Introduction
More than 85% of lung cancer cases are non-small
cell lung cancer (NSCLC), and the mortality rate of NSCLC remains
high all over the world (1). In USA,
NSCLC was the second most prevalent cancer among all new cancer
cases and cancer deaths in 2016 (2).
Traditional chemotherapy regimens for NSCLC have many disadvantages
such as limited efficacy, high recurrence rate, and high toxicity
(3). These disadvantages limit the
efficacy of drug therapy for NSCLC, so an improved understanding of
the exact mechanisms of this disease and developing new, targeted
therapy drugs for NSCLC is urgent.
Transforming growth factor b-induced protein
(TGFBI), known as βIg-h3 or keratoepithelin, contains a
carboxyl-terminal Arg-Gly-Asp (RGD) integrin-binding sequence and
four conserved fasciclin-1 (FAS1) domains (4). TGFBI plays an important role in many
cellular disease processes, for example, tumorigenesis,
progression, and metastasis (5).
TGFBI is down-regulated in many cancer types such as lung cancer
(6), breast cancer (7) and ovary carcinoma (8). TGFBI functions as a suppressive role
via inhibition of cell proliferation, delay of the G1-S phase
transition, and induction of senescence in the prevention of breast
cancer and mesothelioma cells (9).
TGFBI-derived peptides might be used as possible therapeutic
adjuvants to enhance the response to chemotherapy in NSCLC
(10). Recent studies suggested that
loss of TGFBI expression has been described in lung cancer
(11). It has been reported that
downregulation of TGFBI protein is a frequent event and related to
the tumor progression in human lung cancer through comparing 130
primary lung carcinomas to normal lung tissues (6,12).
However, the upstream regulatory mechanism of TGFBI is not fully
understood.
During the past decades, microRNAs (miRNAs, small
non-coding RNA molecules), which is about 22 nucleotides in length,
have been found to act in post-transcriptional regulation and RNA
silencing of gene expression via binding the 3′-untranslated region
(3′-UTR) of target mRNAs (13). Many
reports have indicated miRNAs function as tumor suppressors or
oncogenes in many cancer types (14). Many researches have indicated that
miRNAs regulate many cellular processes including differentiation,
proliferation, migration, and apoptosis (15). Recent researches have indicated
miRNAs also function in the initiation and progression of NSCLC.
For example, miR-455-3p was shown to regulate NSCLC cell
proliferation and migration by downregulation of HOXB5 (16). Cell proliferation and invasion of
NSCLC were inhibited by miR-504 by targeting LOXL2 (17). miR-142-5p inhibited tumorigenesis of
NSCLC by targeting PIK3CA expression (18). Cell survival and metastasis of NSCLC
were promoted by CXCL6 via down-regulation of miR-515-5p (19). Recent studies have reported miR-21-5p
is upregulated in NSCLC patients (20). However, how miR-21-5p regulates cell
proliferation in NSCLC and the involved molecular mechanisms remain
poorly understood.
In our research, we show that miR-21-5p directly
regulated TGFBI in two NSCLC cell lines. Additionally, we found
that miR-21-5p promoted the proliferation of NSCLC cells via
inhibiting TGFBI expression.
Materials and methods
Cell culture
We obtained two human NSCLC cell lines A549 and
H1299 and human lung cell line HLF from the Cell Bank, Chinese
Academy of Sciences (Shanghai, China). We cultured the cells using
10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) in DMEM (HyClone; GE Healthcare Life Sciences,
Logan, UT, USA) at 37°C with a humidified atmosphere of 5%
CO2.
miRNA overexpression and
knockdown
Either a miR-21-5p mimic or a miR-21-5p inhibitor
were transfected into NSCLC cells to overexpress or knockdown
miR-21-5p, respectively. We purchased the synthetic miR-21-5p
mimic, miR-21-5p inhibitor, and negative control mimic and control
inhibitor RNAs from Guangzhou RiboBio Co., Ltd., (Guangzhou,
China). We seeded A549 and H1299 cells in 6-well plates using 10%
FBS in DMEM. When the cells reached 60–70% confluence, the media
was switched to Opti-MEM Reduced Serum Medium (Gibco; Thermo Fisher
Scientific, Inc.) before transfection. Then, we transfected equal
amounts of the miR-21-5p mimic, the control mimic, the miR-21-5p
inhibitor, or the control inhibitor into the cells using
Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocols. After 48 h, the cells
were harvested. The TGFBI expression levels were determined by
western blotting, and the miR-21-5p expression levels were
determined by RT-qPCR.
miR-21-5p expression in NSCLC
We used The Cancer Genome Atlas data (https://cancergenome.nih.gov/) to analyze and compare
the miR-21-5p expression in NSCLC tissues and normal solid tissues.
This database compared the miR-21-5p expression between 91 normal
solid tissues and 792 primary solid tumors of NSCLC.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
We extracted total RNA from transfected cells using
TRIzol Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) under
the guidance of the manufacturer's protocols. We employed TaqMan
miRNA probes (Applied Biosystems; Thermo Fisher Scientific, Inc.)
to detect and quantify the levels of miRNAs under the guidance of
the manufacturer's protocols. We performed RT-qPCR on a 7500
Real-time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) using a TaqMan PCR kit. Each reaction was run in triplicate.
After that, we determined the cycle threshold (Cq) data using fixed
threshold settings, then calculated the average Cq of triplicate
PCR reactions. The relative amount of miRNAs were confirmed by the
comparative Cq method (21). We used
U6 snRNA as an internal control. The relative amount of miR-21-5p,
which was normalized to U6, was calculated with the equation
2−ΔΔCq, in which
ΔΔCq=(CqmiR-21-5p-CqU6)
tumor-(CqmiR-21-5p-CqU6) control.
To quantify the amount of TGFBI mRNA, we used oligo
dT primers (Takara Biotechnology Co., Ltd., Dalian, China) to
reverse transcribe total RNA to cDNA. Next, RT-qPCR was run on the
samples using SYBR Green Dye (Invitrogen; Thermo Fisher Scientific,
Inc.) and specific primers targeting TGFBI and GAPDH (sequences
listed in Table I). When the
reactions were finished, we determined the Cq values with fixed
threshold setting. The relative amount of TGFBI mRNA, which was
normalized to GAPDH, was calculated through a similar method as
described above.
 | Table I.Sequences of primers. |
Table I.
Sequences of primers.
| Primer | Sense | Antisense |
|---|
| TGFBI |
5′-GTGCGGCTAAAGTCTCTCCA-3′ |
5′-AAGCCCTGGAAAACGCTGAT-3′ |
| GAPDH |
5′-CGAGCCACATCGCTCAGACA-3′ |
5′-GTGGTGAAGACGCCAGTGGA-3′ |
Luciferase reporter assay
Direct targeting by miR-21-5p of the TGFBI gene was
tested using luciferase reporter assays (22). Human genomic DNA was used as the
template to amplify the entire 3′-UTR of human TGFBI by PCR. Next,
the amplified PCR product was directly cloned into the
p-MIR-reporter plasmid (Ambion; Thermo Fisher Scientific, Inc.),
then we confirmed the successful insertion via sequencing. We
mutated the sequences (interacted with the miR-21-5p seed region)
from ATAAGCTA to TATTCGAT to test binding specificity, then
inserted the TGFBI 3′-UTR mutant fragment into the p-MIR-reporter
plasmid. Then, we seeded A549 cells into 24-well plates. The next
day, we used Lipofectamine 3000 to transfect equal amounts (0.3 µg)
of β-galactosidase (β-gal) expression plasmid (Ambion; Thermo
Fisher Scientific, Inc.) and the firefly luciferase reporter
plasmid together with 20 pmol of the miR-21-5p mimic, mimic
control, miR-21-5p inhibitor, or inhibitor control. We used the
β-gal plasmid as a control of transfection efficiency. The cells
were assayed after 24 h post-transfection via a luciferase assay
kit (Promega Corporation, Madison, WI, USA).
Plasmid construction and siRNA
interference assays
A plasmid containing the human TGFBI open reading
frame was purchased from Shanghai Genechem Co., Ltd., (Shanghai,
China) and was used to overexpress TGFBI. We used an empty plasmid
as a negative control. To silence TGFBI expression, TGFBI siRNA was
obtained from Guangzhou RiboBio Co., Ltd. We used a scrambled siRNA
sequence as a negative control. We then transfected the TGFBI
overexpressing plasmid and TGFBI siRNA into A549 and H1299 cells
using Lipofectamine 3000. After 48 h of transfection, total protein
and RNA were extracted and evaluated by western blotting and
RT-qPCR, respectively.
Protein extraction and western
blotting
RIPA lysis buffer (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany), which contained a protease inhibitor, was used
to lyse protein from the cells. We separated proteins by 10% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The TGFBI expression levels
were determined by western blotting. We normalized the protein
levels by probing the same blots with a GAPDH antibody and analyzed
the protein bands using Bandscan software Image J. The TGFBI
antibody (A2561) was purchased from ABclonal Biotechnology (Woburn,
MA, USA), and the GAPDH antibody (FL-335, sc-25778) was purchased
from Santa Cruz Biotechnology, Inc., (Dallas, TX, USA).
Cell proliferation assay
For the CCK-8 assay, A549 cells were seeded into
96-well plates at 1×104 cells per well and cultured for
12 h using 10% FBS in DMEM. Cell proliferation was assessed under
the guidance of the manufacturer's protocols of the Cell Counting
Kit-8 (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). After
transfection, we added 10 µl aliquot of CCK-8 solution to the test
wells at 12, 24, 36, 48, and 60 h. Absorbance at 450 nm wavelength
was measured after a 2 h incubation.
For the EdU assay, we seeded A549 cells into 48-well
plates (Corning Incorporated, Corning, NY, USA). Until the
transfected A549 cells reached 80% confluency, we used an EdU assay
kit (Guangzhou RiboBio Co., Ltd.) to measure the cell proliferation
rate. We followed the manufacturer's protocols except the nucleus
staining dye was changed from Hoechst 33342 (supplied with the kit)
to DAPI (Beyotime Institute of Biotechnology, Haimen, China)
(23). When the stain was finished,
the cells were imaged by fluorescence microscopy (BX51; Olympus
Corporation, Tokyo, Japan).
Statistical analysis
Date shown are the mean ± standard deviation (SD) of
at least three independent experiments, with P<0.05 considered
to indicate a statistically significant difference. SPSS software,
v.17.0 (SPSS, Inc., Chicago, IL, USA) was used to perform
statistical analysis. The results were compared using one-way
analysis of variance followed by a post hoc Tukey test for multiple
comparisons.
Results
miR-21-5p expression is upregulated in
NSCLC cell lines and tissues
We have measured miR-21-5p expression in two NSCLC
cell lines (A549 and H1299) and one normal lung cell lines (HLF).
And we found miR-21-5p expression in A549 is upregulated compared
with that in HLF and H1299, and we also found miR-21-5p expression
in H1299 is upregulated compared with that in HLF (Fig. 1A). To investigate miR-21-5p levels
compare between NSCLC vs. normal lung in clinical tissue samples,
the TCGA database was employed to analyze miR-21-5p expression of
91 normal solid tissues and 792 primary solid tumors of NSCLC
(Fig. 1B). The results showed that
miR-21-5p was significantly upregulated in the solid tumors
compared to the normal tissues (fold change=4.24 and P-value
<0.001).
miR-21-5p promotes NSCLC cell
proliferation
To better study the effect of miR-21-5p on the
proliferation of NSCLC cells, we analyzed the role of miR-21-5p on
the proliferation of NSCLC cells (A549) via CCK-8 and EdU assay
methods. The results showed that overexpression of miR-21-5p
resulted in an increase in proliferation of A549 cells, but that
the proliferation of A549 cells was inhibited when the expression
of miR-21-5p was inhibited (Fig. 2).
These results indicate that miR-21-5p induces the proliferation of
NSCLC cells.
Prediction of TGFBI as a miR-21-5p
target gene
In order to elucidate the specific mechanism by
which miR-21-5p induces the proliferation of NSCLC cells, we used
three different computer software programs, TargetScan (24), miRanda (25), and PicTar (26) to predict genes that might bind to
miR-21-5p. Among these candidate genes (zinc finger protein 367, G
protein-coupled receptor 64, SMAD family member 7 and so on), TGFBI
was predicted as miR-21-5p target-gene by all three programs. The
potential for binding of TGFBI and miR-21-5p is shown in Fig. 2A. The binding energy of the hairpin
structure is −20.2 kj/mol, and this binding energy is within the
binding energy range for miRNA and target genes. In addition, the 3
′UTR region of TGFBI binding with miR-21-5p is highly conserved
across species (Fig. 3A).
Identification of the TGFBI as a
miR-21-5p direct target gene
To verify whether miR-21-5p binds to the 3 ′UTR
region of TGFBI, two luciferase reporter gene plasmids, TGFBI
wild-type and TGFBI mutant-type, were constructed. We transfected
these plasmids with miR-21-5p mimic, control mimic, miR-21-5p
inhibitor, or control inhibitor into A549 cells. In wild type, the
activity of luciferase was decreased after the overexpression of
miR-21-5p (Fig. 3B) and the activity
of luciferase was increased after inhibiting the expression of
miR-21-5p (Fig. 3B). In the mutant,
the activity of luciferase did not change after overexpression or
inhibition of miR-21-5p (Fig. 3B).
These findings indicate that the binding sequence is required for
interaction of miR-21-5p and TGFBI mRNA.
To further investigate whether TGFBI is a miR-21-5p
target gene, we measured the expression of TGFBI in NSCLC cell
lines A549 and H1299. To alter the miR-21-5p levels, we transfected
a miR-21-5p mimic or a miR-21-5p inhibitor into A549 and H1299. As
expected, the miR-21-5p expression increased and the TGFBI
expression decreased significantly after miR-21-5p was
overexpressed in A549 and H1299 cell lines (Fig. 4A-C). When we transfected the
miR-21-5p inhibitor in A549 and H1299 cells, the miR-21-5p
expression was decreased, and the TGFBI expression was increased
(Fig. 4A-C). To better explain the
effect of miR-21-5p on TGFBI expression, we measured the expression
of TGFBI mRNA from transfected cells. Neither overexpression nor
inhibition of miR-21-5p affected the mRNA level of TGFBI (Fig. 4D), indicating that miR-21-5p
regulates the expression of the TGFBI gene at the
post-transcriptional level.
miR-21-5p promotes NSCLC cell
proliferation by targeting TGFBI
Because miRNA can act on multiple genes, we studied
whether the regulation of TGFBI expression by miR-21-5p affects the
proliferation of NSCLC. To alter the expression of TGFBI, we
transfected TGFBI siRNA or an overexpression plasmid into A549
cells. The efficiency of over expression and knockdown of TGFBI was
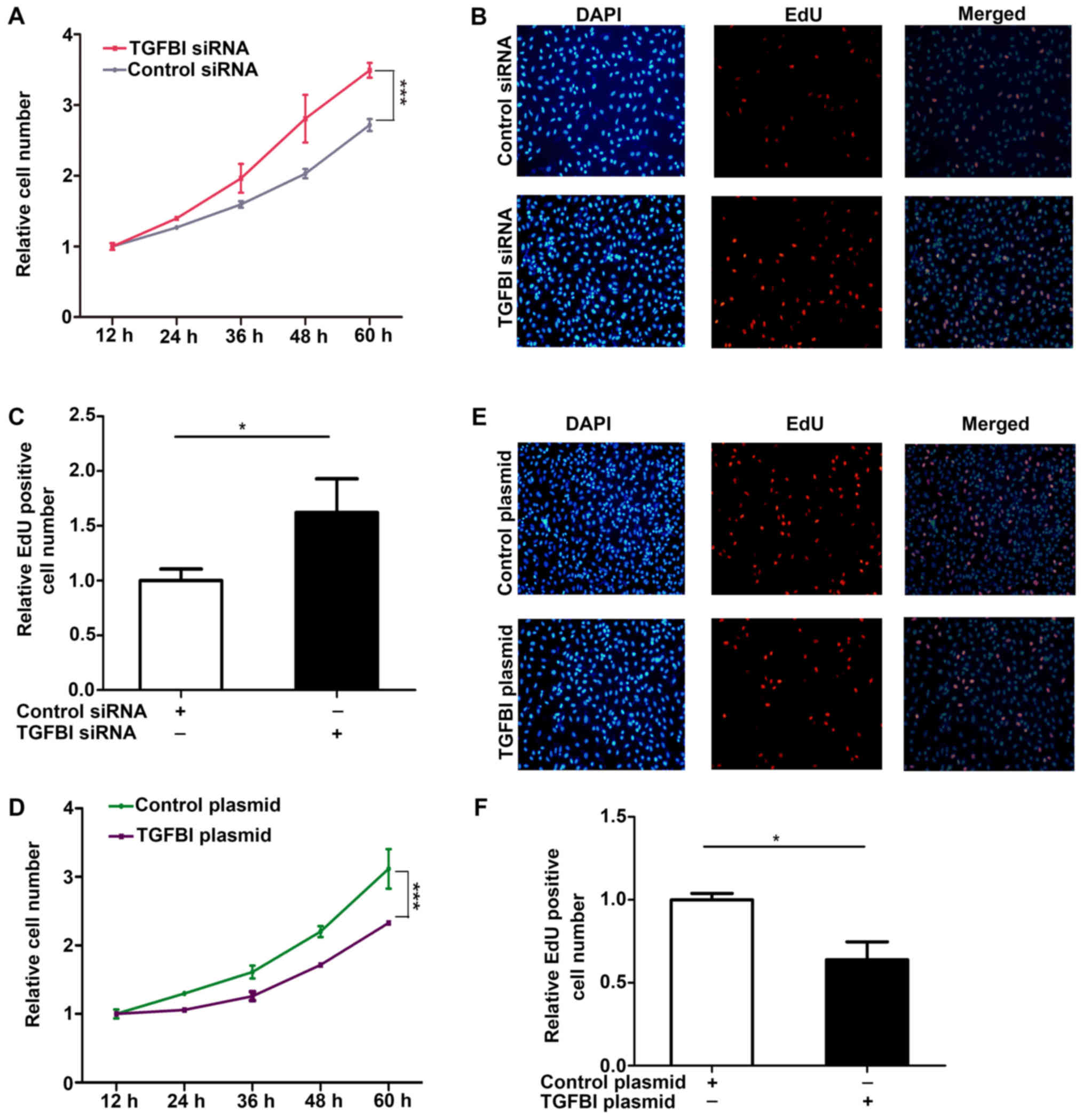
measured and was shown in Fig. 5.
The results showed that the proliferation of A549 cells is promoted
after knockdown of TGFBI (Fig.
6A-C), but overexpression of TGFBI inhibited the proliferation
of A549 cells (Fig. 6D-F). These
results demonstrate that miR-21-5p and TGFBI exhibit opposite
effect on the proliferation of NSCLC cells. Importantly, the
proliferation rates of cells co-transfected with the miR-21-5p
mimic and the TGFBI overexpressing plasmid were significantly lower
than that of cells transfected with the miR-21-5p mimic alone
(Fig. 7). These results indicate
that the additional expression of TGFBI gene can restore the effect
of miR-21-5p on cell proliferation. This result suggests that the
regulation of TGFBI gene expression by miR-21-5p may explain how
miR-21-5p exerts its function as an oncomiR, a microRNA (miRNA)
that is associated with cancer.
Discussion
TGFBI is expressed at low levels in many types of
cancers, including lung cancer (27). There have been many studies of TGFBI
in cancer, but most studies have mainly focused on TGFBI expression
levels, and the specific role of TGFBI in NSCLC is not clear.
Recent studies have shown that TGFBI may serve as a predictive
factor for chemotherapy response and suggest that TGFBI-derived
peptides can be used as adjuvant for the treatment of NSCLC
(10). In this study, silencing
TGFBI by siRNA promoted the proliferation of NSCLC cells. After
overexpression of TGFBI, the opposite results were obtained,
suggesting that TGFBI may be a tumor suppressor gene that acts in
the prevention of NSCLC. The lncRNA H19/miR-675 axis inhibits
metastasis of prostate cancer cells by targeting TGFBI (28). Recent study has shown that in corneal
fibroblasts, TGF-β regulated the expression of the TGFBI protein
through miR-21- and miR-181a-coordinated activity and Smad
signaling (29). Several miRNA can
be highly expressed in cancer, and can act on tumor suppressor
genes, decreasing gene expression. Therefore, we used three
analysis programs to predict miRNAs that could target TGFBI. Among
these candidate miRNAs (such as miR-489-3p, miR-21-5p, miR-590-5p
and miR-9-5p), miR-21-5p has been reported as an oncogene in a
variety of cancers including lung cancer (30). We demonstrated the interaction of
miR-21-5p and TGFBI by luciferase experiment. To verify miR-21-5p
regulation of TGFBI gene expression, we overexpressed miR-21-5p in
NSCLC cell lines A549 and H1299. The expression of the TGFBI
protein decreased significantly and the expression of TGFBI
increased after miR-21-5p was inhibited, but the expression level
of TGFBI mRNA did not change. These results show that miR-21-5p can
inhibit the TGFBI expression by post-transcriptional regulation.
Finally, we indicate that miR-21-5p promotes the proliferation of
NSCLC cells by inhibiting the expression of TGFBI. These findings
reveal the important role of miR-21-5p targeting TGFBI as a novel
regulatory pathway in the development of NSCLC.
Previous researches have shown that miRNAs may be an
important regulator of cancer (31).
The miRNA miR-21-5p is highly expressed in a variety of cancers,
such as laryngeal squamous cell carcinoma (32), gastric cancer (33), colorectal cancer (34), breast cancer (35), and NSCLC (36). Multiple genes can be targeted by one
single miRNA, and the same gene can be regulated by multiple miRNA.
Therefore, miR-21-5p might have different mRNA binding partners
other than TGFBI, and these different target genes may also play an
important role in carcinogenesis. For example, miR-21-5p has been
reported to promote ox-LDL-induced endothelial cell senescence by
down-regulation of the mitochondrial fission protein Drp1 (37). PDCD4 may be a functional target of
miR-21-5p in osteosarcoma cells (38). Therefore, it is important to study
the mechanistic details of this new pathway in the process of NSCLC
cells. In our research, we showed that overexpression of miR-21-5p
promotes the proliferation of NSCLC cells, and the reduction of
TGFBI can mimic the induction of miR-21-5p. Interestingly, although
miR-21-5p has many other targeted genes, overexpression of TGFBI
can successfully counter the role of miR-21-5p to induce the
proliferation of NSCLC cells. Our results suggest targeting TGFBI
may be the important mechanism by which miR-21-5p exhibit its
oncomiR roles. Thus, the reason why the up-regulation of miR-21-5p
in NSCLC can induce cell proliferation might be that TGFBI is
regulated by miR-21-5p.
In summary, this study describes a new regulatory
network in which miR-21-5p and TGFBI can precisely tune the NSCLC
cells proliferation. This study suggest new directions for NSCLC
treatment in the future. Future studies should pay more attention
on the effect of miR-21-5p on other cellular functions of NSCLC
such as apoptosis, migration ability or invasiveness.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
fund for outstanding talented young persons in colleges and
universities of Anhui province (grant no. 2012SQRL119), the natural
science research project in colleges and universities of Anhui
province (grant nos. KJ2016A734, KJ2016SD59 and KJ2017A256), the
National Natural Science Foundation of China (grant nos. 81601380,
81802651 and 31372198), and the Outstanding Young Talent Support
Program Key Projects in Anhui Colleges and Universities (grant no.
gxyqZD2016173).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
LN and YaZ were involved in the study concept and
design, and analysis and interpretation of data; LY, JM, JZ, and YW
were involved in acquisition of data, analysis and interpretation
of data, statistical analysis, and drafting of the manuscript; YuZ
performed bioinformatics analysis; ZW and YiZ performed the
luciferase reporter assay; LC and LX performed RNA isolation and
RT-qPCR; LL and SL performed the cell proliferation assay and
western blotting; ZQ and LX revised the manuscript, and performed
analysis and interpretation of data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hung MS, Chen IC, You L, Jablons DM, Li
YC, Mao JH, Xu Z, Hsieh MJ, Lin YC, Yang CT, et al: Knockdown of
Cul4A increases chemosensitivity to gemcitabine through
upregulation of TGFBI in lung cancer cells. Oncol Rep.
34:3187–3195. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim JE, Kim SJ, Lee BH, Park RW, Kim KS
and Kim IS: Identification of motifs for cell adhesion within the
repeated domains of transforming growth factor-beta-induced gene,
betaig-h3. J Biol Chem. 275:30907–30915. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Y, Wen G, Shao G, Wang C, Lin C,
Fang H, Balajee AS, Bhagat G, Hei TK and Zhao Y: TGFBI deficiency
predisposes mice to spontaneous tumor development. Cancer Res.
69:37–44. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao Y, El-Gabry M and Hei TK: Loss of
Betaig-h3 protein is frequent in primary lung carcinoma and related
to tumorigenic phenotype in lung cancer cells. Mol Carcinog.
45:84–92. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wen G, Partridge MA, Li B, Hong M, Liao W,
Cheng SK, Zhao Y, Calaf GM, Liu T, Zhou J, et al: TGFBI expression
reduces in vitro and in vivo metastatic potential of lung and
breast tumor cells. Cancer Lett. 308:23–32. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ahmed AA, Mills AD, Ibrahim AE, Temple J,
Blenkiron C, Vias M, Massie CE, Iyer NG, McGeoch A, Crawford R, et
al: The extracellular matrix protein TGFBI induces microtubule
stabilization and sensitizes ovarian cancers to paclitaxel. Cancer
Cell. 12:514–527. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li B, Wen G, Zhao Y, Tong J and Hei TK:
The role of TGFBI in mesothelioma and breast cancer: Association
with tumor suppression. BMC Cancer. 12:2392012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Irigoyen M, Pajares MJ, Agorreta J,
Ponz-Sarvisé M, Salvo E, Lozano MD, Pio R, Gil-Bazo I and Rouzaut
A: TGFBI expression is associated with a better response to
chemotherapy in NSCLC. Mol Cancer. 9:1302010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao Y, Shao G, Piao CQ, Berenguer J and
Hei TK: Down-regulation of Betaig-h3 gene is involved in the
tumorigenesis in human bronchial epithelial cells induced by
heavy-ion radiation. Radiat Res. 162:655–659. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shao G, Berenguer J, Borczuk AC, Powell
CA, Hei TK and Zhao Y: Epigenetic inactivation of Betaig-h3 gene in
human cancer cells. Cancer Res. 66:4566–4573. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shimono Y, Zabala M, Cho RW, Lobo N,
Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al:
Downregulation of miRNA-200c links breast cancer stem cells with
normal stem cells. Cell. 138:592–603. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu X, Zhang M, Miao J, Wang X and Huang C:
miRNA-4317 suppresses human gastric cancer cell proliferation by
targeting ZNF322. Cell Biol Int. 42:923–930. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mei Q, Li X, Guo M, Fu X and Han W: The
miRNA network: Micro-regulator of cell signaling in cancer. Expert
Rev Anticancer Ther. 14:1515–1527. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao X, Zhao H, Diao C, Wang X, Xie Y, Liu
Y, Han J and Zhang M: miR-455-3p serves as prognostic factor and
regulates the proliferation and migration of non-small cell lung
cancer through targeting HOXB5. Biochem Biophys Res Commun.
495:1074–1080. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ye MF, Zhang JG, Guo TX and Pan XJ:
MiR-504 inhibits cell proliferation and invasion by targeting LOXL2
in non small cell lung cancer. Biomed Pharmacother. 97:1289–1295.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Z, Liu Z, Fang X and Yang H:
MiR-142-5p suppresses tumorigenesis by targeting PIK3CA in
non-small cell lung cancer. Cell Physiol Biochem. 43:2505–2515.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li J, Tang Z, Wang H, Wu W, Zhou F, Ke H,
Lu W, Zhang S, Zhang Y, Yang S, et al: CXCL6 promotes non-small
cell lung cancer cell survival and metastasis via down-regulation
of miR-515-5p. Biomed Pharmacother. 97:1182–1188. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Y, Sheng B, Xia Q, Guan X and Zhang
Y: Association of long non-coding RNA H19 and microRNA-21
expression with the biological features and prognosis of non-small
cell lung cancer. Cancer Gene The. 24:317–324. 2017. View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen X, Wang K, Chen J, Guo J, Yin Y, Cai
X, Guo X, Wang G, Yang R, Zhu L, et al: In vitro evidence suggests
that miR-133a-mediated regulation of uncoupling protein 2 (UCP2) is
an indispensable step in myogenic differentiation. J Biol Chem.
284:5362–5369. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu Y, Liu R, Yang F, Cheng R, Chen X, Cui
S, Gu Y, Sun W, You C, Liu Z, et al: miR-19a promotes colorectal
cancer proliferation and migration by targeting TIA1. Mol Cancer.
16:532017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mazière P and Enright AJ: Prediction of
microRNA targets. Drug Discov Today. 12:452–458. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ivanov SV, Ivanova AV, Salnikow K,
Timofeeva O, Subramaniam M and Lerman MI: Two novel VHL targets,
TGFBI (BIGH3) and its transactivator KLF10, are up-regulated in
renal clear cell carcinoma and other tumors. Biochem Biophys Res
Commun. 370:536–540. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng
R, Wang Y, Huang J, Xu M, Yan J and Yu J: lncRNA H19/miR-675 axis
represses prostate cancer metastasis by targeting TGFBI. FEBS J.
281:3766–3775. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Choi SI, Jin JY, Maeng YS, Kim TI and Kim
EK: TGF-beta regulates TGFBIp expression in corneal fibroblasts via
miR-21, miR-181a, and Smad signaling. Biochem Biophys Res Commun.
472:150–155. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ,
Wang YK, Zeng F, Zhou JH and Zhang YK: High expression of serum
miR-21 and tumor miR-200c associated with poor prognosis in
patients with lung cancer. Med Oncol. 29:618–626. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ruvkun G: Clarifications on miRNA and
cancer. Science. 311:36–37. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Re M, Magliulo G, Gioacchini FM,
Bajraktari A, Bertini A, Ceka A, Rubini C, Ferrante L, Procopio AD
and Olivieri F: Expression levels and clinical significance of
miR-21-5p, miR-let-7a and miR-34c-5p in laryngeal squamous cell
carcinoma. Biomed Res Int. 2017:39212582017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chan SH, Wu CW, Li AF, Chi CW and Lin WC:
miR-21 microRNA expression in human gastric carcinomas and its
clinical association. Anticancer Res. 28:907–911. 2008.PubMed/NCBI
|
|
34
|
Zhu M, Huang Z, Zhu D, Zhou X, Shan X, Qi
LW, Wu L, Cheng W, Zhu J, Zhang L, et al: A panel of microRNA
signature in serum for colorectal cancer diagnosis. Oncotarget.
8:17081–17091. 2017.PubMed/NCBI
|
|
35
|
Huo D, Clayton WM, Yoshimatsu TF, Chen J
and Olopade OI: Identification of a circulating microRNA signature
to distinguish recurrence in breast cancer patients. Oncotarget.
7:55231–55248. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang K, Chen M and Wu W: Analysis of
microRNA (miRNA) expression profiles reveals 11 key biomarkers
associated with non-small cell lung cancer. World J Surg Oncol.
15:1752017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang JJ, Liu WQ, Peng JJ, Ma QL, Peng J
and Luo XJ: miR-21-5p/203a-3p promote ox-LDL-induced endothelial
cell senescence through down-regulation of mitochondrial fission
protein Drp1. Mech Ageing Dev. 164:8–19. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang R and Xia T: Long non-coding RNA
XIST regulates PDCD4 expression by interacting with miR-21-5p and
inhibits osteosarcoma cell growth and metastasis. Int J Oncol.
51:1460–1470. 2017. View Article : Google Scholar : PubMed/NCBI
|