Introduction
Prostate cancer is the most common cancer in men in
Western countries (1).
Castrate-resistant or androgen-independent prostate cancer (AIPC)
is a more aggressive form seen later in the disease process, and by
definition, is more resistant to therapeutic intervention (2). Many of the general treatment
strategies for this type of prostate cancer involve androgen
deprivation by a variety of strategies such as luteinizing
hormone-releasing hormone agonists, anti-androgens, estrogens,
orchiectomy and drugs preventing both intratumoral and adrenal
gland androgen production (3).
Since almost all prostate cancers eventually develop castrate
resistance it is critically important to understand the mechanisms
leading to the progression to AIPC, with the hope of discovering
new effective therapeutic methods. In that direction, microRNAs and
their regulators have become an attractive area of research.
MicroRNAs are small non-coding molecules of RNA
(4). They have been shown to
regulate gene expression of proteins that participate in
tumorigenesis, cell cycle regulation, stress response,
inflammation, differentiation, apoptosis and metastasis (4). MicroRNAs are conserved from plants to
human and are encoded by their own genes. miRNA genes are localized
in separate gene loci, or they can be found within introns and
exons of other genes. The maturation process of microRNAs
implicates transcription, nuclear export and cleavage leading to
18–22 nucleotide double-stranded RNA molecules that enter a
cytoplasmic protein complex to regulate gene expression at the
post-transcriptional level (5,6).
miRNAs can modulate entire gene programs. They do not intercept a
single target as in the case of selective protein inhibitors
(4). Examinations of the
regulatory mechanism of the genome to discover RNAs that can
interfere between transcription and translation stages of protein
synthesis are necessary to understand the progression of
androgen-independent prostate cancer and equally important to
develop new therapeutic procedures to treat this disease.
The Lin28 protein family acts as RNA binding
proteins and microRNA regulators (7,8). The
genes that code for human Lin28A and Lin28B, the two known members
of this protein family, are localized on different chromosomes,
1p36.1 (Gene ID 79727) and 6q21 (Gene ID 389421), respectively.
Following their discovery, published literature clearly shows that
Lin28A and Lin28B have different cellular functions (9). Lin28B has been shown to be
tumorigenic in a prostate cancer mouse model (10) but the role of Lin28B in
androgen-independent prostate cancer is unknown.
Lin28B is expressed in all grades of prostatic
carcinomas and prostate cancer cell lines, but not in normal
prostate tissue. We found that Lin28B co-localized in the nucleus
and cytoplasm of the DU145 androgen-independent prostate cancer
cells. Also, the expression of Lin28B protein positively correlated
with the expression of the c-Myc protein in prostate cancer cells.
Furthermore, the silencing of Lin28B also correlated with a lower
expression of c-Myc protein, but not with the downregulation of
c-Myc messenger RNA. MiR-212 and miR-2278 seems to be the most
upregulated microRNAs upon Lin28B silencing by siRNA. Prior reports
have found that miR-212 is suppressed in prostate cancer tissues
but not in normal prostate tissues (9). Therefore, our results may suggest
that Lin28B is an oncogene suppressing miR-212 expression in
androgen-independent prostate cancer cells. On the other hand,
miR-2278 has not been study in cancer or other disease states. Our
analysis using the Target Scan shows that only miR-212 could target
the mRNA of Lin28B. Members of the Lin28 protein family are RNA
binding proteins that act as regulators of microRNAs (7) and crystallography and modeling work
in our laboratory suggests that Lin28B has unique and specific
interactions with microRNAs.
Materials and methods
Cells, antibodies and the chemical
inhibitor
Human prostate cancer cell lines (VCaP, vertebral
metastasis from prostate; LNCaP, prostate left supraclavicular
lymph node; PC3, prostate-bone; and DU145, prostate-brain) were
purchased from American Type Culture Collection (ATCC, Manassas,
VA, USA; catalog nos. CRL-2876, CRL-1740, CRL-1435 and HTB-81,
respectively). VCaP, PC3 and DU145 cells were maintained in
Dulbecco’s modified Eagle’s medium (DMEM). RPMI-1640 medium was
used for LNCaP cells. In all cases, the medium was supplemented
with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin
solution.
Primary antibodies used for fluorescent-activated
cell sorting (FACS) and western blotting were human Lin28B and
β-actin (Cell Signaling, Danvers, MA, USA; catalog nos. 4196S,
2496S and 3700, respectively). The human c-Myc antibody was
purchased from Santa Cruz Biotechnology (Dallas, TX, USA; catalog
no. sc-788). Lin28B antibody used for the immunofluorescence study
was purchased from Santa Cruz Biotechnology (catalog no.
sc-130802).
Transfection of prostate cancer cells
using siRNA
For the transient transfection experiments, the
DU145 AIPC cells were transfected with ON-TARGETplus siRNA which is
an exceptional choice for optimal gene silencing and it may reduce
off-targets up to 90% compared to unmodified siRNA. We transfected
DU145 AIPC cells with the siRNA ON-TARGETplus smart pool, human
Lin28B 3ÚTR/ORF [a pool of four different siRNAs
(GGAUAUUCCAGUCGAUGUA, GCCCAUAAGUGUUAAUAGA, CAAGCGUAUUGCAGCAUUA and
CCAGAGAGCUAGAAGUAUU)] or siRNA ON-TARGETplus non-targeting pool
(control for the transfection/silencing experiment), from Thermo
Scientific Rockford, IL, USA; catalog nos. L-028584-01-0005 or
D-001810-10-05, respectively. The Block-it transfection kit
(Invitrogen, Carlsbad, CA, USA; catalog no. 13570-070) was used
according to manufacturer suggestions.
FACS analysis and western blot
examination to evaluate Lin28B expression in prostate cancer cell
lines
FACS and western blot examinations were performed on
LNCaP, VCaP, PC3 and DU145 prostate cancer cell lines to determine
the expression of Lin28B and c-Myc proteins. In addition, FACS and
western blot examinations were made on DU145 cells transfected.
FACS
The transfected DU145 AIPC cells were treated with
trypsin (Sigma-Aldrich, St. Louis, MO, USA) and washed using cold
1× PBS. 500,000 cells were fixed in 2% methanol-free formaldehyde
(Thermo Scientific) and permeabilized with BD IntraSure kit
(Pharmingen, Pasadena, CA, USA; catalog no. 641776). Then, cells
were exposed to primary antibodies (Lin28B and c-Myc) for 1 h at
room temperature, followed by three washes with cold 1× PBS. Next,
cells were stained with Alexa Fluor-488 goat anti-rabbit IgG (H+L)
antibody (Molecular Probe, Carlsbad, CA, USA; catalog no. A11008).
Finally, cells were washed three times with cold 1× PBS and
suspended in 250 μl of 1× PBS. The samples were analyzed by FACS
using the LSR II instrument (BD Biosciences, San Jose, CA, USA) at
the core facility of the University of Kansas Medical Center. All
data were analyzed by FlowJo software.
Western blotting
Cell lysates from the transfected DU145 AIPC cells
were prepared by the addition of lysis buffer, containing 50 mm
Tris-HCl, pH 8.0, 0.1% SDS, 150 mM NaCl, 1% nonidet P-40, and a
protease inhibitor combination including 1 μg/ml aprotinin, 1 μg of
leupeptin and 1.0 mM PMSF. Equal amounts of protein were loaded to
10% SDS-PAGE and transferred to a nitrocellulose membrane.
Membranes were incubated with specific primary antibodies overnight
followed by secondary antibodies. Super Signal ULTRA
Chemiluminescent Substrate (Thermo Scientific) quantitated the
signals by using ID Image Analysis Software Version 3.6 (Eastman
Kodak Co., Rochester, NY, USA).
RNA isolation
Total RNA was extracted from the DU145 AIPC cells
upon transfection with and without Lin28B-siRNA using TRIzol
(Invitrogen; catalog no. 15596018) following the manufacturer’s
protocol.
GeneChip-miRNA 2.0 array
MicroRNA profiling was performed at the Genomics
Facility at the University of Kansas (Lawrence, KS) using
GeneChip-miRNA 2.0 array (Affymetrix, Santa Clara, CA, USA; catalog
no. 901754). For miRNA sample labeling, the Genisphere FlashTag
Biotin HSR RNA Labeling kit, GeneChip Eukaryotic Hybridization
Control kit and GeneChip Hybridization, Wash and Stain kit were
used (Affymetrix; catalog nos. 901910, 900454 and 900720,
respectively).
Real-time PCR
For validation of the overexpression of miR-212, a
Taqman microRNA reverse transcription kit was used to prepare cDNA
from the total RNA. The cDNA was used to run the real-time PCR
using Taqman universal PCR and a Taqman microRNA assay kit (Applied
Biosystem Step One real-time PCR system). PCR was completed for 15
sec at 95°C and 1 min at 60°C for 40 cycles. CT values for miR-212
were normalized to control RNU58A by subtracting the average CT
value for each sample. The relative quantification values for
miR-212 in each sample were determined using the 2−Δ ΔCT
method (11). Each PCR reaction
was performed in at least triplicate (mean ± SD). The fold
expression levels for Lin28B and c-Myc once Lin28B was silencing in
DU145 prostate cancer cells using siRNA was calculated by
ΔΔCT methods using GAPDH as an endogenous control
(following the instruction from Applied Biosystems) (11). Relative quantitation of gene
expression was analyzed with the comparative CT method
(11). Each ΔΔCT was
computed as a contrast in a one-way ANOVA analysis, and the p-value
testing the significance of the contrast was used to test the
significance of the corresponding fold change expression.
The genes targeted by miR-212 were predicted by
software packages available online from TargetScan (www.targetscan.org/) and microRNA (www.microrna.org/microrna/home.do).
Immunofluorescence
Protein expressions were evaluated by
immunofluorescence using Abcan protocols. Briefly,
paraffin-embedded sections from the prostate tissue array T191 and
PR243a (US Biomax, Inc., Rockville, MD, USA) were de-paraffinized
in xylene, hydrated with 100% ethanol and 95% ethanol for 5 and 1
min, respectively. Slides were then rinsed in distilled water and
samples were washed twice with ice cold PBS. For blocking
unspecific binding of the antibodies, samples were incubated with
1% BSA in PBST for 1 h. The anti-Lin28B primary antibody was
diluted in the blocking solution and incubated with the samples
overnight at 4°C. Slides were washed with 1× PBS and the secondary
antibody was added in blocking solution for 1 h at room
temperature, followed by three washes with 1× PBS and then the
addition of the mounting anti-fade solution with DAPI. The slides
were stored in the dark at 4°C. The expression of Lin28B was
evaluated in the DU145 prostate cancer cell line using the same
protocol for labeling. Fixation and permeabilization of the samples
were done using acetone for 10 min at room temperature.
Structure prediction calculations
Structure prediction calculations were performed
using the I-TASSER (12) server
without additional constraints or templates. The following amino
acid sequence of human Lin28B (NCBI Reference: NP_001004317.1) was
submitted online in September 2013:
MAEGGASKGGGEEPGKLPEPAEEESQVLRGTGHCKWFNVRMGFGFISMINREGSPLDIPVDVFVHQSKLFMEGFRSLKEGEPVEFTFKKSSKGLESIRVTGPGGSPCLGSERRPKGKTLQKRKPKGDRCYNCGGLDHHAKECSLPPQPKKCHYCQSIMHMVANCPHKNVAQPPASSQGRQEAESQPCTSTLPREVGGGHGCTSPPFPQEARAEISERSGRSPQEASSTKSSIAPEEQSKKGPSVQKRKKT.
Results
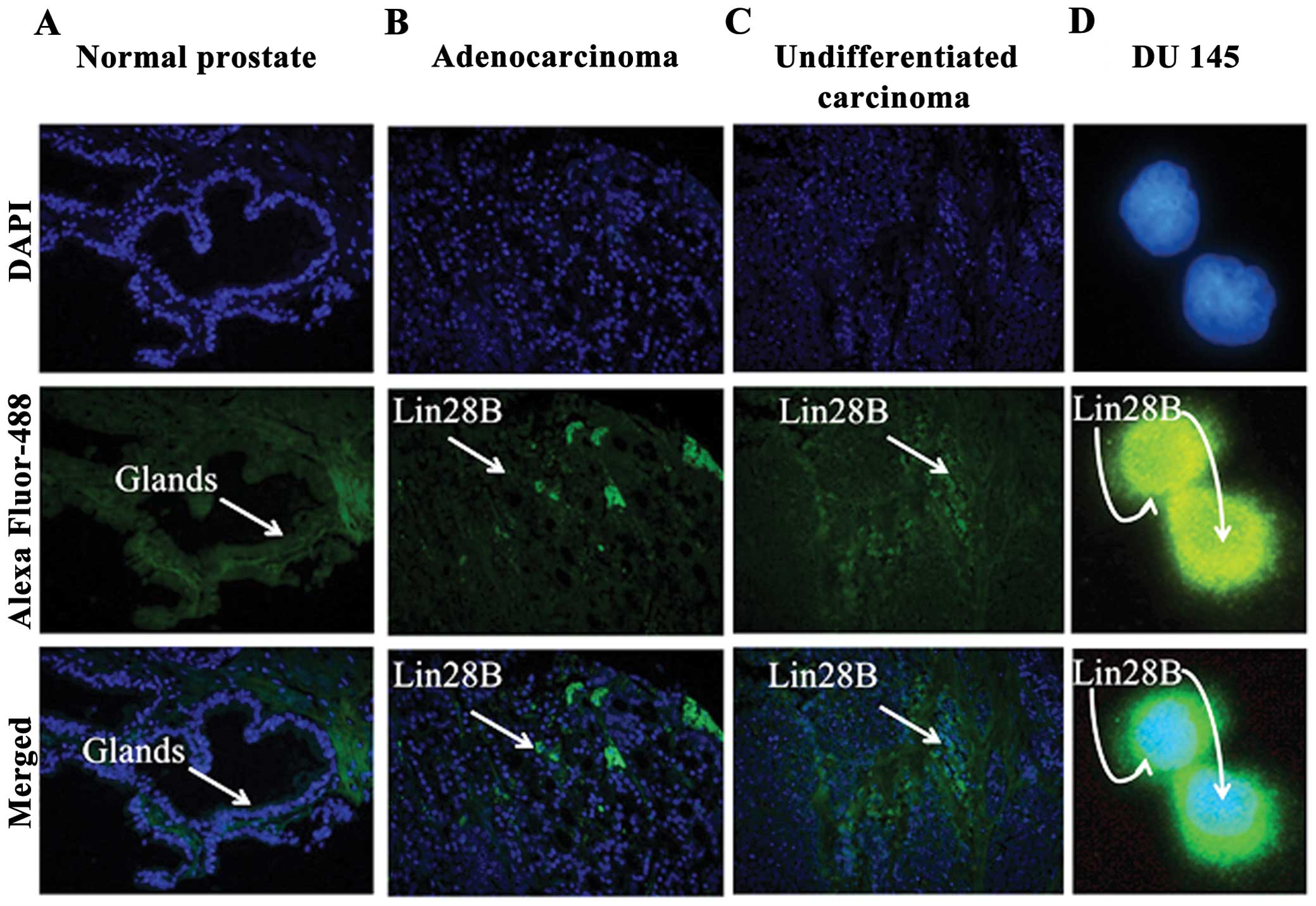
Lin28B is expressed in prostate cancer
tissues
It co-localized in the nucleus and cytoplasm of the
DU145 AIPC cells. Two prostate tissue arrays T191 and PR243a from
US Biomax, Inc., which includes normal prostate control tissues,
well to poorly differentiated adenocarcinomas and undifferentiated
carcinoma were used to identify the expression of Lin28B. Lin28B
expression was not detected in the glands of normal prostate
tissue. Lin28B is expressed in grade 2–3 prostate adenocarcinoma,
and undifferentiated grade 4 prostate carcinomas (Fig. 1A–C). Only selected panels from the
tissue arrays are shown. Lin28B co-localizes in the nucleus and in
the cytoplasm of the DU145 prostate cancer cell line, indicating
possible purpose in both cellular compartments (Fig. 1D).
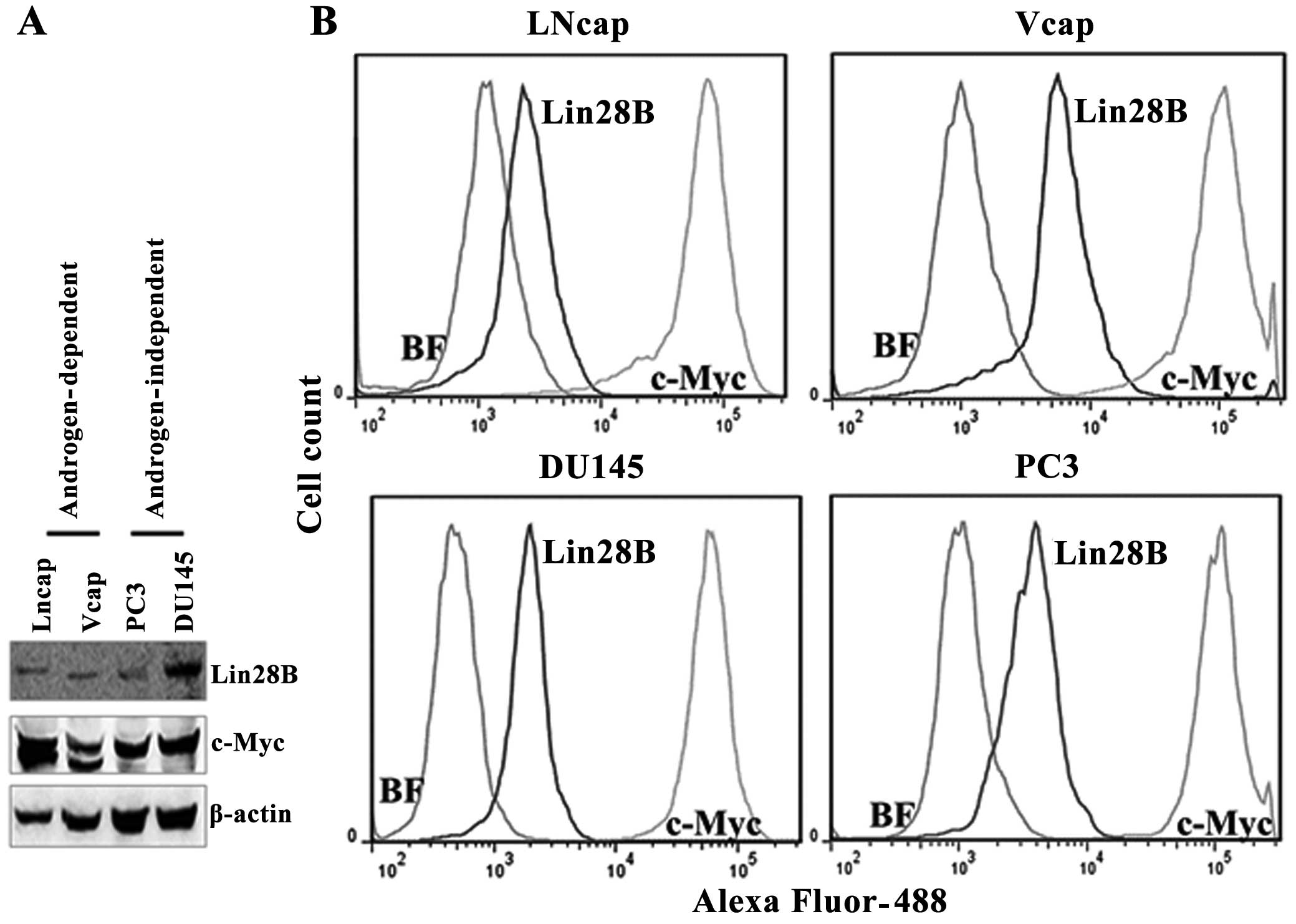
Lin28B protein expression correlates with
c-Myc protein expression in prostate cancer cell lines
Lin28B and c-Myc protein levels were detected by
western blotting in androgen-dependent (LNCaP and VCaP) and
androgen-independent prostate cancer cell lines (DU145 and PC3) by
western blotting, using β-actin to normalize the expression of the
proteins. The expression of Lin28B correlates with the expression
of c-Myc protein levels in these cell lines. But, the c-Myc
antibody detected two bands for c-Myc protein in the two
androgen-dependent cell lines and a single band in the
androgen-independent cell lines. The quantification of the protein
levels for Lin28B and c-Myc was also performed by flow cytometry
because western blotting is not a protein quantification tool. The
flow cytometry data indicated that Lin28B protein levels are 12.8,
84, 80.9 and 63.7% in LnCaP, VCaP, PC3 and DU145 cells,
respectively, in 10,000 cells counted (Fig. 2). The mean fluorescence of cells
expressing Lin28B was low (in the range of
103–104) whereas cells expressing c-Myc were
bright, in the range of 105 for all the cell lines. This
may indicate that copies of Lin28B/cell are lower than c-Myc. The
protein levels of c-Myc were the highest in all the cell lines
analyzed. Lin28B and c-Myc were detected in all the prostate cancer
cells investigated, regardless of androgen-dependence status.
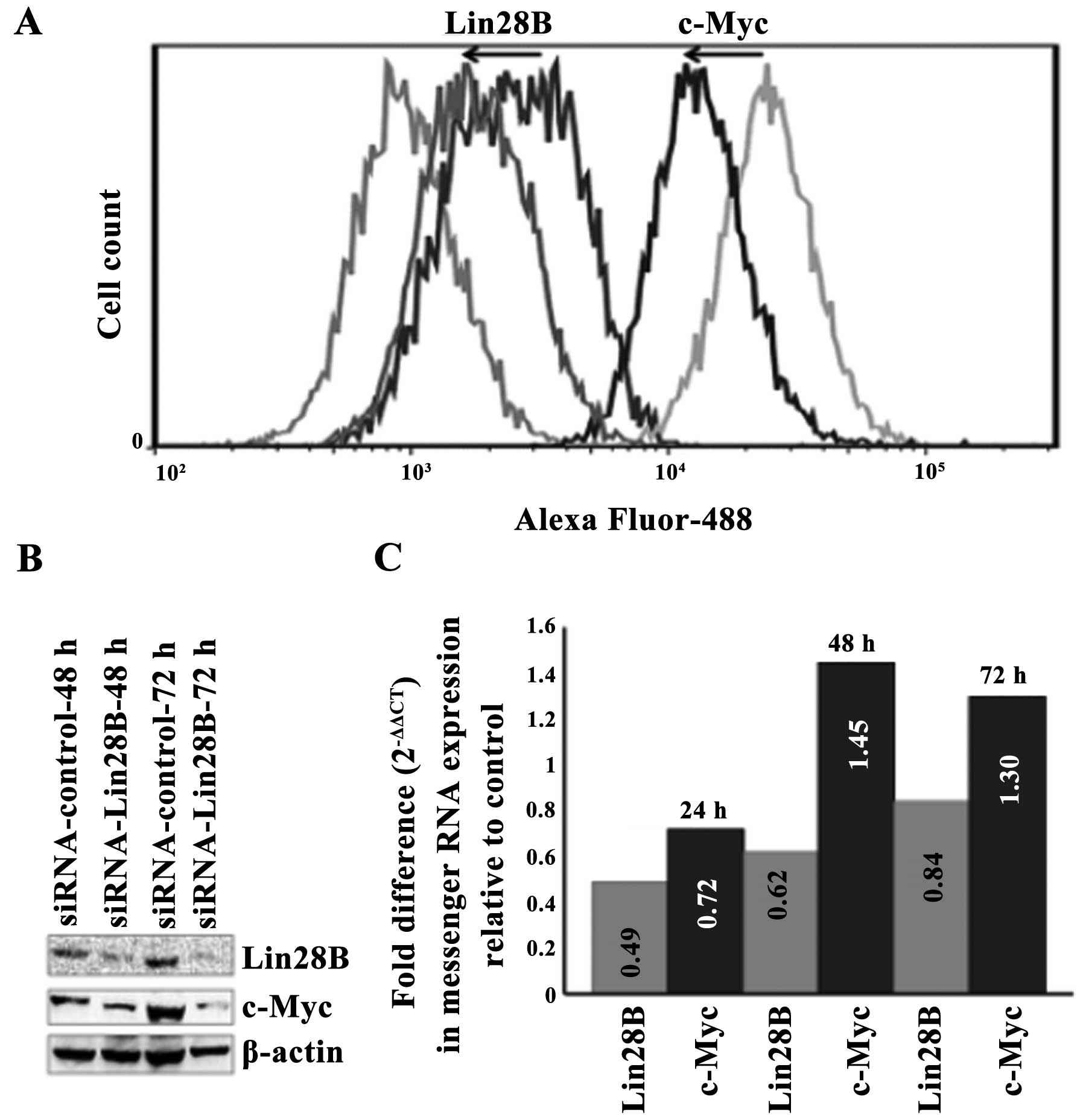
The silencing of Lin28B in the DU145 AIPC
cells correlates with the downregulation of c-Myc protein but not
with the downregulation of c-Myc messenger RNA
Lin28B was transiently silenced in the DU145 AIPC
cell line using a targeting pool of four siRNAs. The silencing of
Lin28B protein correlates with the downregulation of c-Myc protein
at 48 and 72 h as detected by western blotting (Fig. 3A). Quantification using flow
cytometry and data analysis by FlowJo software also show that the
silencing of the Lin28B protein correlates with the downregulation
of c-Myc protein at 48 h. The transient silencing of the Lin28B
gene decreased Lin28B protein by 42.6% and this compares with the
decrease of c-Myc protein by 58.6% at 42 h upon transfection with
Lin28B siRNA (Fig. 3B). The level
of c-Myc messenger RNA decreased at 24 h after Lin28B gene
silencing but it did not reach statistical significance and these
levels after 48 h (Fig. 3C), while
c-Myc protein remained downregulated at 48 and 72 h.
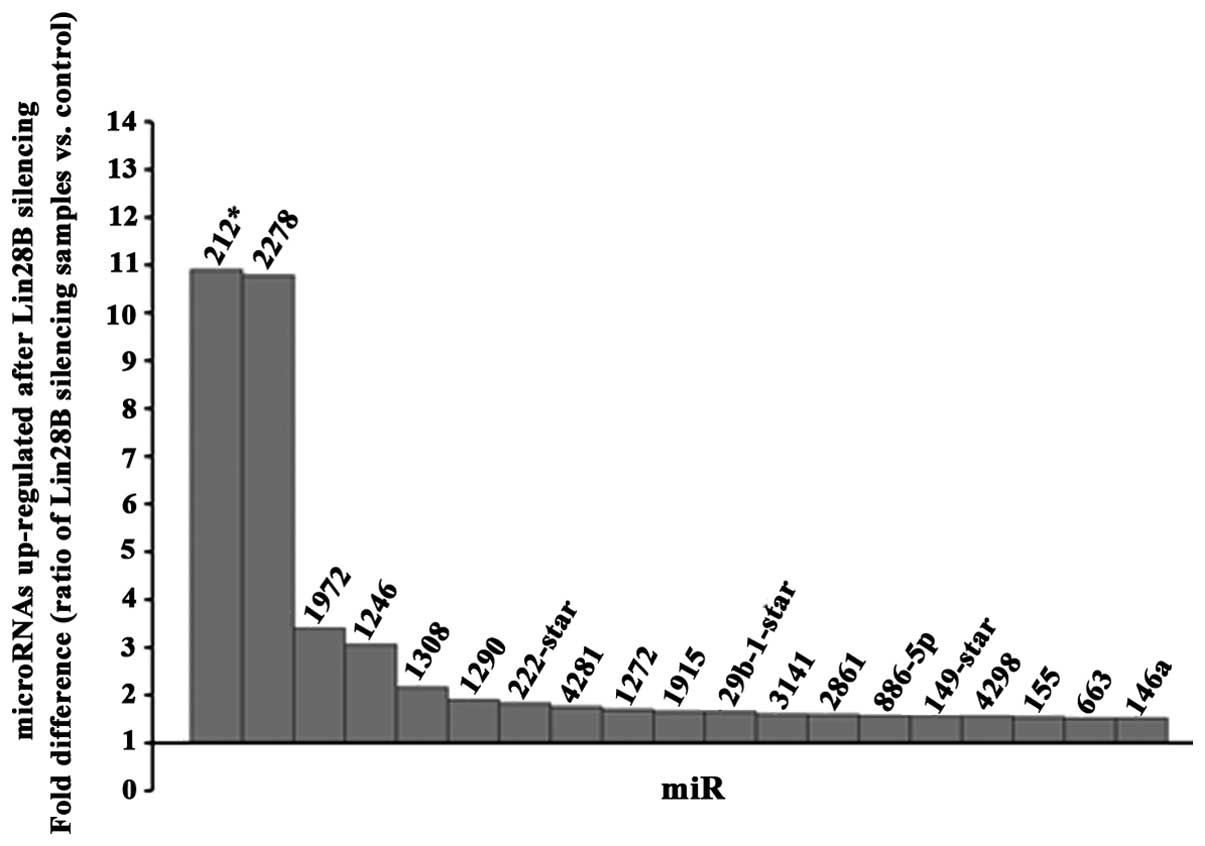
Antagonist effect of the RNA
binding-microRNA regulator Lin28B protein on miR-212 and miR-2278
microRNAs in DU145 AIPC cells
A microRNA profile (GeneChip hybridization) at 48 h
upon transfection of DU145 prostate cancer cells using siRNA
against Lin28B was determined. A threshold of 1.5-fold change up or
down (ratio of silencing samples vs. control) was used for the
final analysis. Among the 19 microRNAs found upregulated (Fig. 4) and 11 microRNAs downregulated
(Fig. 5), miR-212 and miR2278 were
the most upregulated. TargetScan (http://www.targetscan.org/) was used to detect which
of these microRNAs found altered in this study can potentially
target the messenger RNA of Lin28B and it revealed that among all
of these microRNAs found altered; only miR212, miR-181b and
miR-181c could target the messenger RNA of Lin28B. Because miR-212
was the most altered microRNA, it has never been linked
functionally to Lin28B in any cancer, and miR-212 was found
suppressed in prostate cancer tissues (13), we validated the upregulation of
miR-212 by real-time PCR at 24, 48 and 72 h after the Lin28B
silencing and the maximum upregulation of miR-212 was detected at
24 h (fold difference, 440.93). A test for contrast used revealed
that the upregulation of miR-212 was significant at all the time
points analyzed (Table I).
 | Table IValidation of the downregulation of
miR-212 at 24, 48 and 72 h upon Lin28B silencing in the DU145
androgen-independent prostate cancer cell line using real-time
PCR. |
Table I
Validation of the downregulation of
miR-212 at 24, 48 and 72 h upon Lin28B silencing in the DU145
androgen-independent prostate cancer cell line using real-time
PCR.
| Sample | Δ ΔCT = ΔCT treated
- ΔCT untreated | Fold difference:
miR-212 expression relative to control 2(−Δ ΔCT) | Test for
contrast |
|---|
| siLin28B |
| 24 h | −8.78 | 440.93 | F=6178.7; df=1, 12;
p<0.001 |
| 48 h | −3.78 | 13.69 | F=15.65; df=1, 26;
p<0.001 |
| 72 h | −3.73 | 13.269 | F=93.32; df=1, 15;
p<0.005 |
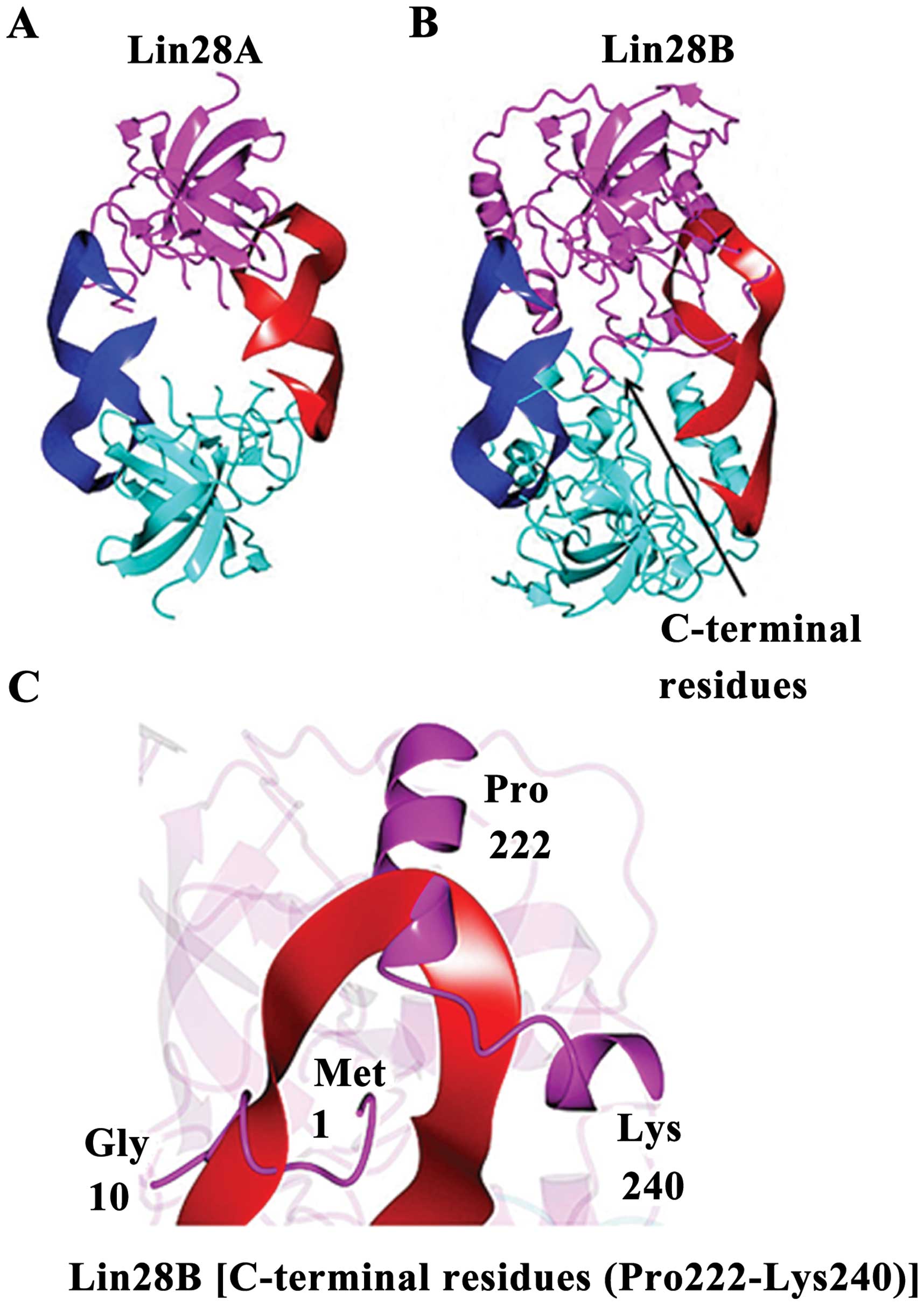
Lin28A and Lin28B have structural
differences
Lin28B shows unique microRNA binding
characteristics. The Lin28B structure predicted from I-TAS-SER is
shown in Fig. 6. A 2-fold
symmetric model was generated by the superposition of the Lin28A
structure (Fig. 6A) onto the
I-TASSER model of Lin28B. If Lin28B were to bind miRNAs in a manner
similar to Lin28A, conformational changes would be necessary,
relative to the I-TASSER model, to prevent steric clashes between
C-terminal residues of the putative Lin28B dimer (Fig. 6B). The C-terminal residues (Pro
222-Lys 240) downstream of the ZNF2 domain as well as the
N-terminal residues (Met1 to Gly 10) would also need to adopt a
different conformation than that observed for the I-TASSER model of
Lin28B in order to accommodate RNA binding or the RNA would need to
adopt a different binding mode relative to the Lin28A structure
(Fig. 6C).
Discussion
We report novel findings regarding the expression,
function and structure of Lin28B and its connection to the microRNA
pathways in AIPC. Lin28B is a member of the Lin28 protein family,
whose gene is localized on chromosome 6. Lin28B expression has
recently been detected in the nuclei and cytosol of prostate cancer
cells; with no substantial changes in its expression with
increasing tumor aggressiveness as measured by Gleason score
(5). Similarly, we found Lin28B is
expressed in prostate adenocarcinomas (irrespective of Gleason
grade) but not in the glands of normal prostate tissue. In
addition, we found strong nuclear staining in the nuclei and in the
cytosol of DU145 prostate cancer cells. Lin28B has been presented
to be tumorigenic in a prostate cancer mouse model (10) but the role of Lin28B in
androgen-independent prostate cancer is unidentified. MicroRNA
profiling in the androgen-independent prostate cancer cell line
DU145 performed after silencing Lin28B by siRNA revealed up- and
downregulations of several microRNAs with these most alterated
being miR-212 and miR-2278. MiR-212 upregulation correlated with a
decreased Lin28B messenger RNA and protein levels upon Lin28B
silencing. This inverse relationship between Lin28B and miR-212 and
its implication in prostate cancer development has not been
previously reported. Our findings correspond with a recent
publication by Walter and colleagues demonstrating the absence of
miR-212 in prostate tumors, as compared to its normal expression in
neighboring normal epithelium and/or stroma (13). In addition, miR-212 down-regulation
has been described in lung cancer and is associated with the
severity of the disease (14).
TargetScan predictions display that Lin28B messenger RNA may be a
target of miR-212 but not a target of miR-2278. Based on our
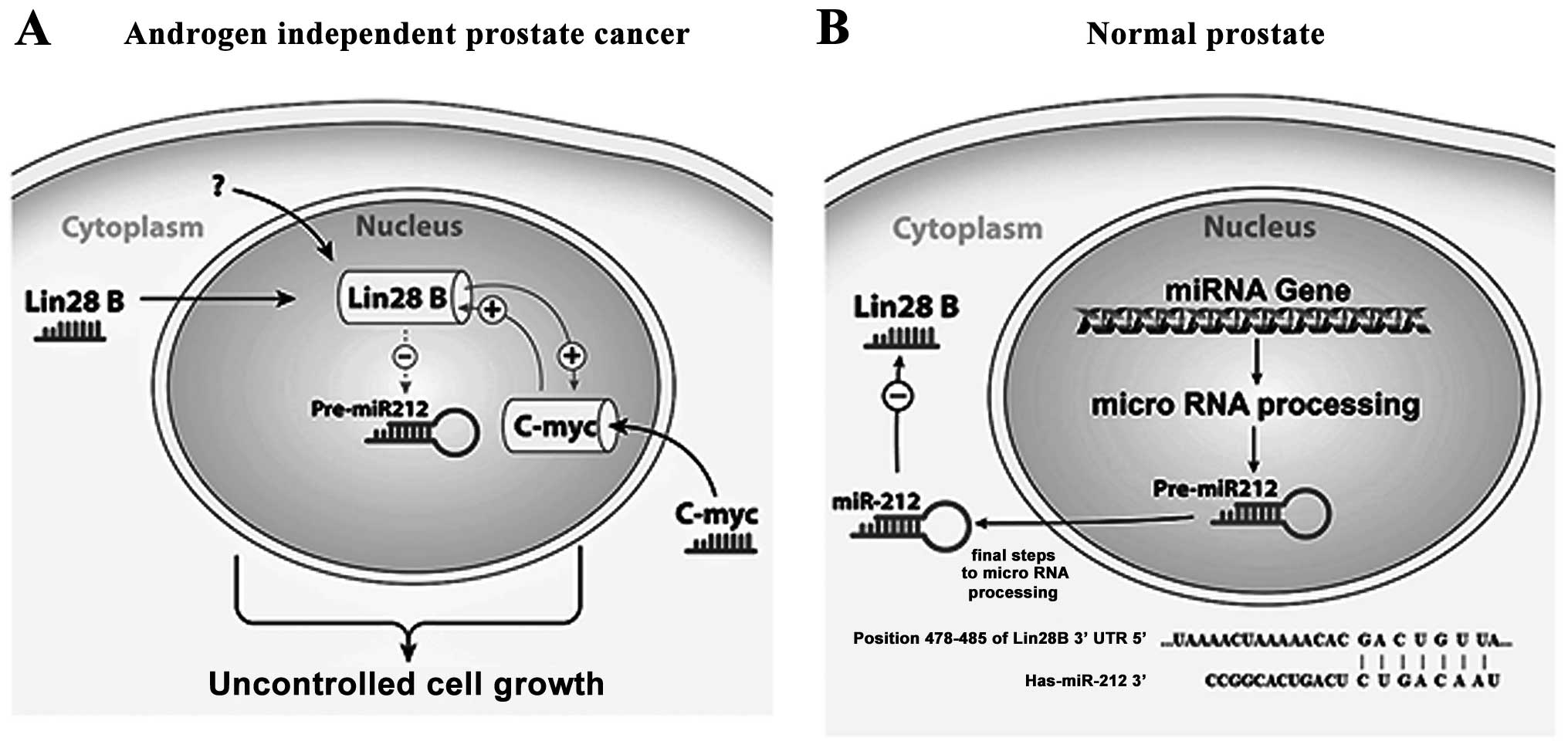
results, we hypothesize that Lin28B inhibits the tumor suppressor
activity of miR-212 in AIPC (Fig.
7A), potentially playing a role in prostate carcinogenesis or
progression, while in normal prostate miR-212 suppresses the
expression of Lin28B by targeting it mRNA (Fig. 7B). The published data obviously
suggest an important role of miR-212 in the carcinogenesis process
in general, but the microRNA profile we analyzed shows Lin28B as an
oncogene regulator involving more than one pathway. Some of the
microRNAs upregulated upon Lin28B silencing have never been
associated with Lin28B. TargetScan did not predict them as
biological Lin28B targets (by searching for the presence of
conserved sites). It is possible that Lin28B knock/down activates
other molecular pathways not yet described, inducing alteration of
these microRNAs in prostate cancer cells, relating them to
Lin28B.
Correlating well with previous data, miRNAs found to
be upregulated upon Lin28B silencing in our present study include
miR-212, downregulated in prostate cancer (13), and miR-146 which has been reported
to suppresses tumor growth and progression in castration-resistant
prostate cancer (15).
Additionally, miR-1246 and miR-1308, are potential diagnostic
biomarkers in multiple myeloma (16), and miR-1290 has been linked with
the estrogen receptor-positive breast cancer phenotype (17).
Differing to our data, miR-4281, miR29b-1, miR-2861
have been reported to be upregulated in malignant melanoma
(18), head and neck cancers
(19) and thyroid cancer (20). Conversely, in our study they were
only upregulated upon Lin28B silencing. This was also true for
miR-1915 and miR-663, which have been associated with drug-therapy
resistance in colorectal carcinoma (21) and breast cancer (22). MiR-4298 has been linked with the
uncontrolled growth of gastric cancer cells (23). MiR-886-5p is an inhibitor of
apoptosis in cervical cancer cells (24). Additionally, miR-149 was linked to
the inhibition of non-small cell lung cancer (25) and miR-155 has been reported as a
prognostic marker in chronic lymphocytic leukemia (25).
Lin28B has been linked to the negative targeting of
the let-7 family of microRNAs in cancer and in prostate cancer
(10,26) but, we did not find any evidence
that the silencing of Lin28B in the DU145 AIPC cells affected any
members of the Let7 microRNA family (data not shown).
In addition, eleven microRNAs were downregulated
after Lin28B was silencing. MiR-335 (13) and miR-181c (27) have been found to be upregulated in
prostate and gastric cancers, respectively. MiR-30e induces
invasiveness of human glioma cells (28). Contradictorily, miR-34a
overexpression was associated with the inhibition of prostate
cancer cell growth (29), and
miR-181b and miR-146b-5p have also been reported to be
downregulated in prostate cancer (30,31).
Also, miR-542-5p, reported as a tumor suppressor in neuroblastoma
(32), was downregulated. A
polymorphism at miR-629 was connected to the increased risk of lung
cancer in Southern and Eastern Chinese population (33). Finally, from the thirty microRNAs
found altered in this study, the biological functions of miR-2278,
miR-1972, miR-1272, miR-3141 and miR-3172 have never been reported.
These results need to be further validated to find each of the
potential oncogenic pathways as it pertains specifically to
prostate cancer development.
Human Lin28B is a 27-kDa protein that has been
implicated in messenger RNA and microRNA binding (7,34).
Lin28B shares highly sequence identity with Lin28A whose structure
has been determined in complex with microRNAs (35). The resulting model using I-TASSER
in our study suggests that Lin28B contains CSD/ZNF domains, similar
to Lin28A, which one would expect to be implicated in the binding
of nucleic acids as well. However, it also shows unique microRNA
binding characteristics for Lin28B. If Lin28B were to bind miRNAs
in a manner similar to Lin28A, conformational changes would be
necessary to prevent steric clashes in the C-terminal and linker
regions between the CSD and ZNF domains.
Our results provide additional data on the function
of Lin28B and its interaction with microRNAs in AIPC. Our work also
suggests structural differences between Lin28A and Lin28B, and a
specific and unique interaction between Lin28B and microRNAs. We
are proposing for the first time a novel oncogenic pathway in
prostate carcinogenesis, involving Lin28B expression, miR-212
downregulation and activation of c-Myc oncogenic programs leading
to the development of androgen-independent prostate cancer. There
are few publications addressing the role of Lin28B in prostate
cancer (10,36–38),
and there are no publications addressing the function of
Lin28B-microRNAs (different than let7)-c-Myc pathway in prostate
cancer. To date, there are no reports in the literature regarding
the function of miR-2278 in cancer or other disease states. But,
our data using target scan shows no reciprocity between
Lin28B:miR-2278. Therefore, for the first time; we are reporting a
potential regulatory loop formed between Lin28B:miR-212 to regulate
c-Myc in AIPC. This work may lead to the identification of a unique
role of these molecules in the prostate cancer. Furthermore, the
elucidation of the Lin28B-miR-212-c-Myc pathway may lead to new
therapeutic approaches in the management of androgen-independent
prostate cancer.
For the first time, we are proposing a unique
nucleotide binding feature for Lin28B. Lin28B protein was found
overexpressed in prostate adenocarcinoma tissue, regardless the
grade or Gleason score, and in prostate cancer cell lines but not
in normal prostate cancer tissues. We are showing an oncogenic
pathway in prostate cancer of Lin28B overexpression involving
miR-212 downregulation and increased levels of c-Myc protein, but
not c-Myc messenger RNA. The messenger RNA of Lin28B was predicted
to be a target of miR-212. Our findings open a new possible avenue
for the study, understanding and treatment of androgen-independent
prostate cancer. More studies are needed to further characterize
this Lin28B-miR-212-c-Myc oncogenic pathway.
Acknowledgements
This study was primarily supported by the Basic
Research Grant (2011–2013) from the KUMC Department of Internal
Medicine with additional support by the Midwest Biomedical Research
Foundation (Cynthia Bruce and Virginia Gross) and the Heartland
Institute for Clinical and Translational Research-Curriculum
Program NIH Clinical and Translational Science Award
(UL1TR000001-02). We acknowledge the support of the Division of
Hematology/Oncology, Department of Internal Medicine, University of
Kansas Medical Center (KUMC) including Chao Huang, Nisreen Haideri
and Bea Colton. We thank Dr Andrew Godwin for the scientific
discussions and manuscript review. We appreciate the support of Dr
Rama Sharma, Dr Mukut Sharma, Sarah Spencer, Margarita Thakur,
Nathan McGee, Nishi Chavarkar and Bob Moreno at the Veteran
Administration Medical Center Kansas City, MO, who all provided
valuable insight and administrative support. We acknowledge the
Flow Cytometry Core Laboratory and Richard Hastings at the
University of Kansas Medical Center, which is sponsored, in part,
by the NIH/NIGMS COBRE grant P30 GM103326. In addition, we
acknowledge Stanton Fernald, KIDDRC Imaging Center, 3019 HLSIC and
University of Kansas Medical Center. Use of the University of
Kansas Protein Structure Laboratory was supported by grants from
the National Center for Research Resources (5P20RR017708-10) and
the National Institute of General Medical Sciences
(8P20GM103420-10).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Crawford ED, Stone NN, Yu EY, et al:
Challenges and recommendations for early identification of
metastatic disease in prostate cancer. Urology. 83:664–669. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Trendel JA: The hurdle of antiandrogen
drug resistance: drug design strategies. Expert Opin Drug Discov.
8:1491–1501. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mirnezami AH, Pickard K, Zhang L, Primrose
JN and Packham G: MicroRNAs: key players in carcinogenesis and
novel therapeutic targets. Eur J Surg Oncol. 35:339–347. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ritchie W, Rasko JE and Flamant S:
MicroRNA target prediction and validation. Adv Exp Med Biol.
774:39–53. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lovat F, Valeri N and Croce CM: MicroRNAs
in the pathogenesis of cancer. Semin Oncol. 38:724–733. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hafner M, Max KE, Bandaru P, et al:
Identification of mRNAs bound and regulated by human LIN28 proteins
and molecular requirements for RNA recognition. RNA. 19:613–626.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou J, Ng SB and Chng WJ: LIN28/LIN28B:
an emerging oncogenic driver in cancer stem cells. Int J Biochem
Cell Biol. 45:973–978. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gaytan F, Sangiao-Alvarellos S,
Manfredi-Lozano M, et al: Distinct expression patterns predict
differential roles of the miRNA-binding proteins, Lin28 and Lin28b,
in the mouse testis: studies during postnatal development and in a
model of hypogonadotropic hypogonadism. Endocrinology.
154:1321–1336. 2013. View Article : Google Scholar
|
|
10
|
Tummala R, Nadiminty N, Lou W, et al:
Lin28 promotes growth of prostate cancer cells and activates the
androgen receptor. Am J Pathol. 183:288–295. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
|
|
12
|
Roy A, Kucukural A and Zhang Y: I-TASSER:
a unified platform for automated protein structure and function
prediction. Nat Protoc. 5:725–738. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Walter BA, Valera VA, Pinto PA and Merino
MJ: Comprehensive microRNA profiling of prostate cancer. J Cancer.
4:350–357. 2013. View
Article : Google Scholar
|
|
14
|
Incoronato M, Urso L, Portela A, et al:
Epigenetic regulation of miR-212 expression in lung cancer. PLoS
One. 6:e277222011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu B, Wang N, Wang X, et al: MiR-146a
suppresses tumor growth and progression by targeting EGFR pathway
and in a p-ERK-dependent manner in castration-resistant prostate
cancer. Prostate. 72:1171–1178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jones CI, Zabolotskaya MV, King AJ, et al:
Identification of circulating microRNAs as diagnostic biomarkers
for use in multiple myeloma. Br J Cancer. 107:1987–1996. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Endo Y, Toyama T, Takahashi S, et al:
miR-1290 and its potential targets are associated with
characteristics of estrogen receptor alpha-positive breast cancer.
Endocr Relat Cancer. 20:91–102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sand M, Skrygan M, Sand D, et al:
Comparative microarray analysis of microRNA expression profiles in
primary cutaneous malignant melanoma, cutaneous malignant melanoma
metastases, and benign melanocytic nevi. Cell Tissue Res.
351:85–98. 2013. View Article : Google Scholar
|
|
19
|
Nurul-Syakima AM, Yoke-Kqueen C, Sabariah
AR, Shiran MS, Singh A and Learn-Han L: Differential microRNA
expression and identification of putative miRNA targets and
pathways in head and neck cancers. Int J Mol Med. 28:327–336.
2011.PubMed/NCBI
|
|
20
|
Wang Z, Zhang H, Zhang P, Li J, Shan Z and
Teng W: Upregulation of miR-2861 and miR-451 expression in
papillary thyroid carcinoma with lymph node metastasis. Med Oncol.
30:5772013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu K, Liang X, Cui D, Wu Y, Shi W and Liu
J: miR-1915 inhibits Bcl-2 to modulate multidrug resistance by
increasing drug-sensitivity in human colorectal carcinoma cells.
Mol Carcinog. 52:70–78. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu H, Li S, Cui X, et al: The
overexpression of hypomethylated miR-663 induces chemotherapy
resistance in human breast cancer cells by targeting heparin
sulfate proteoglycan 2 (HSPG2). J Biol Chem. 288:10973–10985. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
An J, Pan Y, Yan Z, et al: MiR-23a in
amplified 19p13.13 loci targets metallothionein 2A and promotes
growth in gastric cancer cells. J Cell Biochem. 114:2160–2169.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li JH, Xiao X, Zhang YN, et al: MicroRNA
miR-886-5p inhibits apoptosis by down-regulating Bax expression in
human cervical carcinoma cells. Gynecol Oncol. 120:145–151. 2011.
View Article : Google Scholar
|
|
25
|
Ferrajoli A, Shanafelt TD, Ivan C, et al:
Prognostic value of miR-155 in individuals with monoclonal B-cell
lymphocytosis and patients with B chronic lymphocytic leukemia.
Blood. 122:1891–1899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Oh JS, Kim JJ, Byun JY and Kim IA:
Lin28-let7 modulates radiosensitivity of human cancer cells with
activation of K-Ras. Int J Radiat Oncol Biol Phys. 76:5–8. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cui MH, Hou XL, Lei XY, et al:
Upregulation of microRNA 181c expression in gastric cancer tissues
and plasma. Asian Pac J Cancer Prev. 14:3063–3066. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang L, Lin C, Song L, et al:
MicroRNA-30e* promotes human glioma cell invasiveness in
an orthotopic xenotransplantation model by disrupting the
NF-κB/IκBα negative feedback loop. J Clin Invest. 122:33–47.
2012.
|
|
29
|
Kashat M, Azzouz L, Sarkar SH, Kong D, Li
Y and Sarkar FH: Inactivation of AR and Notch-1 signaling by
miR-34a attenuates prostate cancer aggressiveness. Am J Transl Res.
4:432–442. 2012.PubMed/NCBI
|
|
30
|
Schaefer A, Jung M, Mollenkopf HJ, et al:
Diagnostic and prognostic implications of microRNA profiling in
prostate carcinoma. Int J Cancer. 126:1166–1176. 2010.PubMed/NCBI
|
|
31
|
Man YG, Fu SW, Liu AJ, et al: Aberrant
expression of chromogranin A, miR-146a, and miR-146b-5p in prostate
structures with focally disrupted basal cell layers: an early sign
of invasion and hormone-refractory cancer? Cancer Genomics
Proteomics. 8:235–244. 2011.PubMed/NCBI
|
|
32
|
Bray I, Tivnan A, Bryan K, et al:
MicroRNA-542-5p as a novel tumor suppressor in neuroblastoma.
Cancer Lett. 303:56–64. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang L, Li Y, Cheng M, et al: A functional
polymorphism at microRNA-629-binding site in the 3′-untranslated
region of NBS1 gene confers an increased risk of lung cancer in
Southern and Eastern Chinese population. Carcinogenesis.
33:338–347. 2012.PubMed/NCBI
|
|
34
|
Ali PS, Ghoshdastider U, Hoffmann J,
Brutschy B and Filipek S: Recognition of the let-7g miRNA precursor
by human Lin28B. FEBS Lett. 586:3986–3990. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nam Y, Chen C, Gregory RI, Chou JJ and
Sliz P: Molecular basis for interaction of let-7 microRNAs with
Lin28. Cell. 147:1080–1091. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nadiminty N, Tummala R, Lou W, et al:
MicroRNA let-7c is downregulated in prostate cancer and suppresses
prostate cancer growth. PLoS One. 7:e328322012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vencio EF, Nelson AM, Cavanaugh C, et al:
Reprogramming of prostate cancer-associated stromal cells to
embryonic stem-like. Prostate. 72:1453–1463. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kong D, Banerjee S, Ahmad A, et al:
Epithelial to mesenchymal transition is mechanistically linked with
stem cell signatures in prostate cancer cells. PLoS One.
5:e124452010. View Article : Google Scholar : PubMed/NCBI
|