Introduction
Hepatocellular carcinoma (HCC) represents the fifth
most common and aggressive malignancy worldwide and the third in
terms of mortality (1,2). HCC has become a serious threat to
human health due to its rising incidence and high metastatic
recurrence and mortality rates (3,4).
Although patients diagnosed with this malignant disease can benefit
from some of the existing effective treatments, including liver
transplantation, surgical resection, embolization, stereotactic
body radiation therapy, ablation and chemotherapy, which improve
their chances of survival (5),
the current treatment options cannot smeet the requirements for the
survival of HCC patients and the prognosis remains dismal (6). Therefore, the search for novel and
more effective treatment strategies is of particularl importance.
Gene therapy, which is increasingly being tested in clinical trials
and has shown potential in clinical practice, has the advantage of
high specificity, efficiency and security (7), and has been shown to have potential
future perspectives (8).
miR-206, a member of the muscle-specific miR-1
family of muscle-specific microRNAs (myomiRs), is a skeletal
muscle-specific miRNA involved in muscle development (9). However, studies have revealed that
miR-206 is closely related to various tumors. The ectopic
expression of miR-206 has been shown to inhibit the growth of
rhabdomyosarcoma (RMS) (10),
breast cancer (11,12), endometrial endometrioid carcinoma
(EEC) (13), lung cancer
(14) and HeLa cells (15). Furthermore, fluorescence-activated
cell sorting (FACS) has demonstrated that miR-206 activates
apoptosis in lung cancer (14)
and HeLa cells (15) and induces
cell cycle arrest at the G0/G1 phase of the cell cycle in RMS
(10) and EEC cells (13). Cell invasive and migratory ability
has also been shown to be impaired by miR-206 in RMS (10), EEC (13), lung cancer (14) and HeLa cells (15).
Although the multiple anticancer functions of
miR-206 have been confirmed, its underlying anticancer mechanisms
of action are not yet fully understood. However, it is a worth
noting that Song et al first identified an almost perfect
complementarity between miR-206 and the 3′-untranslated regions
(3′-UTRs) of both mouse and human Notch3 and found that the ectopic
expression of miR-206 induced apoptotic cell death in HeLa cells,
which was associated with its inhibition of Notch3 signaling
(15). Early research has
demonstrated that the Notch3 receptor, one of the mammalian Notch
family receptors (Notch1-4), plays an important role in cellular
differentiation (16) and
embryonic development (17). Of
note, a growing body of evidence in recent years has indicated that
Notch3 is also involved in the regulation of cancer development and
progression (18–22). Using immunohistochemistry, Zhou
et al demonstrated that Notch3 had a stronger positive
degree of expression in lung squamous cell carcinoma and
adenocarcinoma compared with the corresponding non-tumor tissue
(P<0.01) (23). Moreover,
Notch3 overexpression has been shown to significantly correlate
with poor prognosis in human non-small cell lung cancer (NSCLC)
(24). By contrast, the
inhibition of Notch3 by γ-secretase inhibitor (GSI) induces
apoptosis and suppresses the proliferation of cancer cells through
the downregulation of the pro-survival proteins, pBcl-2 and
pBcl-xL, and not Bax in NSCLC (25). A decrease in Notch3 expression can
also activate apoptosis by increasing the cleavage of caspase-3 and
poly(ADP-ribose) polymerase (PARP) (21).
Moreover, an increasing number of studies has
indicated that Notch3 contributes to the promotion of HCC
development and progression. Notch3, Jagged1, Delta1 and the
downstream effector gene, hairy and enhancer of split 1 (Hes1), are
highly expressed in the HepG2 tumor cell line, which was thought to
be necessary for malignant liver cell proliferation (19). In addition, by regulating matrix
metalloproteinase (MMP)-2 and MMP-9 through the ERK1/2 pathway,
high Notch3 expression also strongly correlates with HCC metastasis
(26). However, the
downregulation of Notch3 in 2 HCC cell lines has been shown to
result in the downregulation of Hes1, the upregulation of
CDKN1C/p57, and reduced cell growth through the induction of
senescence instead of apoptosis (27).
In this study, we aimed to investigate the potential
function of miR-206 in the development and progression of HCC. It
was hypothesized that Notch3 is a direct target gene of miR-206 in
HCC cells. miR-206 mimics were transiently transfected into HepG2
cells. We found that miR-206 significantly suppressed tumor growth
and metastasis at least in part by targeting the Notch3 signaling
pathway in vitro. To the best of our knowledge, this study
is the first to reveal the function and possible underlying
mechanisms of action of miR-206 in HCC and suggests that miR-206
has the potential for use in the targeted therapy of HCC.
Materials and methods
Immunohistochemistry and evaluation of
immunostaining
Formalin-fixed and paraffin-embedded (FFPE) tissue
samples from HCC and adjacent non-neoplastic tissues (at least 1.5
cm away from the tumor) were collected from 12 patients who were
histopathologically diagnosed with primary HCC and had undergone
surgical treatment at the Second Affiliated Hospital of Nanchang
University (Nanchang, China) in the last 5 years. Sections
(4-μm-thick) mounted on glass slides were processed for
immunohistochemistry. All slides were dewaxed in xylene and
dehydrated in an alcohol gradient; endogenous peroxidase activity
was quenched with 3% hydrogen peroxide for 10 min. Antigen
retrieval was obtained by heating the slides covered with citrate
buffer (pH 6.0) at 95°C for 10 min. Subsequently, 10% goat serum
albumin was used to block non-specific binding by incubating the
sections for 1 h at room temperature. Gently tilting without
washing, the sections were then incubated with anti-Notch3 (1:50
dilution; polyclonal anti-Notch3, M-134: sc-5593; Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) diluted in 1× TBS for 2 h
in a moist box at room temperature. The sections were then
incubated with the secondary antibody at room temperature for 1 h
and rinsed in phosphate-buffered saline (PBS). Diaminobenzidine
(DAB) was used as the chromogen and the sections were
counterstained with hematoxylin. For negative controls, the
sections incubated with PBS instead of the primary antibody. Brown
particles present in the cytoplasm and/or nuclei were considered
positive. Imaging analysis was conducted under a microscope
(Olympus, Tokyo, Japan); the exact location of the measured visual
field was determined and then 3 complete visual fields without
overlap were randomly selected for viewing. A validated
semi-quantitative scale was used to assess the immunostaining: ‘−’
denotes no hepatocyte staining; ‘+/−’ denotes occasional weak
hepatocyte staining; ‘+’ denotes <5% hepatocyte staining; ‘++’
denotes 5–30% hepatocyte staining; and ‘+++’ denotes >30%
hepatocyte staining.
Cell culture and transfection
HepG2 cells were grown in Dulbecco’s modified
Eagle’s medium (DMEM) (Solarbio, Beijing, China) plus 10% fetal
calf serum (FBS; TransGen Biotech, Beijing, China), 2 mM
L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin (all
reagents were from Gibco-BRL Life Technologies, Gaithersburg, MD,
USA) and incubated in a 5% CO2 humidified incubator at
37°C. Cy3-modified miR-206 mimic and Cy3-modified mimic negative
control were purchased from RiboBio Co., Ltd. (Guangzhou, China).
For convenience, Cy3-modified miR-206 mimic and Cy3-modified mimic
negative control were simply referred to as miR-206 and negative
control (NC), respectively. Complete medium without antibiotics was
used to culture the cells at least 24 h prior to transfection. The
cells were washed with PBS and then transiently transfected with
100 nM miR-206 or NC using Lipofectamine™ 2000 (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer’s
instructions.
Cellular proliferation assay
The HepG2 cells (1,000 cells/well) were seeded in a
96-well plate and incubated under normal culture conditions for 24
h prior to transfection. Cell proliferation was measured using the
CellTiter 96® AQueous One Solution Cell Proliferation
Assay (MTS) kit (Promega, Beijing, China) according to the
manufacturer’s instructions. MTS reagent (20 μl) was added to the
cells in each well followed by incubation for 2 h in a 37°C, 5%
CO2 humidified incubator at 0, 24, 48, 72 and 96 h after
transfection. The absorbance (A) of each plate was measured at 490
nm.
Hoechst 33342 staining
The HepG2 cells were plated in 12-well plates at a
density of 1×105 cells/well 1 day prior to transfection.
Forty-eight hours after transfection, the plates were washed twice
with PBS, then 500 μl Hoechst 33342 (Beyotime, Shanghai, China)
were added to each well followed by incubation for 30 min at 37°C
in the dark. Nuclear DNA staining was observed using a fluorescence
microscope (Olympus). A total of 500 cells was counted from 5
random high-power fields and the fluorescence staining percentage
of positive cells was expressed as the ratio of apoptotic cells
with respect to the total amount of cells.
RNA isolation and quantitative reverse
transcription polymerase chain reaction (qRT-PCR)
Total RNA from the cultured cells was extracted
using TRIzol reagent (TransGen Biotech, Beijing, China) according
to the manufacturer’s instructions. miRNA levels were measured by
qRT-PCR. For the qRT-PCR detection of mature miR-206 expression, we
purchased the Bulge-Loop™ miRNA qRT-PCR Primer Set and the miRNA
qRT-PCR Control Primer Set (both from RiboBio). RNA (1 μg) was
converted into cDNA using the PrimeScript™ RT reagent kit with gDNA
Eraser (Takara, Dalian, China) according to the manufacturer’s
instructions. qRT-PCR was performed using SYBR® Premix
Ex Taq™ II (Takara) in the ABI PRISM® 7500 real-time PCR
system (Applied Biosystems, Foster City, CA, USA). β-actin and U6
were used as endogenous controls. In addition, melting curves were
used to evaluate non-specific amplification. The relative
expression level was calculated using the ΔΔCt method. The primer
sequences used in this study are presented in Table I.
 | Table IPrimers used for qRT-PCR. |
Table I
Primers used for qRT-PCR.
| Gene symbol | NCBI RefSeq
no. | Sequence
(5′→3′) | Product length
(bp) |
|---|
| Notch3 | NM_000435 | (F)
GTGTGTGTCAATGGCTGGAC | (R)
GTGACACAGGAGGCCAGTCT | 150 |
| Bax | NM_138763 | (F)
CCCGAGAGGTCTTTTTCCGAG | (R)
CCAGCCCATGATGGTTCTGAT | 155 |
| Bcl-2 | NM_000633 | (F)
CTTTGAGTTCGGTGGGGTCA | (R)
GGGCCGTACAGTTCCACAAA | 162 |
| Hes1 | NM_005524 | (F)
TCAACACGACACCGGATAAAC | (R)
GCCGCGAGCTATCTTTCTTCA | 153 |
| p57 | NM_000076 | (F)
CCCTTCTTCTCGCTGTCCTC | (R)
CTGGTCCTCGGCGTTCA | 231 |
| MMP-9 | NM_004994 | (F)
CTGCAGTGCCCTGAGGACTA | (R)
ACTCCTCCCTTTCCTCCAGA | 135 |
| β-actin | NM_001101 | (F)
TTAGTTGCGTTACACCCTTTC | (R)
GCTGTCACCTTCACCGTTC | 156 |
Western blot analysis
Forty-eight hours after transfection, total protein
was extracted from the HepG2 cells using RIPA cell lysis reagent
containing proteinase and phosphatase inhibitors (Solarbio) at 4°C
for 30 min. Cell lysates were centrifuged at 12,000 × g for 20 min
at 4°C, and the protein concentrations of the supernatants were
determined using the BCA protein assay reagent kit (Beyotime). The
supernatants containing total protein were then mixed with a
corresponding volume of 5× SDS loading buffer and heated at 95°C
for 5 min; 40 μg of total protein from each sample were
concentrated on 5% Tris-glycine SDS gels, separated on 12%
Tris-glycine SDS gels and transferred onto 0.22-μm polyvinylidene
fluoride (PVDF) membranes. The membranes were blocked with 5%
non-fat dry milk in TBST and incubated overnight with the
appropriate primary antibody. The primary antibodies and dilutions
used were as follows: anti-Notch3 (Cat. no. 5276, 1:200), anti-p57
(Cat. no. 2557, 1:500), anti-MMP-9 (Cat. no. 852, 1:200),
anti-caspase-3 (Cat. no. 9662, 1:300) purchased from Cell Signaling
Technology (Beverly, MA, USA), anti-Hes1 (ab71559, 1:300; Abcam),
anti-Bax (Cat. no. 50599-2-Ig; 1:500) and anti-Bcl-2 (Cat. no.
12789-1-AP, 1:500), both from ProteinTech. The membranes were then
incubated with the secondary horseradish peroxidase-conjugated
AffiniPure goat anti-rabbit lgG (H+L) (1:1,000; TransGen Biotech)
or the secondary horseradish peroxidase-conjugated AffiniPure goat
anti-mouse lgG (H+L) (1:1,000; ZSGB-BIO, Beijing, China).
Anti-β-actin monoclonal antibody (1:1,000; ZSGB-BIO) was used as an
endogenous control. The quantification of western blot analyses was
performed using Quality One 4.6.2 software, and the integral
optical density (IOD) of each band was determined. The relative
protein level was used to evaluate the differences in protein
expression between the miR-206 -treated group and the NC group; the
relative protein level = (IOD ratio between the target gene product
bands and the β-actin protein bands in the miR-206-treated
group)/(IOD ratio between the target gene product bands and the
β-actin protein bands in the NC group).
Annexin V-FITC/PI analysis
The HepG2 cells were harvested at 48 h after
transfection. Cell apoptosis was detected using an Annexin
V-FITC/PI apoptosis detection kit (BestBio, Shanghai, China)
following the manufacturer’s instructions, and the percentage of
apoptotic cells was calculated using a Beckman Coulter FACSCalibur
flow cytometer (Beckman Coulter, Inc., Fullerton, CA, USA).
Cell cycle analysis
The HepG2 cells were collected at 48 h after
transfection and fixed with 70% ethanol in PBS at −20°C overnight.
Cell cycle analysis was performed using the cell cycle kit
(BestBio) according to the manufacturer’s specifications, and cell
cycle distribution was analyzed using a Beckman Coulter FACSCalibur
flow cytometer (Beckman Coulter, Inc.).
Wound healing assay in vitro
The HepG2 cells were seeded in 6-well plates and
incubated for 24 h; a linear wound was tehn created by dragging a
1-ml pipette tip through the monolayer prior to transfection.
Cellular debris was removed by gentle washes with culture medium,
following which transfection was performed immediately, and the
cells were allowed to migrate for a further 24 h. The healing
process was dynamically photographed after the wound was introduced
using a microscope (Olympus). The gap size was analyzed using
Image-Pro Plus 6.0 software. The residual gap between the migrating
cells from the opposing wound edge was expressed as a percentage of
the initial gap size.
Statistical analysis
All experiments were repeated 3 times independently.
The results are presented as the means ± standard deviation (SD). A
rwo-tailed paired t-test was performed using SPSS 19.0 software in
order to detect significant differences in measured variables
between groups. A value of P<0.05 was considered to indicate a
statistically significant difference.
Results
Differential expression of Notch3 in HCC
and adjacent non-neoplastic tissues
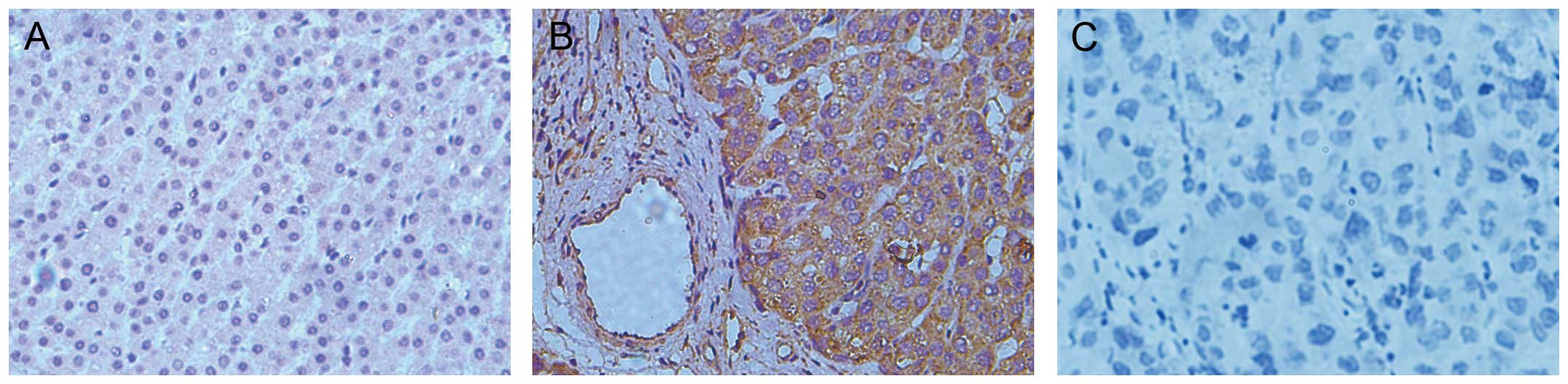
Immunostaining revealed a high Notch3 protein
expression in the cytoplasm of the neoplastic hepatocytes in 12 out
of the 12 (100%) HCC samples compared with occasional weak
hepatocytic staining in their corresponding adjacent non-neoplastic
tissue samples. Representative immunostaining patterns of the
Notch3 expression are shown in Fig.
1.
Inhibition of cell proliferation
following transfection with miR-206
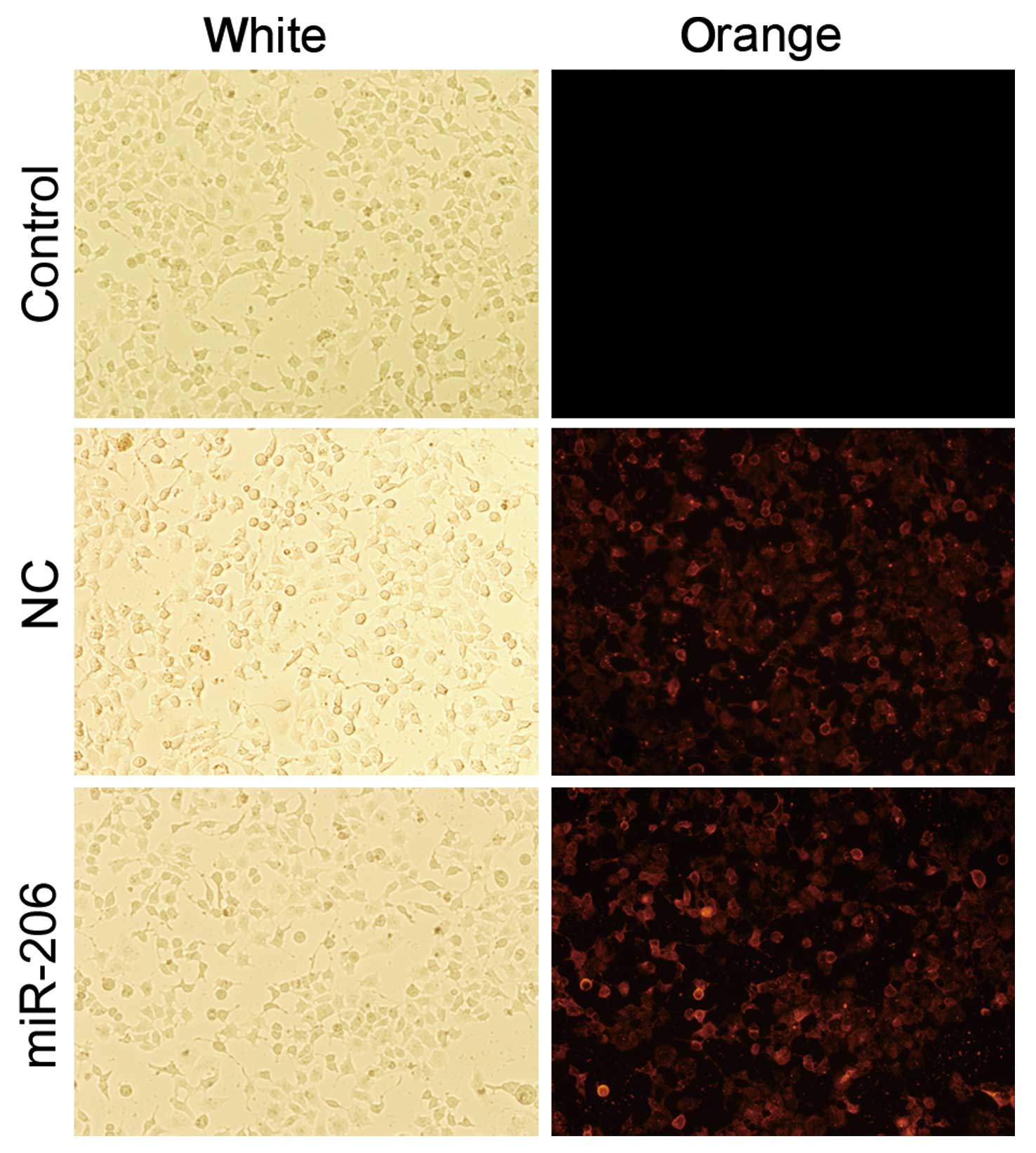
To investigate the functional role of miR-206,
Cy3-modified miR-206 mimic and Cy3-modified mimic negative control
were successfully transiently transfected into the HepG2 cells
(Fig. 2). Furthermore, the mRNA
levels of miR-206 were analyzed by qRT-PCR. We found that the
miR-206 mimic-treated cells had an approximately 60-fold greater
expression of mature miR-206 than the cells transfected with the
negative control mimic (Fig. 3A).
Cell proliferation was significantly decreased in the cells
following 48 h of transfection with miR-206 (Fig. 4). These results indicate that
miR-206 overexpression decreases the proliferation of human
hepatocellular carcinoma HepG2 cells.
miR-206 overexpression promotes apoptotic
cell death in HepG2 cells
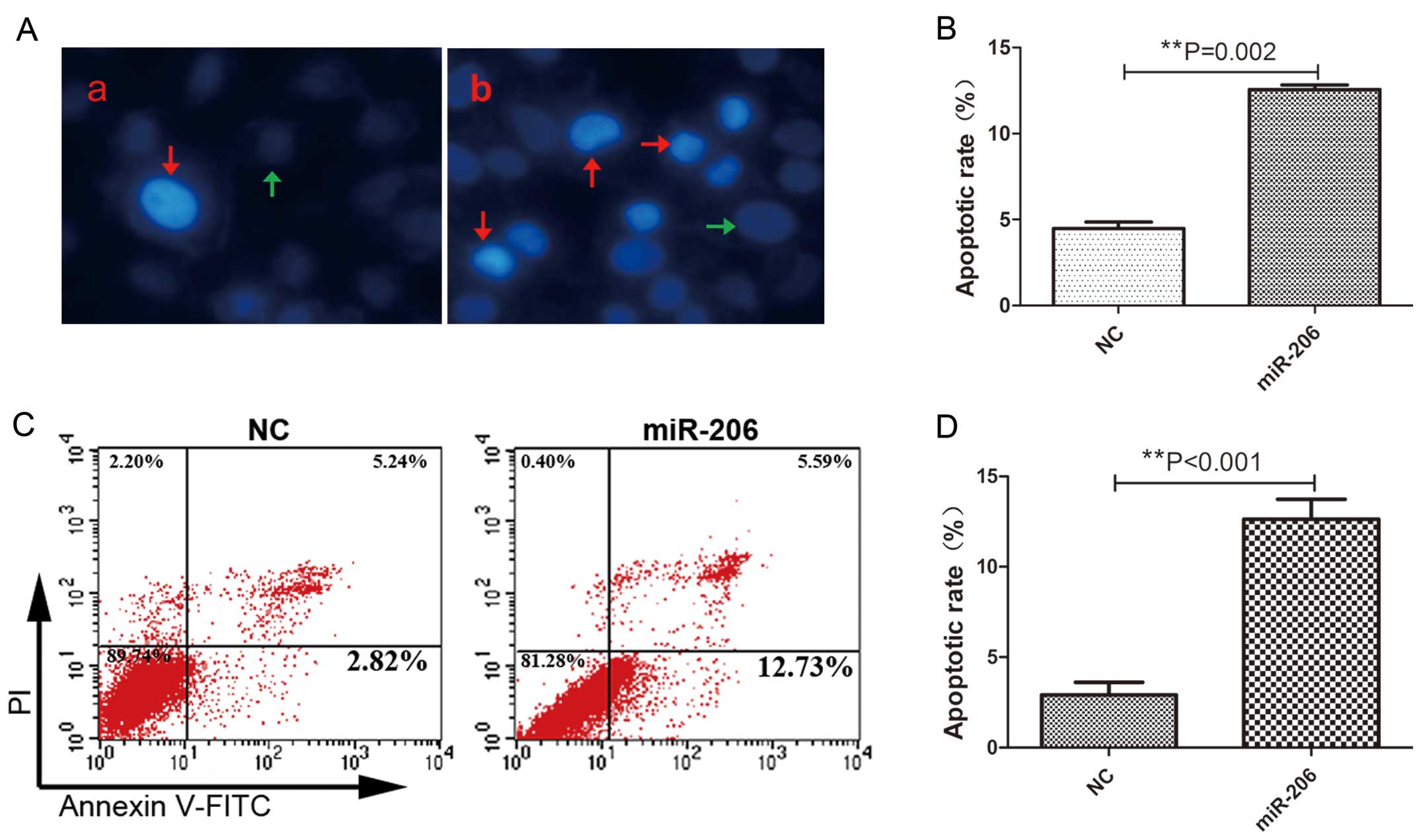
Decreased apoptotic activity in HCC cells is one of
the most important features of HCC (28). In this study, in order to
investigate whether miR-206 induces cellular apoptosis, Hoechst
33342 staining and Annexin V-FITC/PI flow cytometry were conducted.
The miR-206-treated group showed increased numbers of Hoechst 33342
positively stained cells 48 h after transfection, indicating an
enhanced apoptotic activity (Fig.
5A–b). In accordance with Hoechst 33342 staining, FACS analysis
further confirmed that the cells transfected with miR-206 underwent
more apoptosis compared with the miR-206 mimic-treated cells
(Fig. 5C). There was an
approximately 2.0- or 3.0-fold increase in the percentage of
apoptotic cells in the HepG2 cells overexpressing miR-206 (Fig. 5B and D). Moreover, qRT-PCR
analysis revealed that the relative expression of Notch3 and Bcl-2
was markedly reduced, whereas that of Bax was increased at the mRNA
level following transfection with miR-206 (Fig. 3B). In addition, miR-206
overexpression downregulated Notch3 and Bcl-2 expression and
upregulated Bax and caspase-3 expresssion at the protein level, as
shown by western blot analysis (Fig.
6); the protein expression of cleaved caspase-3 (caspase-3 CL)
in particular was markedly increased. Of note, these data support
our hypothesis that Notch3 is likely to be a direct target gene of
miR-206 in HepG2 cells. Furthermore, these results indicate that
the pro-apoptotic effect of miR-206 in HepG2 cells is at least
partially dependent on Notch3-mediated mitochondrial apoptotic
signaling.
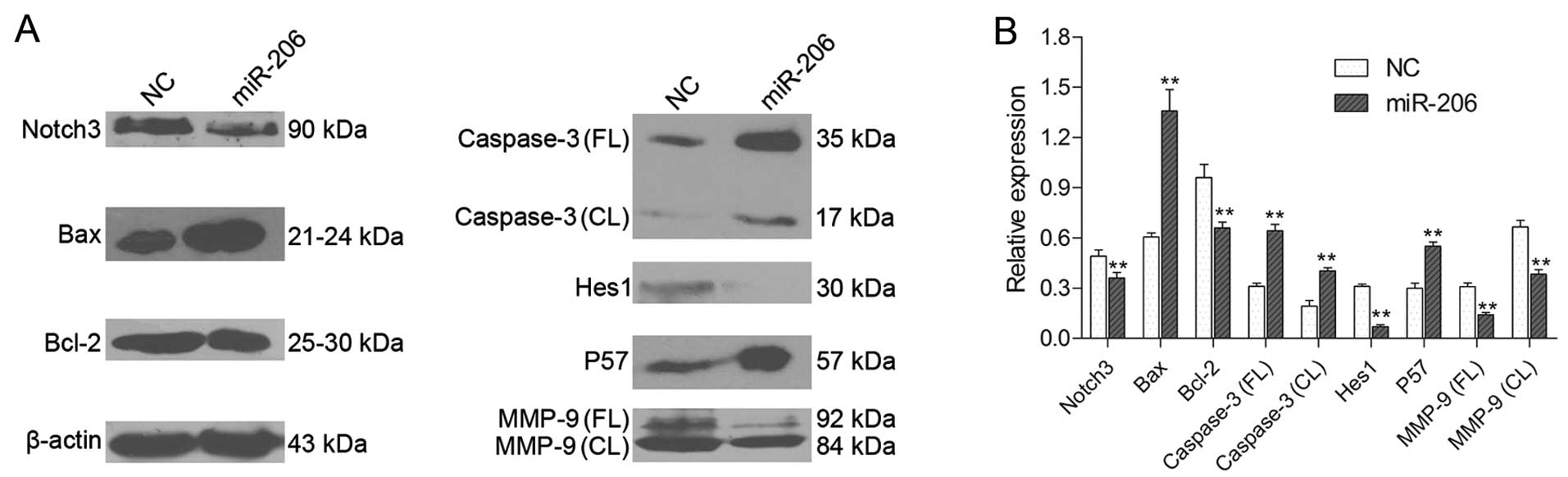 | Figure 6Western blot analysis of Notch3, Bax,
Bcl-2, caspase-3, Hes1, p57 and matrix metalloproteinase-9 (MMP-9)
expression in each group of HepG2 cells. (A) A representative
western blot is shown. (B) Relative protein expression levels of
Notch3, Bax, Bcl-2, full length (FL) caspase-3, cleaved (CL)
caspase-3, Hes1, p57, MMP-9 FL and MMP-9 CL were assessed
calculating the integral optical density (IOD)-values. IOD values
were normalized to those of β-actin protein.
**P<0.01. |
miR-206 induces cell cycle arrest in
HepG2 cells
Flow cytometry was used to investigate the effects
of miR-206 on the cell cycle. Our results revealed that the
proportion of the cells in the G0/G1 and G2 phases was did not
altered in the NC group. By contrast, the transfection of HepG2
cells with miR-206 resulted in an accumulation of cells in the
G0/G1 phase and a decrease in the number of cells in the G2/M phase
compared with the NC group. Flow cytometric analysis indicated that
miR-206 overexpression slowed down cell cycle progression and
caused cell cycle G1 phase blockage in the HepG2 cells (Fig. 7A). The proportion of cells in the
G0/G1 and G2/M phases exhibited significant differences between the
2 groups (Fig. 7B). In order to
discover the probable underlying mechanisms of action of miR-206 in
inducing cell cycle arrest, we further analyzed the expression of
Hes1 and p57 in HepG2 cells at the mRNA and protein level. A
significant inverse correlation between Hes1 and p57 expression was
demonstrated by qRT-PCR (Fig. 3B)
and western blot analysis (Fig.
6), suggesting that Hes1 participates in regulating p57 mRNA
transcription in the HepG2 cells. Moreover, Hes1 is acknowledged to
be a direct target gene of Notch3. Taken together, our results
indicate that the effects of miR-206 on the cell cycle (causing
cell cycle arrest) are possibly mediated through the crosstalk
between these 3 genes (Notch3-Hes1-p57 signaling) in the HepG2
cells.
Cellular migration is impaired following
transfection with miR-206 in HepG2 cells
Cellular migration is an essential process in cancer
metastasis. Thus, we examined the cellular migration ability in
order to explore the potential role of miR-206 in HCC cell
metastasis. The wound healing assay revealed that the cells
transfected with miR-206 healed the wound more slowly than the
NC-transfected cells (Fig. 8).
MMPs may be associated with the impaired migtation of
miR-206-transfected cells. To examine this hypothesis, we detected
MMP-9 expression at the mRNA and protein level. Consistent with the
results of migration assay, the overexpression of miR-206 caused a
significant reduction in MMP-9 expression at the mRNA and protein
level (Figs. 3B and 6); the protein expression level of
cleaved MMP-9 (MMP-9 CL) in particular was downregulated in the
HepG2 cells. Our results indicate that one of the possible
mechanims responsible for the inhibitory effects of miR-206 on the
migration of HepG2 cells is through the Notch3-MMP-9 pathway, at
least through the downregulation of MMP-9.
Discussion
An abundance of in vivo and in vitro
studies has indicated that enhanced cell proliferation, resistance
to apoptosis and the migration state of HCC cells plays an
important role in the progression of HCC (2,8).
Despite increasing evidence pointing to a role for miR-206 as a
tumor suppressor, the tumor suppressive effect of miR-206 has not
been fully elucidated. To the best of our knowledge, the present
study is the first to explore the function and probable underlying
mechanisms of action of miR-206 in HCC HepG2 cells. First, using
immunohistochemistry, we found that Notch3 protein expression was
markedly increased in the HCC tissues compared with the adjacent
normal tissues; these results are consistent with those of previous
studies suggesting that the increased expression levels of Notch3
significantly correlates with HCC progression and unfavorable
prognosis (19,26,29). Secondly, miR-206 mimic and mimic
negative control were successfully transfected into the HepG2
cells. We also found that elevated miR-206 levels inhibited the
growth of HepG2 cells, which was associated with the induction of
apoptosis and cell cycle arrest. In addition, cellular migration
was also impaired following transfection with miR-206 in the HepG2
cells. Our results demonstrated that there are hopeful prospects
for miR-206 gene therapy in HCC; however, the possible underlying
mechanisms require further investigation.
It has been demonstrated that several target mRNAs
are directly regulated by miR-206, including Cdc42, estrogen
receptor α (ERα), Notch3, liver X receptor α (LXRα), high mobility
group box 3-like pseudogene (Hmgb3) and c-Met (12–13,15,30–32), among which Notch3 was hypothesized
to be a direct target gene of miR-206 in this study. Of note, we
found that the enforced overexpression of miR-206 markedly
attenuated Notch3 expression at the mRNA and protein level in the
HepG2 cells, these results are consistent with those of other
studies on other cell lines (1,15).
Therefore, to a certain extent, this result supports our hypothesis
that Notch3 is a direct target gene of miR-206 in HepG2 cells.
An increasing number of studies has suggested that
Notch3 has a potential role in anti-apoptosis (1,15,21,25); however, the underlying mechanisms
of this anti-apoptotic role remain unclear. Wang et al
reported that Notch3 signaling can help smooth muscle cells resist
Fas ligand-induced apoptosis (33). However, our results revealed that
Notch3 was downregulated with the overexpression of miR-206; Bax
expression was increased at the mRNA and protein levels, Bcl-2
expression was significantly reduced, and, finally, caspase-3,
which exhibited a similar effect to GSI (25), was activated. Thus, our results
indicate that the pro-apoptotic effect of miR-206 in HepG2 cells is
at least partially dependent on Notch3-mediated mitochondrial
apoptotic signaling. However, it has been shown that there are 2
different mechanisms associated with cell apoptosis: the extrinsic
receptor-mediated pathway and the intrinsic mitochondrial-dependent
pathway (34). Therefore, the
anti-apoptotic effects of miR-206 in HCC require further
investigation.
Moreover, miR-206 inhibited the growth of HepG2
cells by inducing cell cycle arrest. Our results revealed that the
overexpression of miR-206 markedly downregulated Hes1 expression,
significantly elevated p57 expression, and finally induced cell
cycle G1 phase blockage in the HepG2 cells. These results are in
accordance with those of a previous study (27), which reported that p57 is a target
of transcriptional repression by the Notch3 effector, Hes1. Of
note, the same study also found that the upregulation of p57 by
cDNA transfection decreased tumor growth, as demonstrated by the
growth curve, flow cytometric analysis and cyclin D1
downregulation, without affecting the apoptotic machinery.
Similarly, Chen et al found that miR-206 overexpression
suppressed ERα and induced the cell cycle arrest of ERα-positive
epithelial endometrial cells (EECs) (13). Moreover, it has been demonstrated
that estrogens play an important role in the control of liver cell
proliferation (35). Thus, the
issue of whether miR-206 induces cell cycle arrest in HCC cell
lines by inhibiting ERα, remains to be addressed further.
Cell migration and invasion are involved in a number
of physiological processes as normal events. However, uncontrolled
migration and invasion lead to metastasis, which is the cause of as
high as 90% of human cancer-related deaths (36). Metastasis is a multistep process;
MMPs are involved in cell migration and invasion and are frequently
upregulated in cancer cells (37–39). In the present study, both the mRNA
and protein levels of MMP-9 were downregulated in the
miR-206-transfected cells, which significantly impaired the
migratory capability of HepG2 cells. In a previous study, using
Transwell assay, it was demonstrated that 95D cells transfected
with miR-206 had a decreased invasive capability than the cells
transfected with non-specific control miRNA (14). In the present study, we
demonstrated that miR-206 inhibited cell migration through the
Notch3-MMP-9 pathway, partly due to its effect on MMP-9.
Although we suggested Notch3 is likely to be a
direct target gene of miR-206 in HepG2 cells, further studies are
required to identify any other mRNAs that are directly regulated by
miR-206 in HepG2 cells, as previously reported in other cell lines
(12,13,30–32). In addition, the lack of in
vivo validation of our molecular pathway in HCC cancer is a
limitation of this study. However, to the best of our knowledge,
our study is the first to reveal the function and possible
underlying mechanisms of action of miR-206 in HCC HepG2 cells.
It is worth noting that the modulation of a target
mRNA by several miRNAs and the simultaneous regulation of a variety
of mRNAs by a single mRNA is a normal phenomenon (40). Furthermore, Di Leva et al
suggested a negative transcriptional regulatory loop in which
miR-221 and miR-222 target ERα, which, in turn, suppresses miR-221
and miR-222 expression (11).
There seems to be a doubt as to whether there is a similar
interaction between miR-206 and Notch3. Therefore, further studies
are required to elucidate all the aspects of miR-206 expression in
HCC.
Recent studies (41,42) have shown that miR-206 expression
is significantly downregulated in gastric and breast cancer tissues
when compared with their normal adjacent tissues, which
significantly correlates with tumor progression, suggesting that
miR-206 acts as a tumor suppressor.
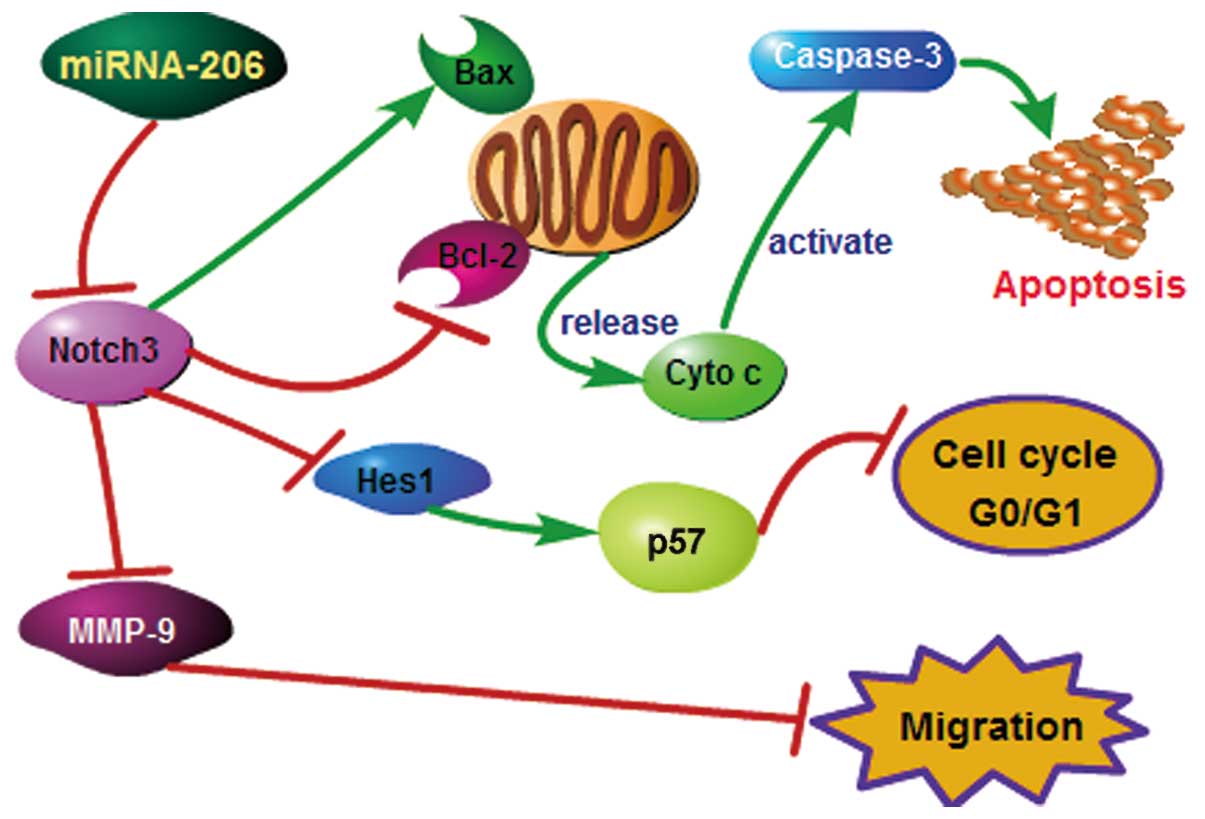
Taken together, our results demonstrate that
miRNA-206 overexpression promotes apoptosis, induces cell cycle
arrest and inhibits the migration of HCC HepG2 cells. The possible
underlying mechanisms of action of miR-206 in HCC are shown in
Fig. 9. In conclusion, this study
suggests that the delivery of miR-206 to HepG2 cells may lead to
the development of novel therapeutic strageties for HCC, and that
miR-206 may be a potential therapeutic agent for human tumors, and
is worthy of further investigation.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (no. 30060029) and the
Natural Science Foundation of Jiangxi Province (no.
2010JXY0237).
References
|
1
|
Jalali S, Ramanathan GK, Parthasarathy PT,
et al: Mir-206 regulates pulmonary artery smooth muscle cell
proliferation and differentiation. PLoS One. 7:e468082012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Subramaniam A, Shanmugam MK, Perumal E, et
al: Potential role of signal transducer and activator of
transcription (STAT)3 signaling pathway in inflammation, survival,
proliferation and invasion of hepatocellular carcinoma. Biochim
Biophys Acta. 1835.46–60. 2013.PubMed/NCBI
|
|
3
|
Sia D and Villanueva A: Signaling pathways
in hepatocellular carcinoma. Oncology. 81(Suppl 1): S18–S23. 2011.
View Article : Google Scholar
|
|
4
|
Qin LX and Tang ZY: Recent progress in
predictive biomarkers for metastatic recurrence of human
hepatocellular carcinoma: a review of the literature. J Cancer Res
Clin Oncol. 130:497–513. 2004.PubMed/NCBI
|
|
5
|
Maluccio M and Covey A: Recent progress in
understanding, diagnosing, and treating hepatocellular carcinoma.
CA Cancer J Clin. 62:394–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Giacomin A, Sergio A, Vanin V, Gazzola A,
Cazzagon N and Farinati F: Molecular targeted therapy in
hepatocellular carcinoma: present achievements and future
challenges. Dig Dis. 30:284–288. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ortiz R, Melguizo C, Prados J, et al: New
gene therapy strategies for cancer treatment: a review of recent
patents. Recent Pat Anticancer Drug Discov. 7:297–312. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Psyrri A, Arkadopoulos N, Vassilakopoulou
M, Smyrniotis V and Dimitriadis G: Pathways and targets in
hepatocellular carcinoma. Expert Rev Anticancer Ther. 12:1347–1357.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McCarthy JJ: MicroRNA-206: the skeletal
muscle-specific myomiR. Biochim Biophys Acta. 1779:682–691. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Missiaglia E, Shepherd CJ, Patel S, et al:
MicroRNA-206 expression levels correlate with clinical behaviour of
rhabdomyosarcomas. Br J Cancer. 102:1769–1777. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Di Leva G, Gasparini P, Piovan C, et al:
MicroRNA cluster 221–222 and estrogen receptor alpha interactions
in breast cancer. J Natl Cancer Inst. 102:706–721. 2010.
|
|
12
|
Liu H, Cao YD, Ye WX and Sun YY: Effect of
microRNA-206 on cytoskeleton remodelling by downregulating Cdc42 in
MDA-MB-231 cells. Tumori. 96:751–755. 2010.PubMed/NCBI
|
|
13
|
Chen X, Yan Q, Li S, et al: Expression of
the tumor suppressor miR-206 is associated with cellular
proliferative inhibition and impairs invasion in ERα-positive
endometrioid adenocarcinoma. Cancer Lett. 314:41–53.
2012.PubMed/NCBI
|
|
14
|
Wang X, Ling C, Bai Y and Zhao J:
MicroRNA-206 is associated with invasion and metastasis of lung
cancer. Anat Rec (Hoboken). 294:88–92. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song G, Zhang Y and Wang L: MicroRNA-206
targets notch3, activates apoptosis, and inhibits tumor cell
migration and focus formation. J Biol Chem. 284:31921–31927. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bellavia D, Campese AF, Vacca A, Gulino A
and Screpanti I: Notch3, another Notch in T cell development. Semin
Immunol. 15:107–112. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hansson EM, Lendahl U and Chapman G: Notch
signaling in development and disease. Semin Cancer Biol.
14:320–328. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Konishi J, Yi F, Chen X, Vo H, Carbone DP
and Dang TP: Notch3 cooperates with the EGFR pathway to modulate
apoptosis through the induction of bim. Oncogene. 29:589–596. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Giovannini C, Lacchini M, Gramantieri L,
Chieco P and Bolondi L: Notch3 intracellular domain accumulates in
HepG2 cell line. Anticancer Res. 26:2123–2127. 2006.PubMed/NCBI
|
|
20
|
Jonusiene V, Sasnauskiene A, Lachej N, et
al: Down-regulated expression of Notch signaling molecules in human
endometrial cancer. Med Oncol. 30:4382013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang X, Samadi AK, Roby KF, Timmermann B
and Cohen MS: Inhibition of cell growth and induction of apoptosis
in ovarian carcinoma cell lines CaOV3 and SKOV3 by natural
withanolide Withaferin A. Gynecol Oncol. 124:606–612. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mizugaki H, Sakakibara-Konishi J, Ikezawa
Y, et al: γ-Secretase inhibitor enhances antitumour effect of
radiation in Notch-expressing lung cancer. Br J Cancer.
106:1953–1959. 2012.
|
|
23
|
Zhou M, Jin WY, Fan ZW and Han RC:
Analysis of the expression of the Notch3 receptor protein in adult
lung cancer. Oncol Lett. 5:499–504. 2013.
|
|
24
|
Ye YZ, Zhang ZH, Fan XY, Xu XL, Chen ML,
Chang BW and Zhang YB: Notch3 overexpression associates with poor
prognosis in human non-small-cell lung cancer. Med Oncol.
30:5952013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Konishi J, Kawaguchi KS, Vo H, Haruki N,
Gonzalez A, Carbone DP and Dang TP: Gamma-secretase inhibitor
prevents Notch3 activation and reduces proliferation in human lung
cancers. Cancer Res. 67:8051–8057. 2007. View Article : Google Scholar
|
|
26
|
Zhou L, Zhang N, Song W, et al: The
significance of Notch1 compared with Notch3 in high metastasis and
poor overall survival in hepatocellular carcinoma. PLoS One.
8:e573822013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Giovannini C, Gramantieri L, Minguzzi M,
Fornari F, Chieco P, Grazi GL and Bolondi L: CDKN1C/p57 is
regulated by the Notch target gene Hes1 and induces senescence in
human hepatocellular carcinoma. Am J Pathol. 181:413–422. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fabregat I: Dysregulation of apoptosis in
hepatocellular carcinoma cells. World J Gastroenterol. 15:513–520.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gramantieri L, Giovannini C, Lanzi A, et
al: Aberrant Notch3 and Notch4 expression in human hepatocellular
carcinoma. Liver Int. 27:997–1007. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhong D, Huang G, Zhang Y, et al:
MicroRNA-1 and microRNA-206 suppress LXRα-induced lipogenesis in
hepatocytes. Cell Signal. 25:1429–1437. 2013.PubMed/NCBI
|
|
31
|
Maciotta S, Meregalli M, Cassinelli L, et
al: Hmgb3 is regulated by MicroRNA-206 during muscle regeneration.
PLoS One. 7:e434642012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yan D, da Dong XE, Chen X, et al:
MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma
development. J Biol Chem. 284:29596–29604. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang W, Prince CZ, Mou Y and Pollman MJ:
Notch3 signaling in vascular smooth muscle cells induces c-FLIP
expression via ERK/MAPK activation. Resistance to Fas
ligand-induced apoptosis. J Biol Chem. 277:21723–21729. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang DH, Hu JR, Wang LY, et al: The
apoptotic function analysis of p53, Apaf1, Caspase3 and Caspase7
during the spermatogenesis of the Chinese fire-bellied newt
Cynops orientalis. PLoS One. 7:e39920,2012. View Article : Google Scholar
|
|
35
|
Chen L, Zheng J, Zhang Y, et al:
Tumor-specific expression of microRNA-26a suppresses human
hepatocellular carcinoma growth via cyclin-dependent and
-independent pathways. Mol Ther. 19:1521–1528. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
37
|
Chen RX, Xia YH, Xue TC, Zhang H and Ye
SL: Down-regulation of osteopontin inhibits metastasis of
hepatocellular carcinoma cells via a mechanism involving MMP-2 and
uPA. Oncol Rep. 25:803–808. 2011.PubMed/NCBI
|
|
38
|
Yeh CB, Hsieh MJ, Hsieh YS, Chien MH, Lin
PY, Chiou HL and Yang SF: Terminalia catappa exerts
antimetastatic effects on hepatocellular carcinoma through
transcriptional inhibition of matrix metalloproteinase-9 by
modulating NF-κB and AP-1 activity. Evid Based Complement Alternat
Med. 2012:5952922012.PubMed/NCBI
|
|
39
|
Li J, Lau G, Chen L, et al: Interleukin 23
promotes hepatocellular carcinoma metastasis via NF-kappa B induced
matrix metalloproteinase 9 expression. PLoS One. 7:e462642012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang S and He X: The role of microRNAs in
liver cancer progression. Br J Cancer. 104:235–240. 2010.
View Article : Google Scholar
|
|
41
|
Li Y, Hong F and Yu Z: Decreased
expression of microRNA-206 in breast cancer and its association
with disease characteristics and patient survival. J Int Med Res.
41:596–602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang Q, Zhang C, Huang B, Li H, Zhang R,
Huang Y and Wang J: Downregulation of microRNA-206 is a potent
prognostic marker for patients with gastric cancer. Eur J
Gastroenterol Hepatol. 25:953–957. 2013. View Article : Google Scholar : PubMed/NCBI
|