1. Cancer and wound healing - similarities
in cellular physiology and pathology
Cellular migration is fundamental to both wound
healing in health and cancer metastasis in disease. What separates
these two diametrically opposed processes is the control and
termination of this movement. When cellular migration is absent,
wounded edges are stationary, resulting in chronic wounds; when
migration is overactive, abnormal scarring results. In cancer,
excessive cellular motility predisposes to metastasis, which is the
cause of 90% of human cancer deaths (1,2).
Epithelial-mesenchymal transition (EMT) refers to the morphological
changes observed during embryogenesis and foetal development,
cutaneous wound repair and neoplastic invasion and metastases
(3,4). In response to wounding,
keratinocytes at the wound edge are transformed from a sedentary to
a migratory phenotype. They lose their cell-cell and
cell-extracellular matrix (ECM) attachments, rearrange their actin
cytoskeleton and express a variety of mesenchymal markers (such as
vimentin) (5). However, in
contrast to cancer progression, the changes observed in these wound
edge keratinocytes are short lived and reversible (4). Maintaining this ‘Goldilocks’ type
requirement of migration (not too much, not too little) is
essential for health, and understanding which factors regulate such
a process is of interest both to cancer biologists and wound
healing scientists.
Historically, it was in the context of excessive
inflammation, rather than excessive cellular migration, that the
cross-over between cancer and wound healing was first recognised.
As early as 1863, Virchow observed that excessive inflammation
predisposed to cancer formation (6). Over 100 years later, Dolberg et
al used an ingenious animal model to demonstrate this link
(7). Chickens injected with the
Rous sarcoma virus were observed to develop cancer not only at the
site of injection, but also at distant body sites which were
wounded. Furthermore, the application of an anti-inflammatory
therapy inhibited cancer generation in these distant wounds
(8). In his landmark study,
Dvorak succinctly suggested that tumours represent ‘wounds that do
not heal’ (9).
Currently, the link between chronic inflammatory
states predisposing to cancer formation is well established
(Table I). Other links between
wound healing and cancer include cell growth, angiogenesis and the
formation of fibrous tissue/ECM (10). These processes are appropriately
regulated during physiological wound healing, whilst being
uncontrolled in cancer. Of interest to our group is the regulation
of cellular movements, in the context of both cancer metastasis and
wound healing. During our search for key molecular markers involved
in wound healing, we noted, as have other authors (11), marked similarities between
molecular effectors of cancer and wound healing. Whilst this
correlation is not absolute, there is a sufficient overlap to
warrant further attention to this association (12). For example, Pedersen et al
performed microarray analysis on keratinocytes during wound healing
and compared this to keratinocytes from squamous cell carcinomas
(SCCs) and normal tissue (13).
They noted that keratinocytes from the wound edge transiently mimic
their SCC transformed counterparts, with the most notable exception
between the groups in gene expression profile relating to control
of growth. Thus, keratinocytes captured whilst undergoing healing
would share a number of similarities with their neoplastic
counterparts. However, this overlap is not just confined to
keratinocytes; fibroblasts also show remarkable parallels with
their response to both wound healing and cancer progression. Chang
et al cultured fibroblasts in serum, recreating a condition
found only in the context of wound healing (14). They analysed the resultant gene
expression profile, and showed a similar profile alteration in
tumour-associated fibroblasts or tumour cells. Furthermore, the
continued expression of this ‘wound healing’ genotype in breast,
lung and gastric carcinomas predicts an increase in metastasis and
death.
 | Table IInflammatory conditions known to
predispose to malignant transformation. |
Table I
Inflammatory conditions known to
predispose to malignant transformation.
| Pre-malignant
inflammatory condition | Cancer-associated
outcome | (Refs.) |
|---|
| Barrett’s
oesophagus | Oesophageal
cancer | (101) |
| Hepatic
cirrhosis | Hepatic cancer | (102) |
| Gastric
ulceration | Gastric cancer | (103) |
| Inflammatory bowel
disease (ulcerative colitis) | Colorectal
cancer | (104) |
| Chronic
pancreatitis | Pancreatic
cancer | (105) |
| Burn wound | Squamous cell
carcinoma (SCC, Marjolin’s ulcer) | (106) |
2. Cellular migration and wound healing
Common to all tissue repair processes is the
migration of cells required for healing into the wound space,
including endothelial cells (15), fibroblasts (16) and epithelial cells (17). Cells migrate through a process of
‘crawling’ motility, consisting of four well conserved steps:
protrusion of the leading edge, adhesion to the substratum,
de-adhesion and retraction of the rear (18). For directional movement, cells
acquire a polarised morphology (19), responding to chemokines
(chemotactic cytokines) or other extracellular signals. At the cell
front (the ‘leading edge’), actin assembles both lamellipodia and
filopodia. It is these lamellipodia that adhere to the substratum
and provide an anchor from which a force can be generated (20). Key to both lamellipodia and
filopodia production is the assembly and protrusion of actin
(20). Once a leading edge has
been created, actin becomes polarised with a forward ‘barbed’ end
and a backward ‘pointed’ end (both named due to myosin decoration
of actin forming an arrow-head type pattern), and ATP fuels the
disassembly of posterior actin filaments, and the assembly of
anterior ones (21). Thus actin
moves forward relative to the cell.
In order for these actin movements to be converted
into cellular locomotion, there needs to exist a link between the
cytoskeleton and the cellular membrane. The protein 4.1R, ezrin,
radixin, moesin (FERM) family of proteins, so named as an acronym
of its founding four members (protein 4.1R, ezrin, radixin and
moesin), is a well described family involved in
membrane-cytoskeletal interactions. We have already noted the
important role FERM family proteins play in cancer progression
(22). Herein we update and
summarise these pertinent findings, and compare the effects of this
protein family on wound healing and tissue repair.
3. The FERM protein superfamily
Members of the FERM superfamily are characterised by
the presence of a conserved FERM domain at the N-terminus of the
molecule, and often a spectrin/actin binding domain (SABD) at the
C-terminus (23). Protein 4.1R,
originally referred to as erythrocyte band protein 4.1, was
identified as an abundant protein in human erythrocyte ghosts,
localising to the cytoskeleton and stabilising its shape, and the
first of the FERM proteins to be identified (24–26). By 1991 three more FERM proteins
were discovered; ezrin (27),
radixin (28) and moesin
(29) (also known as ERM
proteins). Prior to the now commonly accepted title, the FERM
domain has been known by a variety of names, including the 30 kDa
domain, 4.1N30, the membrane-cytoskeletal-linking
domain, the ERM like domain, the ezrin like domain of the band 4.1
superfamily, the conserved N-terminal region, the membrane
attachment domain (23) and the
talin, ezrin, radixin, moesin (TERM) domain (30).
The FERM family has grown considerably since this
initial discovery of protein 4.1R, with more than 40 members
identified to date (31)
(Fig. 1). On the basis of protein
sequence similarity, this superfamily was divided in 1994 into five
subgroups: band 4.1 proteins, ERM-related proteins, talin-related
molecules, protein tyrosine phosphatase (PTP) proteins and novel
band 4.1-like 4 (NBL4) proteins (32).
 | Figure 1Phylogenic tree of the protein 4.1R,
ezrin, radixin, moesin (FERM) family proteins. Phylogenetic
analyses was performed using the ClustalW (http://www.ebi.ac.uk/clustalw/), and the dendrogram
tree was drawn by using the Treeview (version 1.6.6, http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
The bar scale shows the identity shared by the proteins in their
amino acid sequences. NBL4, novel band 4.1-like protein 4; FRMD,
FERM domain-containing protein; Ehm2, expressed in high metastatic
cells 2; FARP, FERM RhoGEF and pleckstrin domain-containing
protein; MYLIP, myosin regulatory light chain interacting protein;
PTPN, protein-tyrosine phosphatase non-receptor-type; FRMPD, FERM
and PDZ domain-containing protein; FAK, focal adhesion kinase; JAK,
janus kinase protein; TYK, non-receptor tyrosine-protein kinase;
FERMT, fermitin family homolog; KRIT, Krev interaction trapped
protein; PLEKHH, pleckstrin homology domain-containing family H
member. |
4. The FERM domain
The FERM domain is a 30 kDa, N-terminal, cysteine
rich, basically charged, globular molecule, initially noted as it
was resistant to mild chymotrypsin treatment (24). It is hydrophobic and contains
numerous β-sheet structures, suggesting a tight, compact structure
which may account for its resistance to proteolytic degradation
(33). The FERM domain has been
shown to consist of three lobes, the F1, F2 and F3 domains (also
known as FERM-N, FERM-M and FERM-C subdomains), in a number of FERM
proteins (34,35). Pearson et al suggested
that, although the FERM domain is composed of three smaller
domains, these three function together as a single unit and are
structurally rigid upon activation (36). The F1 and F2 domains appear to be
the most highly conserved amongst the FERM domain proteins, whilst
F3 encodes for more specific protein binding sites (34,36). The N-terminal F1 lobe is
structurally related to ubiquitin, the central F2 lobe is folded
like an acyl-CoA binding protein and the C-terminal F3 subdomain is
similar to a pleckstrin homology (PH)/phosphotyrosine-binding (PTB)
domain (36). The FERM domain can
bind a variety of proteins, lipids and molecules (Table II). Inactive FERM proteins are
retained in the cytoplasm in a closed conformation, which masks
transmembrane protein binding sites in both the FERM domain and the
C-terminal region (36).
Activation of the protein results in weakening of this interaction
and subsequent exposure of the FERM domain and active C-terminal
region (Fig. 2). For example,
when phosphatidylinositol-4,5-bisphosphate (PIP2) binds to the FERM
domain of ezrin, its conserved C-terminal at Thr567
becomes phosphorylated, thus opening and activating the protein
(37). Ezrin can then translocate
to the membrane/cytoskeleton interface, change its conformation,
and undertake binding with various partners.
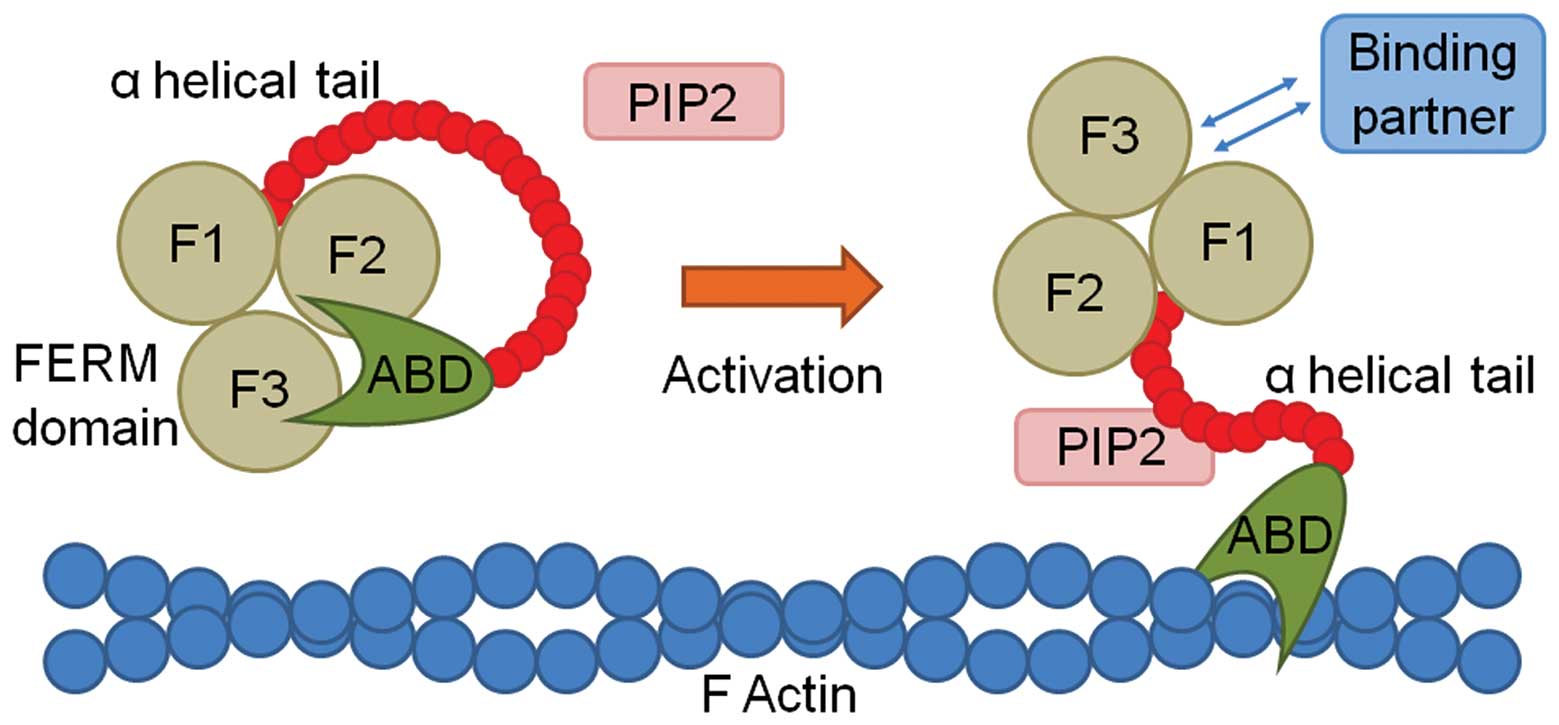 | Figure 2Schematic diagram demonstrating the
activation and binding of protein 4.1R, ezrin, radixin, moesin
(FERM) family proteins. The coiled FERM protein (left) is composed
of three lobes (F1, F2 and F3), an α helical tail, and an actin
binding domain (ABD), which is maintained in a closed conformation,
and thus does not allow binding. On activation, in this instance by
phosphatidyl inositol-4,5-bisphosphate (PIP2), the FERM protein
opens, allowing the ABD to bind to F actin, and the FERM domain to
bind to one of its various binding partners. |
 | Table IIThe various binding partners of the
N-terminal FERM domain. |
Table II
The various binding partners of the
N-terminal FERM domain.
| Class of FERM
domain binding partner | Specific binding
partner | (Refs.) |
|---|
| Membrane
proteins | Band 3 protein | (107) |
| Glycophorin C and
D | (108,109) |
|
Phosphoproteins | p55 | (107,109) |
| ERM-binding
phosphoprotein 50 (EBP50) | (110) |
| Adhesion
molecules | CD44 | (111) |
| Inter-cellular
adhesion molecule 1 (ICAM1) | (112) |
| Inter-cellular
adhesion molecule 2 (ICAM2) | (112) |
|
Na+/H+ exchanger
(NHE1) | (113) |
| Membrane
lipids | PIP2 | (114) |
| Cytoplasmic
proteins | Calmodulin | (115) |
| Other proteins |
Na+/H+ exchanger
regulatory factor (NHERF) | (116) |
Certain FERM proteins have been well characterised
with regards to human disease, cancer progression and wound
healing. The following sections review the effects of certain key
FERM proteins, ezrin, radixin, moesin, protein 4.1R and the novel
protein expressed in highly metastatic cells 2 (Ehm2) in human
disease and wound healing.
5. ERM proteins - background
Ezrin (cytovillin/p81/80k/villin-2), initially
discovered in 1983 (38) as an 81
kDa component of intestinal microvilli, is the most well
characterised of these proteins and considered by some to be the
prototype member of the ERM protein family due to its conserved
wide spread distribution and highly homologous sequences of the
FERM domain. Although ezrin (along with radixin and moesin) is
expressed in most cultured cells, it has been shown to be highly
concentrated in the gastrointestinal tract, lungs and kidneys, and
is expressed in epithelial and mesothelial cells (39–41). Ezrin is localised to the
juxta-membrane region and to soluble cytosolic pools. The dynamic
localization between these two compartments is important in
determining ezrin activity and is responsible for its cellular
action (42,43). Ezrin is involved in a myriad of
cellular functions, such as cell adhesion, motility, apoptosis and
phagocytosis.
Moesin (membrane-organising extension spike protein)
was initially isolated from bovine uterus and characterised as a
possible heparin receptor (29,44). Moesin is concentrated in
specialised microdomains, namely in the intracellular core of
microextensions, such as filopodia, microvilli, microspikes and
retraction fibers (45). Similar
to ezrin, moesin is important in cellular motility and migration.
Moesin is responsible for the formation of cortical actin complexes
when activated (following phosphorylation of Thr558),
and inducing actin depolymerisation and reassembly toward the cell
membrane edge (46). In the
context of cervical cancer, moesin-associated cellular motility has
been shown to be upregulated by vascular endothelial growth factor
C (VEGF-C) and mediated by the RhoA/ROCK-2 signalling pathway
(47).
In 1989, radixin, an 82-kD protein, was purified
from a rat liver and shown to be a barbed end-capping protein
associated with the undercoat of the cell-to-cell adherens junction
(28). Subsequently, it was shown
to be predominantly located to the hepatocyte microvilli (48). It shows high (>84%) levels of
sequence homology with ezrin and moesin (49). Radixin has been shown to interact
with the GABAARα5 subunit, a component of an inhibitory
neurotransmitter receptor in neurons, at extra-synaptic sites
(50). Thus, radixin may be
indirectly important in modulating neuronal network activity.
6. ERM proteins and cancer
Over the years, more data has become available
outlining the role ERM proteins play in cancer biology. Ezrin is
the most studied protein in this context, whilst moderate work has
been performed on moesin and very little on radixin. As FERM family
members, and consistent with their role as membrane cytoskeletal
linkers, ERM proteins typically promote cancer progression and
metastasis (22). For example,
the overexpression of ezrin results in the greater metastatic
potential of osteosarcoma, hepatocellular carcinoma, breast cancer,
lung cancer and pancreatic carcinoma (51–56). In the context of metastatic
melanoma, ezrin deletion reduces important interactions with CD44,
merlin and Lamp-1, and thus their recruitment (57). The resultant aberrant engagement
with the surrounding microenvironment reduces cellular movements,
thereby reducing the metastatic potential of these cells. When
ezrin was knocked down in the CCR9-expressing acute T lymphocytic
cell line, MOLT4, cell polarisation disappeared. Furthermore,
E-cadherin translocation from the cytoplasm to the cell membrane
was reduced, as was the invasive ability of the cells (58).
Ezrin also interacts with other proteins through
which it has an effect on cancer. For example, the
cytoskeletal-related protein, neurofibromatosis type 2 (NF2), acts
as a cancer suppressor in the context of glioblastomas. However,
ezrin has been shown to reduce NF2 expression via aberrant
intracellular recruitment and thus intermolecular association. The
resulting reduced levels of NF2 are observed in approximately
one-third of glioblastoma cell lines and tumours (59).
Moesin has also been shown to promote metastasis in
certain types of cancer. It has already been mentioned that moesin
promotes cervical cancer metastasis through the RhoA/ROCK pathway
(47). Greater levels of moesin
(and also radixin) were found in pancreatic tumours which had
metastasised to regional lymph nodes, compared to non-metastasising
cancers (60). The knockdown of
moesin using siRNA reduces the invasion of melanoma cells into 3D
matrices, in part due to a loss of cellular polarisation (61).
7. ERM proteins and wound healing
Active ERM proteins have been examined using a
phospho-ERM antibody, with affinity for ezrin, radixin and moesin,
by a number of authors. For example, Jensen and Larsson observed
that both CD44 and ERM proteins were co-localised to luminal
F-actin domains in endothelial cells in vitro and in
vivo (62). After wounding of
the confluent monolayer of cells in vitro, focal F-actin
branching points appeared, with CD44 and ERM proteins again
co-localised to this area, and associated with phosphorylated
protein kinase (PK)C. The inhibition of PKC activity resulted in
inhibited wound healing with reduced ERM-F-actin interaction and
associated F-actin branching points. The authors concluded that ERM
proteins, through the stimulation of PKC, play an important role in
endothelial migration and thus, healing. These data compliment the
work by Ng et al who showed that ERM protein phosphorylation
by PKC improved in vitro wound healing in MCF7 breast cancer
cells (63).
It is possible to study neuronal regeneration
following injury in vitro using a neuron transection model.
Haas et al initially showed that phosphorylated ERM proteins
were localised to neuronal processes that formed several hours
after neural transection, but were notably absent from relatively
mature and stable cultured cells (64). They subsequently showed that the
activation of ERM proteins was dependant on Rho kinase, with Rho
kinase inhibition reducing phosphorylated ERM levels and subsequent
response to wounding (65).
Tsuda et al showed that the signalling
adaptor protein Crk associates with ERM proteins and enhances cell
motility in 293T and 3Y1 (fibroblast) cells towards hyaluronic acid
(66). Crk results in the
translocation of ERM proteins to microvilli in the 3Y1 cell line,
although this function was, as with the neuron transection model
above, suppressed by Rho kinase. The authors concluded that Crk
promotes cellular migration and motility in fibroblasts through its
effects on ERM proteins.
Crepaldi et al utilised a kidney-derived
epithelial cell line, LLC-PK1, to analyse the effect of ezrin
overexpression on hepatocyte growth factor (HGF)-induced renal
wound healing and epithelial migration (67). Cells overexpressing ezrin
demonstrated enhanced migration and tubulogenesis in response to
HGF compared to both a wild-type (WT) control, and a control
overexpressing a truncated variant of ezrin, and thus concluded
that ezrin promotes renal wound healing. The effects of moesin and
lung injury were investigated using moesin knockout mice (68). These mice were compared to WT
mice, and both had intra-tracheal bleomycin to simulate widespread
lung injury. Moesin knockout mice developed more pronounced lung
injury and fibrosis, following an abnormal cytokine and chemokine
response, and had lower survival rates when compared to WT
mice.
Moesin knockout mice were also used for
investigating the role of moesin in hepatic injury (69). Hepatic stellate cells (HSCs)
undergo activation in response to injury, and are important in
hepatic wound healing and fibrosis. Response to hepatic wounding,
caused by both common bile duct ligation and focal thermal
denaturation, were compared in moesin knockout mice and WT
controls. In moesin knockout mice inflammatory infiltrate, fibrosis
and collagen deposition at the injury margin was markedly reduced
compared to the WT mice. In vitro migration assay of moesin
knockout HSCs demonstrated a significant reduction in migration
compared to WT controls.
8. Protein 4.1R - background
Protein 4.1R is a 80 kDa protein, initially found
localised to the cytoskeleton on human erythrocytes, which binds to
a variety of other proteins (adducin, tropomyosin, tropomodulin,
dematin and protein p55 (70) and
together stabilises the erythrocyte characteristic biconcave disc
shape (71). Protein 4.1R
consists of four structural domains (72); an N-terminal FERM domain, a 16 kDa
domain of unknown function, SABD and a 22 kDa C-terminal domain
which has been shown to interact with some proteins that form tight
junctions, such as occludin, zonula occludens (ZO)-1 and ZO-2
(73). It is the most studied
member of the protein 4.1 family, which consists of protein 4.1R
(red blood cells), 4.1G (general), 4.1N (neuronal) and 4.1B (brain)
(74).
A decrease in protein 4.1R expression due to
chromosomal deletion results in hereditary elliptocytosis, which is
characterised by abnormally shaped erythrocytes, haemolysis and
splenomegaly (75). Abnormal
erythrocyte architecture has been documented in protein 4.1R null
mice (76). A similar model
suggested that protein 4.1R deficiency results not only in the
diminution of actin, but also in a reduction of various
transmembrane proteins, including glycophorin C, XK, Duffy and Rh,
and results in the conformational change of band 3 and Kell
epitopes (77). Protein 4.1R can
also be deactivated by different methods. For example, in the
presence of calcium, calmodulin decreases the affinity of protein
4.1R for the spectrin-actin complex, thus reducing the mechanical
stability of the cellular membrane (78). Furthermore, protein 4.1R can be
phosphorylated by a number of agents, including casein kinase,
tyrosine kinase, PKA and PKC (79,80). This phosphorylation reduces its
ability to associate with spectrin approximately five-fold, with an
increase in the relaxation of the cytoskeleton and associated
cellular flexibility (81).
9. Protein 4.1R in human disease and wound
healing
Although initially found to localise to human
erythrocytes, it subsequently became clear that protein 4.1R is
expressed in a variety of nucleated cells, is located in several
different subcellular compartments, and is required for other
diverse functions (82–85). Protein 4.1R has been shown to link
with β-catenin in gastric epithelial cells, and such cells lacking
protein 4.1R show impaired cell-cell contacts and disordered
gastric glands (86).
The role of protein 4.1R in wound healing was
recently investigated by Chen et al using protein
4.1R−/− mice (and their cultured keratinocytes)
(87). Protein 4.1R deficient
mice displayed delayed dermal wound healing. In vitro,
protein 4.1R was shown to be present in the cytoplasm and the
leading edge of moving cells. The absence of protein 4.1R resulted
in reduced adhesion, spreading, migration and motility of
keratinocytes. Focal adhesion complexes failed to localise in the
absence of protein 4.1R. Furthermore, β1 integrin levels were
reduced, and shown to directly link to this protein. Of note,
protein 4.1R deficient keratinocytes showed greater levels of 4.1G
and 4.1N, without any change in 4.1B levels, suggesting that these
former two proteins compensate in part for the absence of protein
4.1R (87). Protein 4.1R has also
shown to be important in the migration of endothelial cells.
Ruiz-Sáenz et al showed that protein 4.1R interacts with Ras
GTPase-activating-like protein 1 (IQGAP1), which acts as a
scaffolding protein near the cell cortex at the leading edge of
migrating cells, and both binds and cross-links actin filaments
(88). The authors demonstrated
that protein 4.1R was recruited to the leading edge of migrating
cells, where it recruits and binds IQGAP1 through its FERM domain.
The lack of protein 4.1R significantly slowed cellular
migration.
10. Ehm2 - background
In 1996, Hashimoto et al identified four
novel proteins in both high and low metastatic murine melanoma
cells. Ehm2 was the second novel protein to be found in the highly
metastatic (and notably absent in the low metastatic) K-1735 murine
melanoma cell line, with an mRNA size of 4.0kb (89). By the year 2000, the same team had
cloned Ehm2 (90). The authors
demonstrated that the Ehm2 gene encodes a protein with 527 amino
acids, with a 41% amino acid identity with the FERM domain. Further
analysis revealed that Ehm2 showed the greatest homology with NBL4,
and was thus classified in the NBL4 subfamily. A human homologue,
with significant homology (83% identity, mapped to chromosome 9)
was identified, along with seven rat clones and one pig clone,
again with significant homology (83–92%), suggesting that Ehm2 is a
highly conserved gene (90).
In 2003, Chauhan et al characterised Ehm2 in
a human fibrosarcoma cell line model (HT-AR1 cells) of
steroid-induced cytoskeletal reorganisation, and demonstrated that
Ehm2 was androgen (dihydrotestosterone)-, but not
dexamethasone-regulated (91).
They analysed Ehm2 mRNA levels in a variety of human tissues, and
found the expression of a 3.8 kb transcript (isoform 1), which was
expressed in greater levels in the testis, with a lower expression
in prostate and breast tissue. An additional 2.3 kb Ehm2 transcript
was also found in these three tissues, with their transcripts only
differing in the length of the 3′ untranslated region (3′UTR). In
addition, the human brain contained high levels of a 5.6 kb Ehm2
transcript, encoding a protein of 913 amino acids (isoform 2), with
an additional 409 amino acids of unknown function. Further analysis
of the Ehm2 protein revealed that it appeared to encode only an
extended FERM domain located 70 amino acids from the N-terminus.
Unlike other FERM domain proteins, the C terminus did not appear to
encode any additional protein function. The authors subsequently
hypothesised that Ehm2 either encodes a novel activity, or
possibly, functions as a dominant negative modulator of other FERM
proteins (92). Phylogenetic tree
analysis revealed that Ehm2 belongs to a subfamily consisting of at
least nine members from seven different species, including the
Drosophilia Yurt gene (93).
11. Ehm2 in human disease and wound
healing
Wang et al investigated the role of Ehm2 in
prostate cancer. Ehm2 mRNA and protein levels were higher in
immortalised prostate cancer cell lines (LNCaP, PC-3 and LAPC4)
compared with the immortalised prostate epithelial cell line
(PNT1a) (94). Similarly, Ehm2
mRNA levels, as analysed by qRT-PCR, were significantly greater in
prostate cancer tissues compared to benign controls.
Immunohistochemical (IHC) analysis confirmed a significantly
greater expression of Ehm2 protein in prostate cancer tissues
compared with benign controls, with cancers associated with PSA
recurrence following radical prostatectomy having a significantly
greater staining compared to those without recurrence. Using
transient transfection and siRNA knockdown in PNT1 and LAPC4 cells
respectively, the authors demonstrated that increased Ehm2 levels
were associated with decreased adhesion to collagen IV. Schulz
et al subsequently confirmed that Ehm2 mRNA levels were
significantly increased in cancerous compared to non-cancerous
prostate samples, and also showed that higher levels of Ehm2
expression (mRNA) significantly increased the likelihood of
biochemical recurrence (95).
Other authors have used microarray technology to show that Ehm2
transcript levels are greater in neoplastic prostate samples
(96–98).
Yu et al recently demonstrated that elevated
levels of Ehm2 mRNA and protein were found in breast cancer tissues
compared to controls, analysed with qRT-PCR and IHC (99). Higher levels were observed in
node-positive disease (vs. node-negative), more advanced disease
[TNM (tumour, node, metastasis)-4 disease vs. TNM1-3], higher
levels of the Nottingham prognostic index (a marker of prognosis)
and evidence of metastasis. There was a significant reduction in
disease-free survival in patients with high levels of Ehm2. In
vivo data using MCF7 Ehm2 knockdown cells showed Ehm2 enhanced
growth and invasion; however, there was no effect on motility or
adhesion. Ehm2 knockdown was associated with the reduced mRNA and
protein expression of MMP9.
We recently demonstrated that Ehm2 is upregulated in
samples obtained from the periphery of acute healing wounds
compared to chronic non-healing wounds (100). Furthermore, chronic wounds which
subsequently healed displayed a greater cellular staining for Ehm2
than those which failed to heal, which was noted particularly at
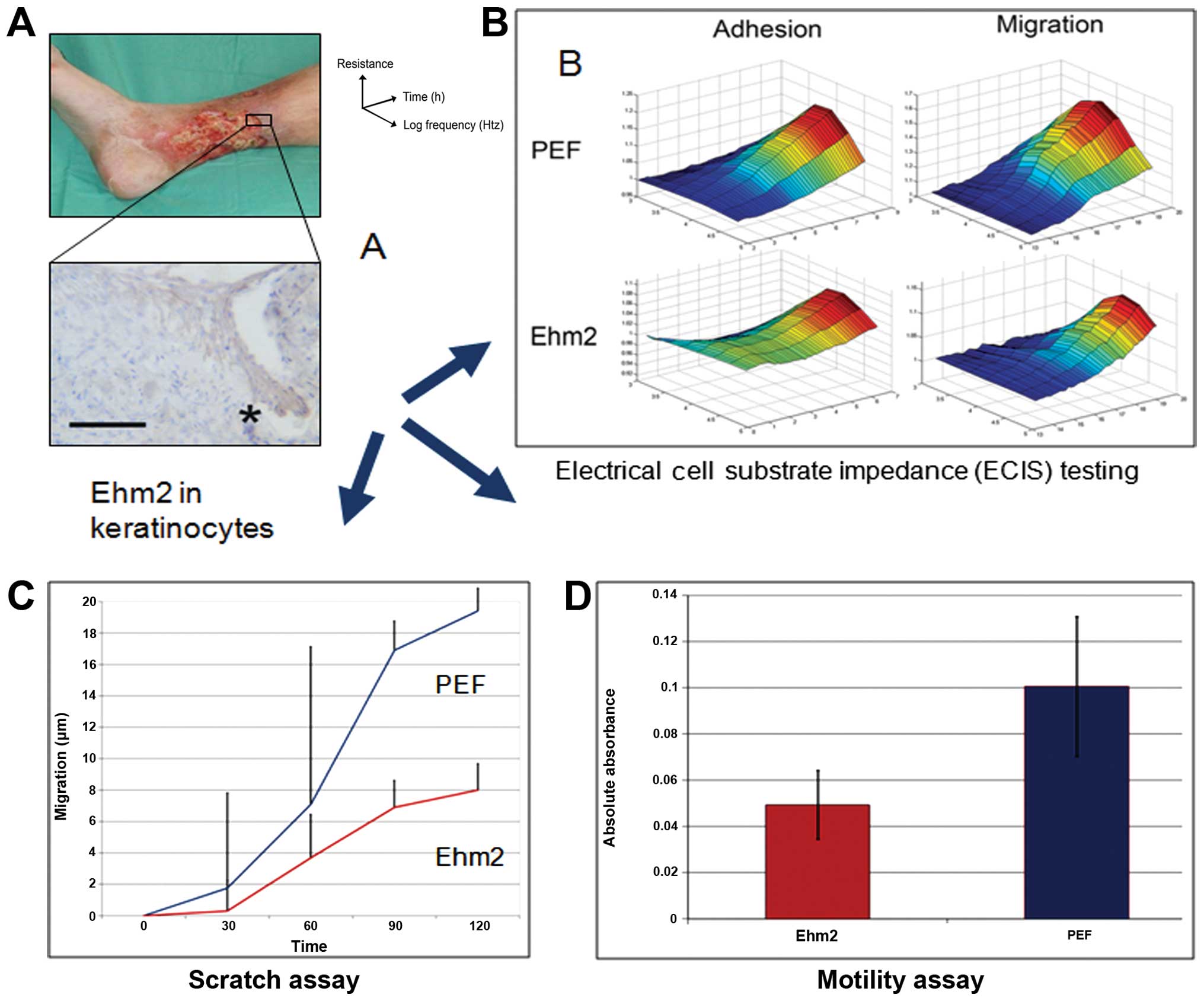
the leading keratinocyte edge of the healing wounds (Fig. 3A). In order to investigate the
mechanisms behind this observation of improved wound healing, we
created a stable Ehm2 knockdown in the HaCaT keratinocyte cell
line. In vitro experiments revealed that Ehm2 knockdown
slowed cellular adhesion, migration and motility (Fig. 3B–D), without affecting cellular
growth, apoptosis or cell cycle rates. These data suggest that Ehm2
acts as a pro-migratory protein, in common with other FERM family
proteins, and promotes wound healing through the process of
re-epithelialisation.
12. Conclusion
The migration of cells is essential not only for
wound healing, but also for embryonic development, neuronal
development and immune responses, and is also crucial for cancer
metastasis. This brief review highlights the role that certain FERM
proteins, the ERM proteins, protein 4.1R and Ehm2, play in wound
healing and other diseases. Although there is some heterogeneity
amongst this family, the FERM proteins generally promote the
migration of cells, such as keratinocytes and endothelial cells,
through their role as mediators between the cytoskeleton and the
cell membrane, linking to a large number of different receptors. It
is suggested that it is through these various mechanisms (i.e.,
re-epithelialisation and angiogenesis) that wound healing
occurs.
This positive wound healing effect has been noted
across a variety of models and in differing cell types. However,
whilst in vivo results can be suggestive of an important
role within wound healing, evidence corroborated with in
vivo tissue data, or animal models, are often lacking. Whilst
this does not necessarily detract from the conclusions, it
highlights the need for further studies to fully assess the
significance of FERM protein alterations in human tissue repair and
regeneration.
Acknowledgements
The authors wish to thank the Welsh Government (A4B
scheme) for supporting this study.
References
|
1
|
Weigelt B, Peterse JL and van ’t Veer LJ:
Breast cancer metastasis: markers and models. Nat Rev Cancer.
5:591–602. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mehlen P and Puisieux A: Metastasis: a
question of life or death. Nat Rev Cancer. 6:449–458. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Arnoux V, Come C, Kusewitt D, Hudson L and
Savagner P: Cutaneous Wound Reepithelializaton: A partial and
reversible EMT. Rise and Fall of Epithelial Phenotype: Concepts of
Epithelial-Mesenchymal Transition. Savagner P: Springer; Berlin:
pp. 111–134. 2005, View Article : Google Scholar
|
|
5
|
Yan CL, Grimm WA, Garner WL, et al:
Epithelial to mesenchymal transition in human skin wound healing is
induced by tumor necrosis factor-alpha through bone morphogenic
protein-2. Am J Pathol. 176:2247–2258. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Virchow R: Aetiologie der neoplastischen
Geschwulste/ Pathogenie der neoplastischen Geschwulste. Die
Krankhaften Geschwülste. Verlag von August Hirschwald; Berlin: pp.
57–101. 1863
|
|
7
|
Dolberg DS, Hollingsworth R, Hertle M and
Bissell MJ: Wounding and its role in RSV-mediated tumor formation.
Science. 230:676–678. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Martinsgreen M, Boudreau N and Bissell MJ:
Inflammation is responsible for the development of wound-induced
tumors in chickens infected with Rous sarcoma virus. Cancer Res.
54:4334–4341. 1994.PubMed/NCBI
|
|
9
|
Dvorak HF: Tumors: wounds that do not
heal. Similarities between tumor stroma generation and wound
healing. N Engl J Med. 315:1650–1659. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schafer M and Werner S: Cancer as an
overhealing wound: an old hypothesis revisited. Nat Rev Mol Cell
Bio. 9:628–638. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Antsiferova M and Werner S: The bright and
the dark sides of activin in wound healing and cancer. J Cell Sci.
125:3929–3937. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Grose R: Common ground in the
transcriptional profiles of wounds and tumors. Genome Biol.
5:2282004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pedersen TX, Leethanakul C, Patel V, et
al: Laser capture microdissection-based in vivo genomic profiling
of wound keratinocytes identifies similarities and differences to
squamous cell carcinoma. Oncogene. 22:3964–3976. 2003. View Article : Google Scholar
|
|
14
|
Chang HY, Sneddon JB, Alizadeh AA, et al:
Gene expression signature of fibroblast serum response predicts
human cancer progression: similarities between tumors and wounds.
PLoS Biol. 2:72004. View Article : Google Scholar
|
|
15
|
Eming SA, Brachvogel B, Odorisio T and
Koch M: Regulation of angiogenesis: wound healing as a model. Prog
Histochem Cytochem. 42:115–170. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Midwood KS, Williams LV and Schwarzbauer
JE: Tissue repair and the dynamics of the extracellular matrix. Int
J Biochem Cell Biol. 36:1031–1037. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Martin P: Wound healing--aiming for
perfect skin regeneration. Science. 276:75–81. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pollard TD and Borisy GG: Cellular
motility driven by assembly and disassembly of actin filaments.
Cell. 112:453–465. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mellman I and Nelson WJ: Coordinated
protein sorting, targeting and distribution in polarized cells. Nat
Rev Mol Cell Biol. 9:833–845. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Small JV, Stradal T, Vignal E and Rottner
K: The lamellipodium: where motility begins. Trends Cell Biol.
12:112–120. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bugyi B and Carlier MF: Control of actin
filament treadmilling in cell motility. Annu Rev Biophys.
39:449–470. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu H, Zhang Y, Ye L and Jiang WG: The FERM
family proteins in cancer invasion and metastasis. Frontiers in
bioscience: a journal and virtual library. 16:1536–1550. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chishti AH, Kim AC, Marfatia SM, et al:
The FERM domain: a unique module involved in the linkage of
cytoplasmic proteins to the membrane. Trends Biochem Sci.
23:281–282. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Leto TL and Marchesi VT: A structural
model of human erythrocyte protein 4.1. J Biol Chem. 259:4603–4608.
1984.PubMed/NCBI
|
|
25
|
Tyler JM, Hargreaves WR and Branton D:
Purification of two spectrin-binding proteins: biochemical and
electron microscopic evidence for site-specific reassociation
between spectrin and bands 2.1 and 4.1. Proc Natl Acad Sci USA.
76:5192–5196. 1979. View Article : Google Scholar
|
|
26
|
Shiffer KA and Goodman SR: Protein 4.1:
its association with the human erythrocyte membrane. Proc Natl Acad
Sci USA. 81:4404–4408. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bretscher A: Purification of the
intestinal microvillus cytoskeletal proteins villin, fimbrin, and
ezrin. Methods Enzymol. 134:24–37. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tsukita S and Hieda Y: A new 82-kD barbed
end-capping protein (radixin) localized in the cell-to-cell
adherens junction: purification and characterization. J Cell Biol.
108:2369–2382. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lankes WT and Furthmayr H: Moesin: a
member of the protein 4.1-talin-ezrin family of proteins. Proc Natl
Acad Sci USA. 88:8297–8301. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang WG, Hiscox S, Singhrao SK, et al:
Induction of tyrosine phosphorylation and translocation of ezrin by
hepatocyte growth factor/scatter factor. Biochem Biophys Res
Commun. 217:1062–1069. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun CX, Robb VA and Gutmann DH: Protein
4.1 tumor suppressors: getting a FERM grip on growth regulation. J
Cell Sci. 115:3991–4000. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Takeuchi K, Kawashima A, Nagafuchi A and
Tsukita S: Structural diversity of band 4.1 superfamily members. J
Cell Sci. 107:1921–1928. 1994.
|
|
33
|
Conboy J, Kan YW, Shohet SB and Mohandas
N: Molecular cloning of protein 4.1, a major structural element of
the human erythrocyte membrane skeleton. Proc Natl Acad Sci USA.
83:9512–9516. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Smith WJ, Nassar N, Bretscher A, Cerione
RA and Karplus PA: Structure of the active N-terminal domain of
Ezrin. Conformational and mobility changes identify keystone
interactions. J Biol Chem. 278:4949–4956. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shimizu T, Seto A, Maita N, Hamada K,
Tsukita S and Hakoshima T: Structural basis for neurofibromatosis
type 2. Crystal structure of the merlin FERM domain. J Biol Chem.
277:10332–10336. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pearson MA, Reczek D, Bretscher A and
Karplus PA: Structure of the ERM protein moesin reveals the FERM
domain fold masked by an extended actin binding tail domain. Cell.
101:259–270. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gautreau A, Louvard D and Arpin M: ERM
proteins and NF2 tumor suppressor: the Yin and Yang of cortical
actin organization and cell growth signaling. Curr Opin Cell Biol.
14:104–109. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bretscher A: Purification of an
80,000-dalton protein that is a component of the isolated
microvillus cytoskeleton, and its localization in nonmuscle cells.
J Cell Biol. 97:425–432. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Franck Z, Gary R and Bretscher A: Moesin,
like ezrin, colocalizes with actin in the cortical cytoskeleton in
cultured cells, but its expression is more variable. J Cell Sci.
105:219–231. 1993.PubMed/NCBI
|
|
40
|
Sato N, Funayama N, Nagafuchi A, Yonemura
S and Tsukita S and Tsukita S: A gene family consisting of ezrin,
radixin and moesin. Its specific localization at actin
filament/plasma membrane association sites. J Cell Sci.
103:131–143. 1992.PubMed/NCBI
|
|
41
|
Louvet-Vallee S: ERM proteins: From
cellular architecture to cell signaling. Biol Cell. 92:305–316.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nowak D, Mazur AJ, Popow-Wozniak A,
Radwanska A, Mannherz HG and Malicka-Blaszkiewicz M: Subcellular
distribution and expression of cofilin and ezrin in human colon
adenocarcinoma cell lines with different metastatic potential. Eur
J Histochem. 54:142010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sarrio D, Rodriguez-Pinilla SM, Dotor A,
Calero F, Hardisson D and Palacios J: Abnormal ezrin localization
is associated with clinicopathological features in invasive breast
carcinomas. Breast Cancer Res Tr. 98:71–79. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lankes W, Griesmacher A, Grunwald J,
Schwartzalbiez R and Keller R: A heparin-binding protein involved
in inhibition of smooth-muscle cell proliferation. Biochem J.
251:831–842. 1988.PubMed/NCBI
|
|
45
|
Amieva MR and Furthmayr H: Subcellular
localization of moesin in dynamic filopodia, retraction fibers, and
other structures involved in substrate exploration, attachment, and
cell-cell contacts. Exp Cell Res. 219:180–196. 1995. View Article : Google Scholar
|
|
46
|
Lallemand D and Arpin M: Moesin/ezrin: a
specific role in cell metastasis? Pigm Cell Melanoma Res. 23:6–7.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
He M, Cheng Y, Li W, et al: Vascular
endothelial growth factor C promotes cervical cancer metastasis via
up-regulation and activation of RhoA/ROCK-2/moesin cascade. BMC
Cancer. 10:2010.PubMed/NCBI
|
|
48
|
Amieva MR, Wilgenbus KK and Furthmayr H:
Radixin is a component of hepatocyte microvilli in situ. Exp Cell
Res. 210:140–144. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hamada K, Shimizu T, Matsui T, Tsukita S
and Hakoshima T: Structural basis of the membrane-targeting and
unmasking mechanisms of the radixin FERM domain. Embo J.
19:4449–4462. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Loebrich S, Bahring R, Katsuno T, Tsukita
S and Kneussel M: Activated radixin is essential for GABAA receptor
alpha5 subunit anchoring at the actin cytoskeleton. EMBO J.
25:987–999. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Elliott BE, Meens JA, SenGupta SK, Louvard
D and Arpin M: The membrane cytoskeletal crosslinker ezrin is
required for metastasis of breast carcinoma cells. Breast Cancer
Research. 7:365–373. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Khanna C, Wan XL, Bose S, et al: The
membrane-cytoskeleton linker ezrin is necessary for osteosarcoma
metastasis. Nat Med. 10:182–186. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yu YL, Khan J, Khanna C, Helman L, Meltzer
PS and Merlino G: Expression profiling identifies the cytoskeletal
organizer ezrin and the developmental homeoprotein Six-1 as key
metastatic regulators. Nat Med. 10:175–181. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kang YK, Hong SW, Lee H and Kim WH:
Prognostic implications of ezrin expression in human hepatocellular
carcinoma. Mol Carcinog. 49:798–804. 2010.PubMed/NCBI
|
|
55
|
Deng XY, Tannehill-Gregg SH, Nadella MVP,
et al: Parathyroid hormone-related protein and ezrin are
up-regulated in human lung cancer bone metastases. Clin Exp
Metastas. 24:107–119. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Meng YX, Lu ZH, Yu SN, Zhang QA, Ma YH and
Chen J: Ezrin promotes invasion and metastasis of pancreatic cancer
cells. J Transl Med. 8:2010.
|
|
57
|
Federici C, Brambilla D, Lozupone F, et
al: Pleiotropic function of ezrin in human metastatic melanomas.
Int J Cancer. 124:2804–2812. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhou BB, Leng J, Hu M, et al: Ezrin is a
key molecule in the metastasis of MOLT4 cells induced by
CCL25/CCR9. Leuk Res. 34:769–776. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Morales FC, Molina JR, Hayashi Y and
Georgescu MM: Overexpression of ezrin inactivates NF2 tumor
suppressor in glioblastoma. Neuro Oncol. 12:528–539. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cui YZ, Wu JM, Zong MJ, et al: Proteomic
profiling in pancreatic cancer with and without lymph node
metastasis. Int J Cancer. 124:1614–1621. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Estecha A, Sanchez-Martin L, Puig-Kroger
A, et al: Moesin orchestrates cortical polarity of melanoma tumour
cells to initiate 3D invasion. J Cell Sci. 122:3492–3501. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Jensen PV and Larsson LI: Actin
microdomains on endothelial cells: association with CD44, ERM
proteins, and signaling molecules during quiescence and wound
healing. Histochem Cell Biol. 121:361–369. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ng T, Parsons M, Hughes WE, et al: Ezrin
is a downstream effector of trafficking PKC-integrin complexes
involved in the control of cell motility. EMBO J. 20:2723–2741.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Haas MA, Vickers JC and Dickson TC:
Binding partners L1 cell adhesion molecule and the
ezrin-radixin-moesin (ERM) proteins are involved in development and
the regenerative response to injury of hippocampal and cortical
neurons. Eur J Neurosci. 20:1436–1444. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Haas MA, Vickers JC and Dickson TC: Rho
kinase activates ezrin-radixin-moesin (ERM) proteins and mediates
their function in cortical neuron growth, morphology and motility
in vitro. J Neurosci Res. 85:34–46. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Tsuda M, Makino Y, Iwahara T, et al: Crk
associates with ERM proteins and promotes cell motility toward
hyaluronic acid. J Biol Chem. 279:46843–46850. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Crepaldi T, Gautreau A, Comoglio PM,
Louvard D and Arpin M: Ezrin is an effector of hepatocyte growth
factor-mediated migration and morphogenesis in epithelial cells. J
Cell Biol. 138:423–434. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hashimoto S, Amaya F, Matsuyama H, et al:
Dysregulation of lung injury and repair in moesin-deficient mice
treated with intratracheal bleomycin. Am J Physiol Lung Cell Mol
Physiol. 295:L566–L574. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Okayama T, Kikuchi S, Ochiai T, et al:
Attenuated response to liver injury in moesin-deficient mice:
impaired stellate cell migration and decreased fibrosis. Biochim
Biophys Acta. 1782:542–548. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Takakuwa Y: Regulation of red cell
membrane protein interactions: implications for red cell function.
Curr Opin Hematol. 8:80–84. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Holzwarth G, Yu J and Steck TL:
Heterogeneity in the conformation of different protein fractions
from the human erythrocyte membrane. J Supramol Struct. 4:161–168.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Diakowski W, Grzybek M and Sikorski AF:
Protein 4.1, a component of the erythrocyte membrane skeleton and
its related homologue proteins forming the protein 4.1/FERM
superfamily. Folia Histochem Cyto. 44:231–248. 2006.PubMed/NCBI
|
|
73
|
Mattagajasingh SN, Huang SC, Hartenstein
JS and Benz EJ: Characterization of the interaction between protein
4.1R and ZO-2. A possible link between the tight junction and the
actin cytoskeleton. J Biol Chem. 275:30573–30585. 2000. View Article : Google Scholar
|
|
74
|
Yamakawa H, Ohara R, Nakajima D, Nakayama
M and Ohara O: Molecular characterization of a new member of the
protein 4.1 family (brain 4.1) in rat brain. Mol Brain Res. 74:247.
1999.
|
|
75
|
Tchernia G, Mohandas N and Shohet SB:
Deficiency of skeletal membrane protein band 4.1 in homozygous
hereditary elliptocytosis. Implications for erythrocyte membrane
stability. J Clin Invest. 68:454–460. 1981. View Article : Google Scholar
|
|
76
|
Shi ZT, Afzal V, Coller B, et al: Protein
4.1R-deficient mice are viable but have erythroid membrane skeleton
abnormalities. J Clin Invest. 103:331–340. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Salomao M, Zhang XH, Yang Y, et al:
Protein 4.1R-dependent multiprotein complex: New insights into the
structural organization of the red blood cell membrane. Proc Natl
Acad Sci USA. 105:8026–8031. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Nunomura W and Takakuwa Y: Regulation of
protein 4.1R interactions with membrane proteins by Ca2+ and
calmodulin. Front Biosci. 11:1522–1539. 2006.
|
|
79
|
Pinder JC, Gardner B and Gratzer WB:
Interaction of protein 4.1 with the red cell membrane: effects of
phosphorylation by protein kinase C. Biochem Biophys Res Commun.
210:478–482. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Horne WC, Prinz WC and Tang EK:
Identification of two cAMP-dependent phosphorylation sites on
erythrocyte protein 4.1. Biochim Biophys Acta. 1055:87–92. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Eder PS, Soong CJ and Tao M:
Phosphorylation reduces the affinity of protein 4.1 for spectrin.
Biochemistry. 25:1764–1770. 1986. View Article : Google Scholar
|
|
82
|
Krauss SW, Larabell CA, Lockett S, et al:
Structural protein 4.1 in the nucleus of human cells: dynamic
rearrangements during cell division. J Cell Biol. 137:275–289.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Mattagajasingh SN, Huang SC, Hartenstein
JS, Snyder M, Marchesi VT and Benz EJ: A nonerythroid isoform of
protein 4.1R interacts with the nuclear mitotic apparatus (NuMA)
protein. J Cell Biol. 145:29–43. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Perez-Ferreiro CM, Luque CM and Correas I:
4.1R proteins associate with interphase microtubules in human T
cells: a 4.1R constitutive region is involved in tubulin binding. J
Biol Chem. 276:44785–44791. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Krauss SW, Heald R, Lee G, et al: Two
distinct domains of protein 4.1 critical for assembly of functional
nuclei in vitro. J Biol Chem. 277:44339–44346. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Yang SM, Guo XH, Debnath G, Mohandas N and
An XL: Protein 4.1R links E-cadherin/beta-catenin complex to the
cytoskeleton through its direct interaction with beta-catenin and
modulates adherens junction integrity. Biochim Biophys Acta.
1788:1458–1465. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Chen L, Hughes RA, Baines AJ, Conboy J,
Mohandas N and An X: Protein 4.1R regulates cell adhesion,
spreading, migration and motility of mouse keratinocytes by
modulating surface expression of beta1 integrin. J Cell Sci.
124:2478–2487. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ruiz-Sáenz A, Kremer L, Alonso MA, Millan
J and Correas I: Protein 4.1R regulates cell migration and IQGAP1
recruitment to the leading edge. J Cell Sci. 124:2529–2538.
2011.PubMed/NCBI
|
|
89
|
Hashimoto Y, Shindo-Okada N, Tani M,
Takeuchi K, Toma H and Yokota J: Identification of genes
differentially expressed in association with metastatic potential
of K-1735 murine melanoma by messenger RNA differential display.
Cancer Res. 56:5266–5271. 1996.PubMed/NCBI
|
|
90
|
Shimizu K, Nagamachi Y, Tani M, et al:
Molecular cloning of a novel NF2/ERM/4.1 superfamily gene, ehm2,
that is expressed in high-metastatic K1735 murine melanoma cells.
Genomics. 65:113–120. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Chauhan S, Pandey R, Way JF, et al:
Androgen regulation of the human FERM domain encoding gene EHM2 in
a cell model of steroid-induced differentiation. Biochem Biophys
Res Commun. 310:421–432. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Cress AE and Nagle RB: Cell Adhesion and
Cytoskeletal Molecules in Metastasis. (Series: Cancer Metastasis -
Biology and Treatment). 9. Springer; Dordrecht: 2006, View Article : Google Scholar
|
|
93
|
Hoover KB and Bryant PJ: Drosophila
Yurt is a new protein-4.1-like protein required for epithelial
morphogenesis. Dev Genes Evol. 212:230–238. 2002. View Article : Google Scholar
|
|
94
|
Wang J, Cai Y, Penland R, Chauhan S,
Miesfeld RL and Ittmann M: Increased expression of the
metastasis-associated gene Ehm2 in prostate cancer. Prostate.
66:1641–1652. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Schulz WA, Ingenwerth M, Djuidje CE, Hader
C, Rahnenfuhrer J and Engers R: Changes in cortical cytoskeletal
and extracellular matrix gene expression in prostate cancer are
related to oncogenic ERG deregulation. BMC Cancer. 10:5052010.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Dhanasekaran SM, Barrette TR, Ghosh D, et
al: Delineation of prognostic biomarkers in prostate cancer.
Nature. 412:822–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Luo JH, Yu YP, Cieply K, et al: Gene
expression analysis of prostate cancers. Mol Carcinog. 33:25–35.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Luo J, Duggan DJ, Chen Y, et al: Human
prostate cancer and benign prostatic hyperplasia: molecular
dissection by gene expression profiling. Cancer Res. 61:4683–4688.
2001.PubMed/NCBI
|
|
99
|
Yu H, Ye L, Mansel RE, Zhang Y and Jiang
WG: Clinical implications of the influence of Ehm2 on the
aggressiveness of breast cancer cells through regulation of matrix
metalloproteinase-9 expression. Mol Cancer Res. 8:1501–1512. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Bosanquet DC, Ye L, Harding KG and Jiang
WG: Expressed in high metastatic cells (Ehm2) is a positive
regulator of keratinocyte adhesion and motility: The implication
for wound healing. J Dermatol Sci. 71:115–121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Reid BJ, Li X, Galipeau PC and Vaughan TL:
Barrett’s oesophagus and oesophageal adenocarcinoma: time for a new
synthesis. Nat Rev Cancer. 10:87–101. 2010.
|
|
102
|
De Minicis S, Marzioni M, Saccomanno S, et
al: Cellular and molecular mechanisms of hepatic fibrogenesis
leading to liver cancer. Transl Gastrointest Cancer. 1:88–94.
2011.
|
|
103
|
Mountford RA, Brown P, Salmon PR,
Alvarenga C, Neumann CS and Read AE: Gastric cancer detection in
gastric ulcer disease. Gut. 21:9–17. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Jess T, Rungoe C and Peyrin-Biroulet L:
Risk of colorectal cancer in patients with ulcerative colitis: a
meta-analysis of population-based cohort studies. Clin
Gastroenterol Hepatol. 10:639–645. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Malka D, Hammel P, Maire F, et al: Risk of
pancreatic adenocarcinoma in chronic pancreatitis. Gut. 51:849–852.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Kerr-Valentic MA, Samimi K, Rohlen BH,
Agarwal JP and Rockwell WB: Marjolin’s ulcer: modern analysis of an
ancient problem. Plast Reconstr Surg. 123:184–191. 2009.
|
|
107
|
Pasternack GR, Anderson RA, Leto TL and
Marchesi VT: Interactions between protein 4.1 and band 3. An
alternative binding site for an element of the membrane skeleton. J
Biol Chem. 260:3676–3683. 1985.PubMed/NCBI
|
|
108
|
Hemming NJ, Anstee DJ, Mawby WJ, Reid ME
and Tanner MJ: Localization of the protein 4.1-binding site on
human erythrocyte glycophorins C and D. Biochem J. 299:191–196.
1994.
|
|
109
|
Marfatia SM, Leu RA, Branton D and Chishti
AH: Identification of the protein 4.1 binding interface on
glycophorin C and p55, a homologue of the Drosophila
discs-large tumor suppressor protein. J Biol Chem. 270:715–719.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Reczek D, Berryman M and Bretscher A:
Identification of EBP50: A PDZ-containing phosphoprotein that
associates with members of the ezrin-radixin-moesin family. J Cell
Biol. 139:169–179. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Nunomura W, Takakuwa Y, Tokimitsu R,
Krauss SW, Kawashima M and Mohandas N: Regulation of CD44-protein
4.1 interaction by Ca2+ and calmodulin. Implications for
modulation of CD44-ankyrin interaction. J Biol Chem.
272:30322–30328. 1997. View Article : Google Scholar
|
|
112
|
Heiska L, Alfthan K, Gronholm M, Vilja P,
Vaheri A and Carpen O: Association of ezrin with intercellular
adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by
phosphatidylinositol 4, 5-bisphosphate. J Biol Chem.
273:21893–21900. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Darmellah A, Rucker-Martin C and Feuvray
D: ERM proteins mediate the effects of Na+/H+
exchanger (NHE1) activation in cardiac myocytes. Cardiovasc Res.
81:294–300. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Niggli V, Andreoli C, Roy C and Mangeat P:
Identification of a phosphatidylinositol-4,5-bisphosphate-binding
domain in the N-terminal region of ezrin. FEBS Lett. 376:172–176.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Tanaka T, Kadowaki K, Lazarides E and
Sobue K: Ca2(+)-dependent regulation of the spectrin/actin
interaction by calmodulin and protein 4.1. J Biol Chem.
266:1134–1140. 1991.
|
|
116
|
Weinman EJ, Steplock D, Wade JB and
Shenolikar S: Ezrin binding domain-deficient NHERF attenuates
cAMP-mediated inhibition of Na(+)/H(+) exchange in OK cells. Am J
Physiol Renal Physiol. 281:F374–F380. 2001.PubMed/NCBI
|