreover, none
of these genes were common to either group. This suggested poor
repeatability between the two studies or a substantial loss of
information due to stringent criterion. Therefore, when the
threshold was set as P<10−2, we identified 4,703 and
5,863 candidate genes in the European and American studies,
respectively, and 1,098 genes (12%) were common in the two gene
lists. Wang et al performed a similar analysis with low
significance standard to identify novel susceptibility genes in the
coronary artery disease (27).
Moreover, the independent GSEA for European and American studies
showed 47% (20/43) common pathways between the two studies. Taken
together, these results suggest the feasibility of adopting a less
stringent significance level combined with pathway analysis to
identify molecular mechanisms of polygenic diseases.
Based on this strategy, we found high degree of
repeatability in the genes identified by the two studies. Thus, we
identified 20 candidate pathways associated with SCZ in the two
independent SCZ GWAS databases (European and American) from NCBI.
We also verified the top three significantly enriched pathways
(P<10−4) through pathway analysis of SCZ
meta-analysis dataset.
The relationship of the axon guidance pathway with
SCZ has not been well studied. The aging of human cerebral cortex
depends on the proliferation and differentiation of neural
progenitor cells, and proper migration and positioning behavior of
neurons (28). Disruption in any of
these processes can result in aberrant development of the cerebral
cortex and is central to many mental illnesses such as SCZ.
However, further investigations are necessary to prove this aspect
beyond doubt.
In our study, we identified two core genes,
ROBO2 and DCC, in the axon guidance pathway. Axons
integrate directional information multiple guidance cues and their
receptors during development. ROBO2 and DCC are axon guidance
receptors that can function individually or in combination with
other guidance receptors to regulate downstream effectors (29,30).
DCC is a membrane receptor for Netrin-1, which is
mainly expressed in various types of axon beams of the developing
central nervous system (CNS) (31).
DCC regulates axon guidance, projections, branching and a number of
important physiological processes of the CNS neurons after binding
to Netrin-1 (32–34).
ROBO2 is a membrane protein that belongs to a class
of conserved and development-related transmembrane receptor family
roundabout, which promotes axon guidance of embryonic CNS,
dendritic branching, axonal growth and neuronal cell migration
(35,36). ROBO2 cooperates with Slit proteins to
guide the major forebrain projections and mainly regulates axon
guidance (37).
DCC and ROBO2 as well as their respective ligands,
Netrin-1 and Slits, guide the callosum projection neurons across
the midline (38–42). Glial wedge and indusium griseum
represent the main glial cell populations located near the center,
which secrete nerve guide factors such as Netrin-1 and Slits. These
neural guidance factors interact with their corresponding membrane
receptors, DCC and ROBO2, on the neuronal axons projected by the
corpus callosum. These processes ensure the axons project correctly
and finally form corpus callosum (38–42).
In this study, we demonstrated that rs9944880
polymorphism in DCC gene on chromosome 18q21.2 was
associated with SCZ. From a mechanistic point of view, this
polymorphism probably altered the axon guidance pathway resulting
in SCZ. In conclusion, our study showed that aberrant axon guidance
pathway was central to SCZ pathogenesis by combining pathway
analysis with less stringent significance criterion in analyzing
gene sets from two GWAS. This study needs to be confirmed in large
cohorts of SCZ patients.
Acknowledgements
We are grateful to all SCZ participants who
generously contributed their time and materials for this research.
This study was supported by the Natural Science Foundation of
Heilongjiang, China (H201497).
Funding
This study was supported by the Natural Science
Foundation of Heilongjiang, China (H201497) and Science and
Technology Research Project of Education Department of Heilongjiang
Provience (2016-KYYWF-0883).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors contributions
ZW drafted the manuscript. ZW and PL were mainly
devoted to collecting and interpreting the general data. TW and SZ
analyzed the European and American SCZ GWAS datasets. LD and GC
were responsible for the analysis of SCZ-related pathways. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Qiqihar Medical University (Qiqihar, China) and in accordance with
the Helsinki Declaration. Signed informed consents were obtained
from the patients or the guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
SCZ
|
schizophrenia
|
|
GWAS
|
genome-wide association studies
|
|
ARVC
|
arrhythmogenic right ventricular
cardiomyopathy
|
|
DCC
|
deleted in colorectal cancer
|
|
GSEA
|
Gene Set Enrichment Analysis
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
EFNA5
|
ephrin-A5
|
|
EPHA4
|
EPH receptor A4
|
|
EPHA5
|
EPH receptor A5
|
|
EPHA6
|
EPH receptor A6
|
|
FYN
|
FYN proto-oncogene, Src family
tyrosine kinase
|
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
PPP3CA
|
protein phosphatase 3 catalytic
subunit α
|
|
ROBO2
|
roundabout guidance receptor 2
|
|
SEMA6D
|
semaphorin 6D
|
|
SLIT1
|
slit guidance ligand 1
|
|
SLIT3
|
slit guidance ligand 3
|
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating
protein 3
|
|
UNC5C
|
unc-5 netrin receptor C
|
|
CNS
|
central nervous system
|
|
CCMD-3
|
Chinese Classification of Mental
Disorders version 3
|
|
DSM-IV
|
Diagnostic and Statistical Manual of
Mental Disorders-IV
|
|
SCID-NP
|
Structured Clinical Interview for
DSM-IV-TR Axis I Disorders-Nonpatient Edition
|
|
RAS
|
relative allele signal.
|
References
|
1
|
Giusti-Rodríguez P and Sullivan PF: The
genomics of schizophrenia: Update and implications. J Clin Invest.
123:4557–4563. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ruzicka WB, Subburaju S and Benes FM:
Circuit- and diagnosis-specific DNA methylation changes at
γ-aminobutyric acid-related genes in postmortem human hippocampus
in schizophrenia and bipolar disorder. JAMA Psychiatry. 72:541–551.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Insel TR: Rethinking schizophrenia.
Nature. 468:187–193. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Harrison PJ: Recent genetic findings in
schizophrenia and their therapeutic relevance. J Psychopharmacol.
29:85–96. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Quednow BB, Brzózka MM and Rossner MJ:
Transcription factor 4 (TCF4) and schizophrenia: Integrating the
animal and the human perspective. Cell Mol Life Sci. 71:2815–2835.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hosák L, Silhan P and Hosáková J:
Genome-wide association studies in schizophrenia, and potential
etiological and functional implications of their results. Acta Med
(Hradec Kralove). 55:3–11. 2012. View Article : Google Scholar
|
|
7
|
Sandin S, Lichtenstein P, Kuja-Halkola R,
Larsson H, Hultman CM and Reichenberg A: The familial risk of
autism. JAMA. 311:1770–1777. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roth TL, Lubin FD, Sodhi M and Kleinman
JE: Epigenetic mechanisms in schizophrenia. Biochim Biophys Acta.
1790:869–877. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ripke S, Neale BM, Corvin A, Walters JT,
Farh KH, Holmans PA, Lee P, Buliksullivan B, Collier DA and Huang
H; Schizophrenia Working Group of the Psychiatric Genomics
Consortium, : Biological insights from 108 schizophrenia-associated
genetic loci. Nature. 511:421–427. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Goodbourn PT, Bosten JM, Bargary G, Hogg
RE, Lawrance-Owen AJ and Mollon JD: Variants in the 1q21 risk
region are associated with a visual endophenotype of autism and
schizophrenia. Genes Brain Behav. 13:144–151. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ripke S, O'Dushlaine C, Chambert K, Moran
JL, Kähler AK, Akterin S, Bergen SE, Collins AL, Crowley JJ, Fromer
M, et al: Multicenter Genetic Studies of Schizophrenia Consortium;
Psychosis Endophenotypes International Consortium; Wellcome Trust
Case Control Consortium 2: Genome-wide association analysis
identifies 13 new risk loci for schizophrenia. Nat Genet.
45:1150–1159. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schizophrenia Psychiatric Genome-Wide
Association Study (GWAS) Consortium, . Genome-wide association
study identifies five new schizophrenia loci. Nat Genet.
43:969–976. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ruderfer DM, Fanous AH, Ripke S, McQuillin
A, Amdur RL, Gejman PV, O'Donovan MC, Andreassen OA, Djurovic S,
Hultman CM, et al: Schizophrenia Working Group of the Psychiatric
Genomics Consortium; Bipolar Disorder Working Group of the
Psychiatric Genomics Consortium; Cross-Disorder Working Group of
the Psychiatric Genomics Consortium: Polygenic dissection of
diagnosis and clinical dimensions of bipolar disorder and
schizophrenia. Mol Psychiatry. 19:1017–1024. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Salavati B, Rajji TK, Price R, Sun Y,
Graff-Guerrero A and Daskalakis ZJ: Imaging-based neurochemistry in
schizophrenia: A systematic review and implications for
dysfunctional long-term potentiation. Schizophr Bull. 41:44–56.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shi Y, Li Z, Xu Q, Wang T, Li T, Shen J,
Zhang F, Chen J, Zhou G, Ji W, et al: Common variants on 8p12 and
1q24.2 confer risk of schizophrenia. Nat Genet. 43:1224–1227. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sleiman P, Wang D, Glessner J, Hadley D,
Gur RE, Cohen N, Li Q, Hakonarson H, Sleiman P, Glessner J, et al:
Janssen-CHOP Neuropsychiatric Genomics Working Group: GWAS meta
analysis identifies TSNARE1 as a novel schizophrenia/bipolar
susceptibility locus. Sci Rep. 3:30752013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stefansson H, Ophoff RA, Steinberg S,
Andreassen OA, Cichon S, Rujescu D, Werge T, Pietiläinen OP, Mors
O, Mortensen PB, et al: Genetic Risk and Outcome in Psychosis
(GROUP): Common variants conferring risk of schizophrenia. Nature.
460:744–747. 2009.PubMed/NCBI
|
|
18
|
Wang KS, Liu XF and Aragam N: A
genome-wide meta-analysis identifies novel loci associated with
schizophrenia and bipolar disorder. Schizophr Res. 124:192–199.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Betcheva ET, Yosifova AG, Mushiroda T,
Kubo M, Takahashi A, Karachanak SK, Zaharieva IT, Hadjidekova SP,
Dimova II, Vazharova RV, et al: Whole-genome-wide association study
in the Bulgarian population reveals HHAT as schizophrenia
susceptibility gene. Psychiatr Genet. 23:11–19. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yue WH, Wang HF, Sun LD, Tang FL, Liu ZH,
Zhang HX, Li WQ, Zhang YL, Zhang Y, Ma CC, et al: Genome-wide
association study identifies a susceptibility locus for
schizophrenia in Han Chinese at 11p11.2. Nat Genet. 43:1228–1231.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Edwards YJK, Beecham GW, Scott WK, Khuri
S, Bademci G, Tekin D, Martin ER, Jiang Z, Mash DC, ffrench-Mullen
J, et al: Identifying consensus disease pathways in Parkinson's
disease using an integrative systems biology approach. PLoS One.
6:e169172011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Eleftherohorinou H, Wright V, Hoggart C,
Hartikainen AL, Jarvelin MR, Balding D, Coin L and Levin M: Pathway
analysis of GWAS provides new insights into genetic susceptibility
to 3 inflammatory diseases. PLoS One. 4:e80682009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang K, Edmondson AC, Li M, Gao F, Qasim
AN, Devaney JM, Burnett MS, Waterworth DM, Mooser V, Grant SFA, et
al: Pathway-wide association study implicates multiple sterol
transport and metabolism genes in HDL cholesterol regulation. Front
Genet. 2:412011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang J, Duncan D, Shi Z and Zhang B:
WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013.
Nucleic Acids Res. 41(W1): W77–W83. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Purcell SM, Moran JL, Fromer M, Ruderfer
D, Solovieff N, Roussos P, O'Dushlaine C, Chambert K, Bergen SE,
Kähler A, et al: A polygenic burden of rare disruptive mutations in
schizophrenia. Nature. 506:185–190. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Manolio TA, Collins FS, Cox NJ, Goldstein
DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR,
Chakravarti A, et al: Finding the missing heritability of complex
diseases. Nature. 461:747–753. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang F, Xu CQ, He Q, Cai JP, Li XC, Wang
D, Xiong X, Liao YH, Zeng QT, Yang YZ, et al: Genome-wide
association identifies a susceptibility locus for coronary artery
disease in the Chinese Han population. Nat Genet. 43:345–349. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi Y, Kirwan P, Smith J, Robinson HP and
Livesey FJ: Human cerebral cortex development from pluripotent stem
cells to functional excitatory synapses. Nat Neurosci. 15:477–486,
S1. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu Y, Taru H, Jin Y and Quinn CC: SYD-1C,
UNC-40 (DCC) and SAX-3 (Robo) function interdependently to promote
axon guidance by regulating the MIG-2 GTPase. PLoS Genet.
11:e10051852015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tang X and Wadsworth WG: SAX-3 (Robo) and
UNC-40 (DCC) regulate a directional bias for axon guidance in
response to multiple extracellular cues. Plos One. 9:e1100312014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shu T, Valentino KM, Seaman C, Cooper HM
and Richards LJ: Expression of the netrin-1 receptor, deleted in
colorectal cancer (DCC), is largely confined to projecting neurons
in the developing forebrain. J Comp Neurol. 416:201–212. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shi M, Guo C, Dai JX and Ding YQ: DCC is
required for the tangential migration of noradrenergic neurons in
locus coeruleus of mouse brain. Mol Cell Neurosci. 39:529–538.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cotrufo T, Pérez-Brangulí F, Muhaisen A,
Ros O, Andrés R, Baeriswyl T, Fuschini G, Tarrago T, Pascual M,
Ureña J, et al: A signaling mechanism coupling netrin-1/deleted in
colorectal cancer chemoattraction to SNARE-mediated exocytosis in
axonal growth cones. J Neurosci. 31:14463–14480. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Teichmann HM and Shen K: UNC-6 and UNC-40
promote dendritic growth through PAR-4 in Caenorhabditis elegans
neurons. Nat Neurosci. 14:165–172. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Brose K, Bland KS, Wang KH, Arnott D,
Henzel W, Goodman CS, Tessier-Lavigne M and Kidd T: Slit proteins
bind Robo receptors and have an evolutionarily conserved role in
repulsive axon guidance. Cell. 96:795–806. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chédotal A, Kerjan G and Moreau-Fauvarque
C: The brain within the tumor: New roles for axon guidance
molecules in cancers. Cell Death Differ. 12:1044–1056. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
López-Bendito G, Flames N, Ma L, Fouquet
C, Di Meglio T, Chedotal A, Tessier-Lavigne M and Marín O: Robo1
and Robo2 cooperate to control the guidance of major axonal tracts
in the mammalian forebrain. J Neurosci. 27:3395–3407. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Marcos-Mondéjar P, Peregrín S, Li JY,
Carlsson L, Tole S and López-Bendito G: The lhx2 transcription
factor controls thalamocortical axonal guidance by specific
regulation of robo1 and robo2 receptors. J Neurosci. 32:4372–4385.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Long H, Sabatier C, Ma L, Plump A, Yuan W,
Ornitz DM, Tamada A, Murakami F, Goodman CS and Tessier-Lavigne M:
Conserved roles for Slit and Robo proteins in midline commissural
axon guidance. Neuron. 42:213–223. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Finci L, Zhang Y, Meijers R and Wang JH:
Signaling mechanism of the netrin-1 receptor DCC in axon guidance.
Prog Biophys Mol Biol. 118:153–160. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang Y, Lee WS, Tang X and Wadsworth WG:
Extracellular matrix regulates UNC-6 (netrin) axon guidance by
controlling the direction of intracellular UNC-40 (DCC) outgrowth
activity. PLoS One. 9:e972582014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kulkarni G, Xu Z, Mohamed AM, Li H, Tang
X, Limerick G and Wadsworth WG: Experimental evidence for UNC-6
(netrin) axon guidance by stochastic fluctuations of intracellular
UNC-40 (DCC) outgrowth activity. Biol Open. 2:1300–1312. 2013.
View Article : Google Scholar : PubMed/NCBI
|