Introduction
Gastric adenocarcinoma (GC) is still one of the most
common and aggressive carcinomas worldwide, especially in China
where it ranks third in incidence and also third in mortality rate
in malignant tumors, with 420,489 new cases and 297,496 deaths
including 206,704 males and 90,792 females in 2011 (1). In the United States, the estimated
new cases and deaths of gastric cancer are 24,590 and 10,720,
respectively, in 2015 (2). Hence,
the burden of gastric cancer is increasing. However, the mechanisms
of the gastric cancer tumorigenesis is still unclear, to explore
the molecular mechanism and therefore the potential therapeutic
targets is of great importance for gastric cancer patients.
Epigenetic modifications have been found to be
involved in the tumorigenesis, development and progression of
gastric cancer, such as promoter DNA hypermethylation of tumor
suppressors as well as post-translational alterations of histones
(3). Previous studies showed that
depletion of histone deacetylase is an effective way to inhibit the
proliferation, promote cell cycle arrest and induce apoptosis of
multiple malignant tumors (4),
including breast (5), pancreatic
(6), prostate (7), colorectal (8), hepatocarcinoma (9), lung cancer (10), leuchemia (11), gastric cancer (12), gliomas (13), and cervical cancer (14) which suggested that HDACs are the
potential therapeutic targets for treatment of cancer.
The histone deacetylase (HDAC) family consists of 18
members (15), which are grouped
into four separate classes, class I, HDAC1, HDAC2, HDAC3 and HDAC8;
class II, HDAC4, HDAC5, HDAC6, HDAC7, HDAC9 and HDAC10; class IV,
HDAC11; and 7 sirtuins (4,16). The class I HDACs (HDAC1, 2, 3 and
8) are most closely related to the yeast (Saccharomyces
cerevisiae) transcriptional regulator RPD3. HDACs function as
key regulators of chromatin structure and post-translational
modifiers of numerous key proteins in any cell type and tissue
(4,17,18).
Although HDAC1-3 and 6 were highlighted in most disease-oriented
research (19), lately HDAC8 has
become increasingly important as a drug target (20,21).
HDAC8 is expressed in multiple adult malignant tumor entities,
including lung, colon, pancreas and breast as well as in childhood
tumor entities such as neuroblastoma (19). Depletion of HDAC8 by small
interference RNA (siRNA) transfection inhibits proliferation of
human lung, colon, and cervical cancer cell lines, moreover, the
forced expression of HDAC8 promotes proliferation and inhibits
apoptosis in hepatocellular carcinoma (22,23).
It has been reported that HDAC8 mediated regulation of
Bcl-2-modifying factor (BMF) via cooperation with STAT3 (24). BMF plays an important role in the
execution of apoptosis triggered by the metabolite
methylselenopyruvate, which is an inhibitor of HDAC8 (25). However, the expression, function
and mechanism of HDAC8 in GC remain unclear.
In the present study, we found that the expression
of HDAC8 was significantly upregulated both in GC cell lines and
tumor tissues. HDAC8 knockdown significantly inhibited GC cell
proliferation, induced cell cycle arrest and cell apoptosis.
Furthermore, forced expression of HDAC8 promoted GC cell
proliferation, inhibited cell cycle arrest and cell apoptosis. In
addition, suppression of HDAC8 also resulted in the upregulation of
BMF transcription followed by increased expression of cleaved
caspase-3 and caspase-6.
Materials and methods
Ethics statement
Written informed consent was obtained from patients
before obtaining tissue samples. The procedures used in the present
study were approved by the Institutional Review Board of the First
Affiliated Hospital, Henan University of Science and Technology and
conformed to the Helsinki Declaration and to local legislation.
Patients and samples
GC tissues and their corresponding normal gastric
tissues from 51 GC patients treated with surgery in 2008 in our
hospital (First Affiliated Hospital, Henan University of Science
and Technology, Henan, China) were enrolled in the present study.
GC was confirmed by expert histopathologist examination in all
these patients. The clinicopathological characteristics of the 57
patients with gastric cancer are shown in Table I.
 | Table IThe relationship between the clinical
parameters and thHDAC8 (mean ± SEM) mRNA expression in primary
gastric adenocarcinoma. |
Table I
The relationship between the clinical
parameters and thHDAC8 (mean ± SEM) mRNA expression in primary
gastric adenocarcinoma.
| Clinical
parameters | N (%) | Relative expression
(mean ± SEM) | P-value |
|---|
| Age (years) | | | |
| ≥60 | 31 (60.8) | 4.04±0.27 | 0.63 |
| <60 | 20 (39.2) | 3.94±0.19 | |
| Gender | | | |
| Male | 36 (70.6) | 3.86±0.20 | 0.58 |
| Female | 16 (29.4) | 4.06±0.28 | |
| Size (cm) | | | |
| ≥5 | 29 (56.7) | 5.20±0.24 | <0.0001 |
| <5 | 22 (43.3) | 2.42±0.027 | |
| Histological
differentiation | | | |
| Well | 17 (33.3) | 2.12±0.11 | <0.0001 |
| Moderately | 17 (33.3) | 4.03±0.23 | |
| Poorly | 17 (33.3) | 5.89±0.29 | |
| Lymph node
metastasis | | | |
| No | 14 (27.5) | 2.08±0.14 | <0.0001 |
| Yes | 37 (72.5) | 4.72±0.26 | |
| TNM stage | | | |
| I | 12 (23.5) | 2.04±0.14 | <0.0001 |
| II | 18 (35.3) | 3.84±0.17 | |
| III | 21 (41.2) | 5.32±0.26 | |
Cell lines and culture
GC cell lines SGC7901, MKN45, MKN28, BGC823, AGS and
GC9811, and the immortalized normal gastric epithelial cell line
GES-1 were kindly donated by Professor Daiming Fan (The Fourth
Military Medical University) and NCI-N87 were purchased from
Shanghai Bioleaf Biotech Co., Ltd. (Shanghai, China). All cell
lines were maintained in our institute according to the recommended
protocols. Cells were cultured in RPMI-1640 medium (Invitrogen,
Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS;
Invitrogen) at 37°C in a 5% CO2 incubator (Thermo
Scitific Forma).
Real-time quantitative
reverse-transcription (qRT-PCR)
Total RNA was extracted from all the cell lines
using TRIzol (Invitrogen) according to the manufacturer's protocol.
RNA was stored at −80°C prior to qRT-PCR analysis. HDAC8 expression
was detected by PCR amplification of a 210-bp product with the
primer pair: forward, 5′-TGCAGCATCTCC AGAAGGTC-3′ and reverse,
5′-TCTTTGCATGATGCCC CCT-3′. PCR conditions were: 30 cycles and the
temperature 60°C. GAPDH was used as the internal control. PCR
products were separated on a 1.5% agarose gel stained with ethidium
bromide and visualized with UV.
RNA interference
siRNAs and negative control (Shanghai GenePharma
Co., Ltd., Shanghai, China) were used to downregulate HDAC8 in
BGC-823 and MKN28 cells using Lipofectamine 2000 (Life
Technologies) according to the manufacturer's instructions. siRNAs
sequences are as follows: siRNA-2 forward,
5′-UUGAGAUAACAAAAACCAGAU-3′ and reverse,
5′-CUGGUUUUUGUUAUCUCAAUG-3′; NC siRNA forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′.
Cells containing siRNA constructions were named
siHDAC8 and siCtrl. Cells containing plasmid-HDAC8 (Guangzhou
RiboBio Co., Ltd., Guangzhou, China) and the negative control (NC)
(Guangzhou RiboBio) plasmid were named as the cell line-HDAC8 and
the cell line-NC, respectively. Cells were plated into 6-well
plates and the siHDAC8 and siCtrl were transfected into cells using
transfection reagents when cells were 40% confluent. These cells
were used for western blot analysis, qRT-PCR and in vitro
experiments.
MTT assay
Cells were transfected with 100 nM miR-216b
inhibitor (Shanghai GenePharma), mimics (Guangzhou RiboBio), or 100
nM siRNA-HDAC8 (Guangzhou RiboBio) as previously described
(26), then seeded in 96-well
plates (2×103/well) 24 h later. Cell viability was
examined by the 3-2,5-diphenyl tetrazolium bromide (MTT) assay
(27–29) according to the manufacturer's
instructions (Sigma, St. Louis, MO, USA) at designated times.
Colony formation assay
BGC-823 and MKN28 cells with or without HDAC8
knockdown or forced expression were seeded into 100-mm dishes at a
density of 5,000 cells/well. After incubation for an additional 7
days, the cells were fixed with methanol for 15 min, and then
stained with 0.1% crystal violet for another 15 min, and colonies
of >50 cells were manually counted (30,31).
The experiments were performed in independent triplicates.
Cell cycle and apoptosis assay
To detect cell cycle and apoptosis alterations,
cells were grown in 6-well plates and treated with siHDAC8, siCtrl,
HDAC8 or NC for 48 h. For cell cycle analysis, cells were harvested
and fixed with ice-cold 75% ethyl alcohol at 4°C overnight and
incubated with DNA Prep kit (Beckman Coulter, Fullerton, CA, USA)
in the dark for 30–60 min. For apoptosis analysis, cells were
harvested, washed twice using phosphate-buffered saline, and fixed
in 70% ethanol at 4°C overnight. They were then incubated with
propidium iodide at room temperature for 1 h and analyzed by flow
cytometry using a FACScan flow cytometer (BD Biosciences, Mountain
View, CA, USA). All the results were from 3 independent
experiments.
Western blot analysis
Proteins from GC cell lines and paired fresh GC and
tumor-adjacent normal gastric tissues were extracted with RIPA
(Beyotime Institute of Biotechnology, Shanghai, China). The
following process was carried out as described (15,32).
The primary antibodies used for western blot analysis: HDAC8
(1:600, ab18968; Abcam), Bmf (1:500, ab9655; Abcam), caspase-3
(1:500, 710431; Thermo Fisher Scientific), caspase-6 (1:500,
MA5-11527; Thermo Fisher Scientific).
Immunohistochemistry
Paraffin sections, 4-μm, were baked for 2 h at 65°C
and deparaffinized. Antigen retrieval was performed using citrate
sodium buffer (pH 7.2) at 95°C for 15 min and then slides were
cooled at room temperature for 30 min. After being treated with 3%
hydrogen peroxide for 15 min to block the endogenous peroxidase,
the sections were treated with normal goat serum confining liquid
for 30 min to reduce non-specific binding and then rabbit
polyclonal anti-HDAC8 (1:200, ab18968; Abcam) was incubated the
sections for 12 h at 4°C. After re-warming for 1 h and washing 5
times, sections were incubated with secondary antibody for 30 min
at room temperature. Diaminobenzidine (DAB) was used for color
reactions. Subsequent immunohistochemical staining was scored as
previously described (33,34).
Statistical analysis
Data were expressed as means ± standard errors of
three independent experiments. For statistical tests, we used the
SPSS statistical software package, version 17.0 (SPSS, Inc.,
Chicago, IL, USA). The Student's t-test and the one-way analysis of
variance (ANOVA) were performed to analyze relative band densities
in western blotting and MTT optical density values. P-values
<0.05 were considered statistically significant.
Results
HDAC8 is upregulated in gastric cancer
cell lines
To further examine these findings, using western
blotting experiments, we found that HDAC8 protein expression was
also upregulated in 7 gastric cancer cell lines compared to GES-1
(P<0.0001), and was significantly higher in BGC823, AGS and
GC9811 cell lines than in NCI-N87, SGC7901, MKN45 and MKN28
(Fig. 1A). To further investigate
the expression pattern of HDAC8 in GC, we then performed qRT-PCR in
these gastric cancer cell lines and GES-1. We found that HDAC8 mRNA
expression was also significantly upregulated in all the gastric
cancer cell lines compared to GES-1 (P<0.0001). Moreover, HDAC8
mRNA expression was significantly higher in poorly-differentiated
cell lines BGC823, AGS and GC9811 (mean ± SEM, 8.43±0.9821),
compared to other moderately and well-differentiated cell lines,
SGC7901, MKN45, MKN28 and BGC823 (mean ± SEM, 3.5±0.5788)
(P=0.0114) (Fig. 1B).
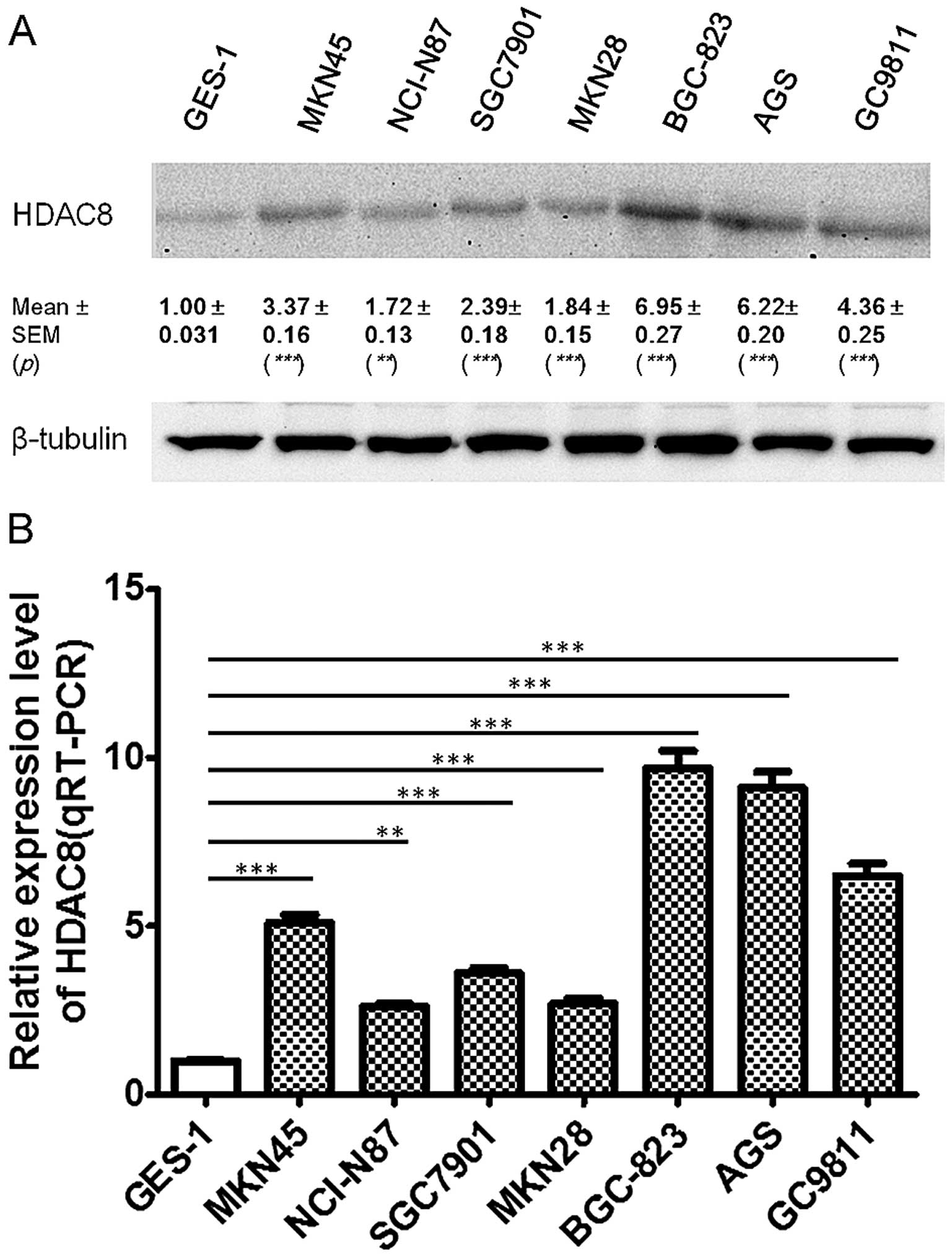 | Figure 1HDAC8 was upregulated in gastric
cancer cell lines. HDAC8 expression in seven gastric cancer cell
lines and the immortalized normal gastric epithelial cell line
GES-1, were quantified by western blotting (A, n=3, paired t-test,
***P<0.0001, **P<0.01) and quantitative
RT-PCR (qRT-PCR) (B, n=3, paired t-test, ***P<0.0001,
**P<0.001). Results in A and B are representative
findings from three or more independent experiments, and all the
values are shown as mean ± SEM, tubulin served as a loading
control. |
HDAC8 is upregulated in gastric cancer
tissues
To examine HDAC8 expression in GC tissues, 51 cancer
tissues, including 17 well, 17 moderately and 17 poorly
differentiated tissues and matched non-tumor tissues were used. By
qRT-PCR, we found that HDAC8 was significantly upregulated in 47
(92.2%) gastric cancer clinical tissues, compared with
non-cancerous tissues (Fig. 2A).
Then, HDAC8 expression was examined by immunohistochemistry and
western blotting, which indicated that 45 and 44 patients had
significantly upregulated HDAC8 expression, as detected by
immunohistochemistry (Fig. 2B) and
western blot analysis, respectively (Fig. 2C). To gain further insight into
this observation, we examined the relationship between HDAC8
expression and the clinical parameters of the patients, and found
that HDAC8 expression positively correlated with lymph node
metastasis, tumor size, TNM stage and negatively with the
histological differentiation (Table
I), but did not correlate with age or gender.
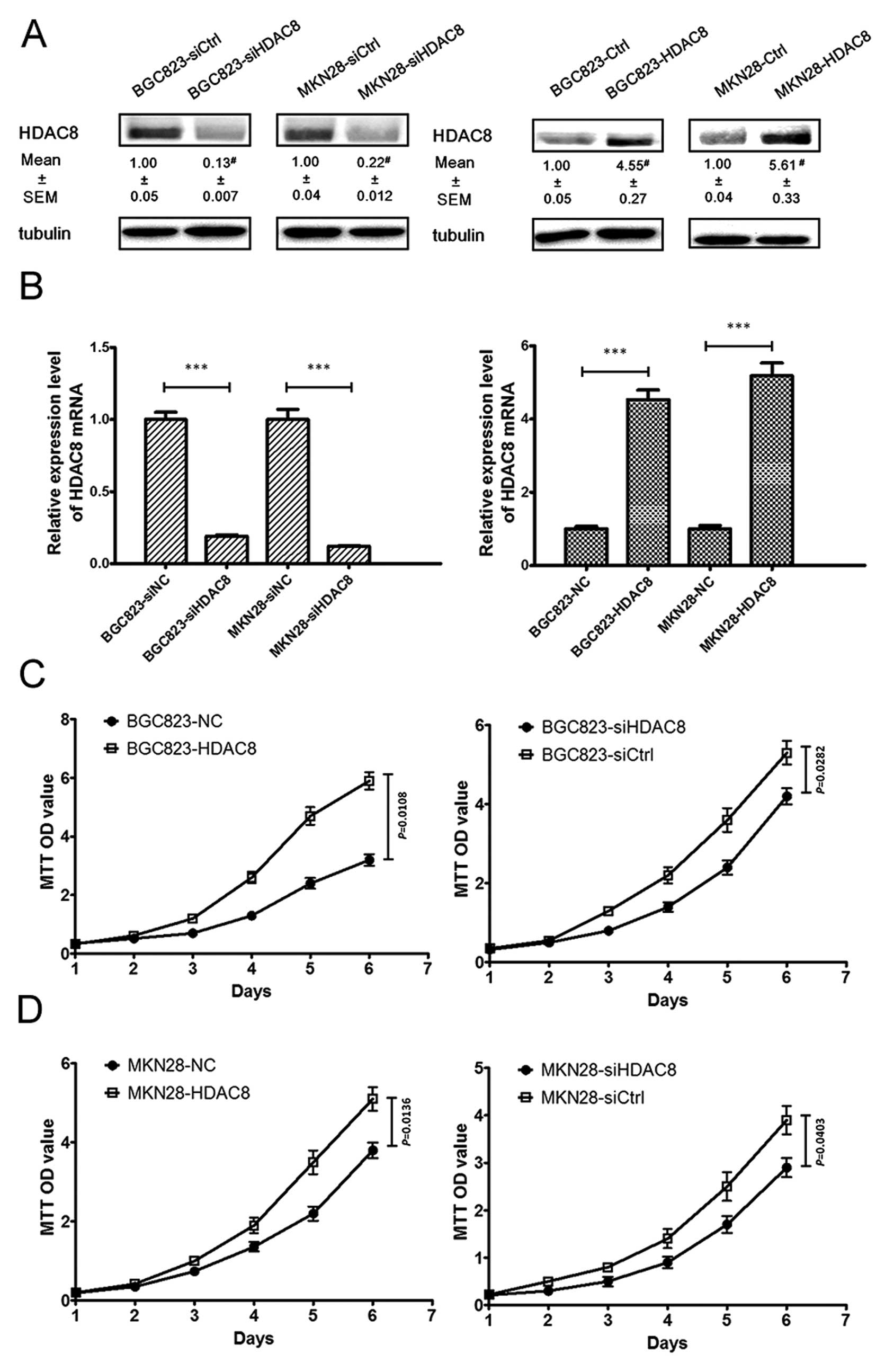
Depletion of HDAC8 inhibits proliferation
in human gastric cancer cells in vitro
To investigate the role of HDAC8 in the
proliferation of human gastric cancer cells, we used BGC823 and
MKN28 cells to knockdown and overexpress HDAC8. Using siHDAC8 or
pcDNA3.1(+)-HDAC8 transfection which was followed with antibiotic
selection to gain stable clones, we knocked down or upregulated
HDAC8 expression. The expression levels were examined via western
blotting (Fig. 3A) and qRT-PCR
(Fig. 3B). As shown in Fig. 3C and D, using MTT method, we found
that HDAC8 overexpression led to a significant increase in cell
proliferation, while HDAC8 knockdown led to a significant decrease
in cell proliferation. To further validate the function of HDAC8 in
the GC proliferation, we performed plate colony experiments to
assess the effect of depletion and upregulation of HDAC8 on the GC
cell proliferation. We found that results of the plate colony
experiments were in line with that of the MTT assay (Fig. 3E and F).
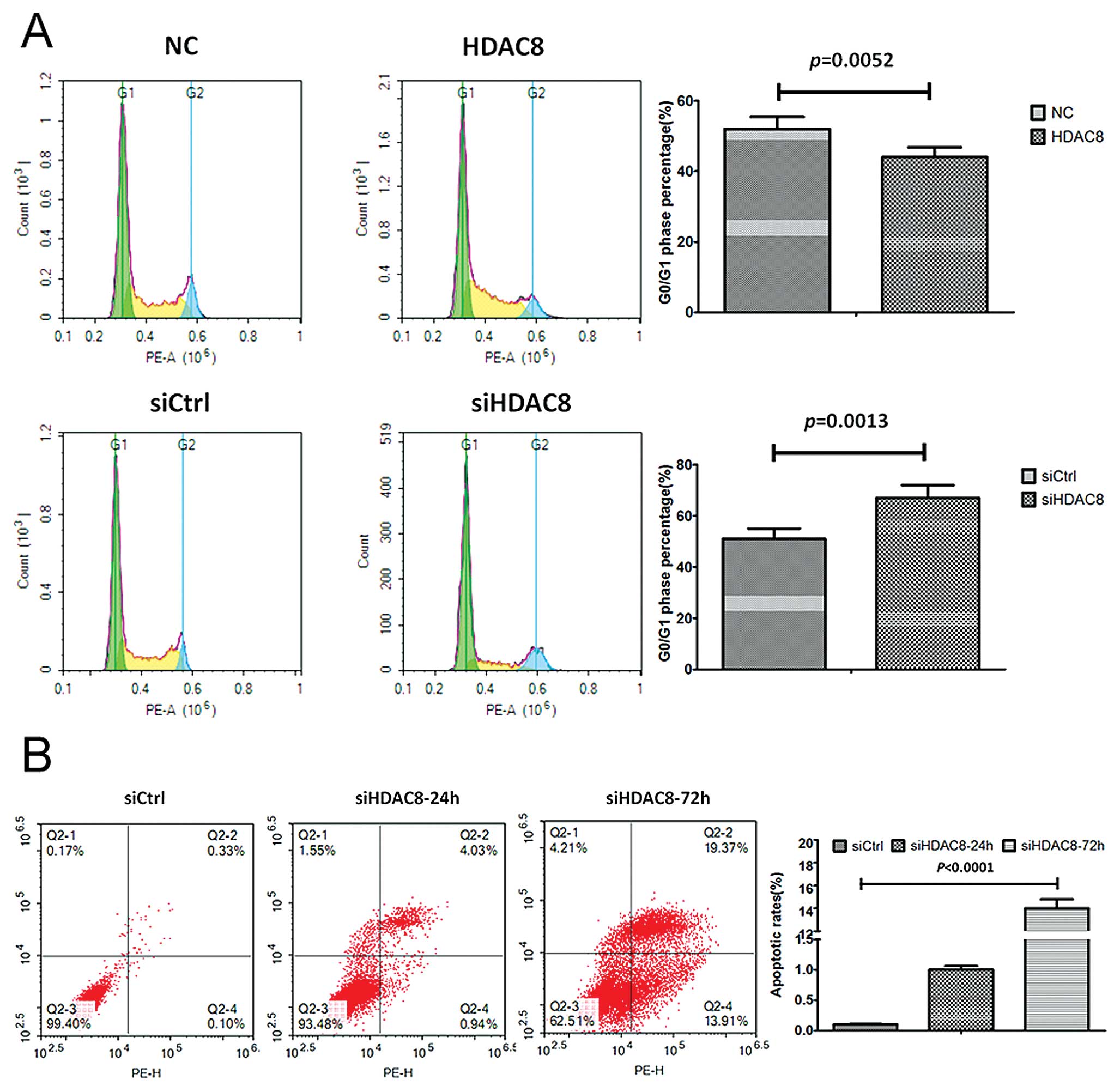
Knockdown of HDAC8 promoted G0/G1 arrest
and apoptosis of gastric cancer
To further study the mechanism by which HDAC8
knockdown or overexpression affected proliferation, cell cycle
progression and apoptosis were analyzed using flow cytometry. The
MKN28-siHDAC8 cells showed a delayed, while MKN28-HDAC8 exhibited a
shortened G1 phase compared with corresponding control groups
(Fig. 4A). Moreover, we also found
that the apoptotic rates increased significantly in the
MKN28-siHDAC8 group after siHDAC8 transfection was performed for 48
h, compared to the control group (Fig.
4B).
Suppression of the HDAC8-induced
upregulation of proteins involved Bmf-mediated apoptosis
Previous studies showed that the HDAC8 inhibitor MSP
triggers Bmf-mediated apoptosis independent of p21 induction via
inducing of pro-apoptotic BMF expression (24). To investigate the apoptotic
mechanism by which siHDAC8 induced apoptosis in more detail,
several factors that are pro-apoptotic or indicative apoptotic were
examined. In the present study, we first assessed Bmf, activated
caspase-3, and activated caspase-6 using western blotting and found
that Bmf, activated caspase-3, and activated caspase-6 were
significantly upregulated when HDAC8 was downregulated (Fig. 5A). The qRT-PCR results were in line
with that of western blotting (Fig.
5B).
Discussion
Epigenetics is the study of heritable alterations in
gene expression that are not accompanied by the corresponding
change in DNA sequence. There are three interlinked epigenetic
processes which regulate gene expression at the level of chromatin,
that is, DNA methylation, nucleosomal remodeling and histone
covalent modifications. Post-translational modifications that occur
on certain amino acid residues of the tails of histone proteins
modify the chromatin structure and form the basis for ‘histone
code’. The level of acetylation of histones and then the gene
expression is controlled by the enzymes histone acetyl transferase
(HAT) and histone deacetylase (HDAC). It was shown that the balance
between HAT and HDAC was altered in many cancers (35).
HDACs are widely involved in cellular processes,
ranging from cell differentiation to proliferation, senescence, and
apoptosis; in particular, protecting a telomerase activator from
ubiquitin-mediated degradation. Studies have shown that HDAC8 could
serve as the prognostic biomarker, promote tumorigenesis and
progress in multiple tumors. For example, Wilmott et al
(36) found that HDAC8 may be a
prognostic biomarker in melanoma, and also provide important data
regarding the regulation of HDACs in melanoma and a rational basis
for targeting them therapeutically; Wu et al (22) reported that HDAC8 was overexpressed
in HCC and HDAC8 knockdown could suppress tumor growth and enhance
apoptosis in HCC via elevating the expression of p53 and
acetylation of p53 at Lys382, indicating that HDAC8 might serve as
a potential therapeutic target in HCC (37); Oehme et al (16) found that the knockdown of HDAC8
resulted in the inhibition of proliferation, reduced clonogenic
growth, cell cycle arrest and differentiation in cultured
neuroblastoma cells; Balasubramanian et al (38) reported that HDAC8-selective
inhibitors had a unique mechanism of action involving PLCγ1
activation and calcium-induced apoptosis, and could offer benefits
including a greater therapeutic index for treating T-cell
malignancies. In the present study, we also found that depletion of
HDAC8 using small interference RNA promoted apoptosis and cell
cycle arrest in gastric cancer cells, moreover, forced expression
of HDAC8 inhibited cell apoptosis and promoted cell proliferation,
which suggested that HDAC8 may be a potential therapeutic target of
gastric cancer.
Bmf, Bcl-2 modifying factor, is the closest relative
of Bim (Bcl-2 interacting mediator of cell death) and functions as
a tumor suppressor. A number of groups have demonstrated that
overexpression of prosurvival Bcl-2 family members significantly
reduces HDACi-mediated tumor cell death and therapeutic efficacy in
preclinical models. In many cases, HDACi activate the intrinsic
pathway via upregulation of a number of proapoptotic BH3-only Bcl-2
family genes including Bim, Bid and Bmf (39). Loss of bmf has been shown to
accelerate the development of thymic lymphoma in a γ-irradiation
carcinogenesis protocol in mice (40); BMF gene silencing in HT29 cells
lead to a decrease in oxaliplatin-induced cell death (41); Graab et al (42) found that GLI1/2 inhibitor GANT61
and PI3K/mTOR inhibitor PI103 cotreatment could increase mRNA and
protein expression of NOXA and BMF, which is required for
apoptosis, since knockdown of NOXA or BMF significantly reduces
GANT61/PI103-induced apoptosis. It has been reported that HDAC8
mediated regulation of Bcl-2-modifying factor (BMF) via cooperation
with STAT3. Here, we reported that inhibition of HDAC8 led to
increased apoptosis rate of gastric cancer cells companied by the
enhanced expression of Bmf, activated caspase-3 and activated
caspase-6 (Fig. 6).
In summary, in the present study, we demonstrated
that HDAC8 was significantly upregulated in GC tissues and gastric
cancer cells, and inhibition of HDAC8 inhibited cell proliferation
and enhanced apoptosis. Moreover, we found that depletion of HDAC8
enhanced expression of Bmf, which is a tumor suppressor via
inducing apoptosis. Our novel evaluation of HDAC8 in gastric
adenocarcinoma may suggest new effective therapeutic strategies in
GC.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (no. 81301763 and no. 81572849)
and the Henan Provincial Key Scientific and Technological Projects
(no. 142102310473).
Abbreviations:
|
HDAC
|
histone deacetylase
|
|
GC
|
gastric cancer
|
|
siRNA
|
small interfere RNA
|
|
qRT-PCR
|
reverse transcription polymerase chain
reaction
|
References
|
1
|
Chen W, Zheng R, Zeng H, Zhang S and He J:
Annual report on status of cancer in China, 2011. Chin J Cancer
Res. 27:2–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Ruijter AJ, van Gennip AH, Caron HN,
Kemp S and van Kuilenburg AB: Histone deacetylases (HDACs):
Characterization of the classical HDAC family. Biochem J.
370:737–749. 2003. View Article : Google Scholar
|
|
4
|
Witt O, Deubzer HE, Milde T and Oehme I:
HDAC family: What are the cancer relevant targets? Cancer Lett.
277:8–21. 2009. View Article : Google Scholar
|
|
5
|
Hait NC, Avni D, Yamada A, Nagahashi M,
Aoyagi T, Aoki H, Dumur CI, Zelenko Z, Gallagher EJ, Leroith D, et
al: The phosphorylated prodrug FTY720 is a histone deacetylase
inhibitor that reactivates ERα expression and enhances hormonal
therapy for breast cancer. Oncogenesis. 4:e1562015. View Article : Google Scholar
|
|
6
|
Seicean A, Petrusel L, Seicean R, To C,
Seicean A and Street C: New targeted therapies in pancreatic
cancer. World J Gastroenterol. 21:6127–6145. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Eigl BJ, North S, Winquist E, Finch D,
Wood L, Sridhar SS, Powers J, Good J, Sharma M, Squire JA, et al: A
phase II study of the HDAC inhibitor SB939 in patients with
castration resistant prostate cancer: NCIC clinical trials group
study IND195. Invest New Drugs. 33:969–976. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang L, Liang Q, Shen K, Ma L, An N, Deng
W, Fei Z and Liu J: A novel class I histone deacetylase inhibitor,
I-7ab, induces apoptosis and arrests cell cycle progression in
human colorectal cancer cells. Biomed Pharmacother. 71:70–78. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bishayee K, Khuda-Bukhsh AR and Huh SO:
PLGA-loaded gold-nanoparticles precipitated with quercetin
downregulate HDAC-Akt activities controlling proliferation and
activate p53-ROS crosstalk to induce apoptosis in hepatocarcinoma
cells. Mol Cells. 38:518–527. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pai JT, Hsu CY, Hua KT, Yu SY, Huang CY,
Chen CN, Liao CH and Weng MS: NBM-T-BBX-OS01, semisynthesized from
osthole, induced G1 growth arrest through HDAC6 inhibition in lung
cancer cells. Molecules. 20:8000–8019. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bian J and Zhang L, Han Y, Wang C and
Zhang L: Histone deacetylase inhibitors: Potent anti-leukemic
agents. Curr Med Chem. 22:2065–2074. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu L, Yang J, Zhao L, Yu X, Wang L, Wang
F, Cai Y and Jin J: Expression of hMOF, but not HDAC4, is
responsible for the global histone H4K16 acetylation in gastric
carcinoma. Int J Oncol. 46:2535–2545. 2015.PubMed/NCBI
|
|
13
|
Dali-Youcef N, Froelich S, Moussallieh
F-M, Chibbaro S, Noël G, Namer IJ, Heikkinen S and Auwerx J: Gene
expression mapping of histone deacetylases and co-factors, and
correlation with survival time and 1H-HRMAS metabolomic profile in
human gliomas. Sci Rep. 5:90872015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khan MA, Hussain A, Sundaram MK, Alalami
U, Gunasekera D, Ramesh L, Hamza A and Quraishi U:
(−)-Epigallocatechin-3-gallate reverses the expression of various
tumor-suppressor genes by inhibiting DNA methyltransferases and
histone deacetylases in human cervical cancer cells. Oncol Rep.
33:1976–1984. 2015.PubMed/NCBI
|
|
15
|
Gregoretti IV, Lee YM and Goodson HV:
Molecular evolution of the histone deacetylase family: Functional
implications of phylogenetic analysis. J Mol Biol. 338:17–31. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Oehme I, Deubzer HE, Wegener D, Pickert D,
Linke JP, Hero B, Kopp-Schneider A, Westermann F, Ulrich SM, von
Deimling A, et al: Histone deacetylase 8 in neuroblastoma
tumorigenesis. Clin Cancer Res. 15:91–99. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Marks P, Rifkind RA, Richon VM, Breslow R,
Miller T and Kelly WK: Histone deacetylases and cancer: Causes and
therapies. Nat Rev Cancer. 1:194–202. 2001. View Article : Google Scholar
|
|
18
|
Pandey R, Müller A, Napoli CA, Selinger
DA, Pikaard CS, Richards EJ, Bender J, Mount DW and Jorgensen RA:
Analysis of histone acetyltransferase and histone deacetylase
families of Arabidopsis thaliana suggests functional
diversification of chromatin modification among multicellular
eukaryotes. Nucleic Acids Res. 30:5036–5055. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Falkenberg KJ and Johnstone RW: Histone
deacetylases and their inhibitors in cancer, neurological diseases
and immune disorders. Nat Rev Drug Discov. 13:673–691. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mottamal M, Zheng S, Huang TL and Wang G:
Histone deacetylase inhibitors in clinical studies as templates for
new anticancer agents. Molecules. 20:3898–3941. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chakrabarti A, Oehme I, Witt O, Oliveira
G, Sippl W, Romier C, Pierce RJ and Jung M: HDAC8: A multifaceted
target for therapeutic interventions. Trends Pharmacol Sci.
36:481–492. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu J, Du C, Lv Z, Ding C, Cheng J, Xie H,
Zhou L and Zheng S: The up-regulation of histone deacetylase 8
promotes proliferation and inhibits apoptosis in hepatocellular
carcinoma. Dig Dis Sci. 58:3545–3553. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vannini A, Volpari C, Filocamo G, Casavola
EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco
R, Gallinari P, et al: Crystal structure of a eukaryotic
zinc-dependent histone deacetylase, human HDAC8, complexed with a
hydroxamic acid inhibitor. Proc Natl Acad Sci USA. 101:15064–15069.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kang Y, Nian H, Rajendran P, Kim E,
Dashwood WM, Pinto JT, Boardman LA, Thibodeau SN, Limburg PJ, Löhr
CV, et al: HDAC8 and STAT3 repress BMF gene activity in colon
cancer cells. Cell Death Dis. 5:e14762014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nian H, Bisson WH, Dashwood WM, Pinto JT
and Dashwood RH: α-keto acid metabolites of organoselenium
compounds inhibit histone deacetylase activity in human colon
cancer cells. Carcinogenesis. 30:1416–1423. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sargent JM and Taylor CG: Appraisal of the
MTT assay as a rapid test of chemosensitivity in acute myeloid
leukaemia. Br J Cancer. 60:206–210. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
van de Loosdrecht AA, Beelen RH,
Ossenkoppele GJ, Broekhoven MG and Langenhuijsen MM: A
tetrazolium-based colorimetric MTT assay to quantitate human
monocyte mediated cytotoxicity against leukemic cells from cell
lines and patients with acute myeloid leukemia. J Immunol Methods.
174:311–320. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Twentyman PR and Luscombe M: A study of
some variables in a tetrazolium dye (MTT) based assay for cell
growth and chemo-sensitivity. Br J Cancer. 56:279–285. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tolosa L, Donato MT and Gómez-Lechón MJ:
General cytotoxicity assessment by means of the MTT assay. Methods
Mol Biol. 1250:333–348. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Y, Zhang D, Wu K, Zhao Q, Nie Y and
Fan D: Long noncoding RNA MRUL promotes ABCB1 expression in
multidrug-resistant gastric cancer cell sublines. Mol Cell Biol.
34:3182–3193. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guzmán C, Bagga M, Kaur A, Westermarck J
and Abankwa D: ColonyArea: An ImageJ plugin to automatically
quantify colony formation in clonogenic assays. PLoS One.
9:e924442014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Feng X, Wang Y, Ma Z, Yang R, Liang S,
Zhang M, Song S, Li S, Liu G, Fan D, et al: MicroRNA-645,
up-regulated in human adencarcinoma of gastric esophageal junction,
inhibits apoptosis by targeting tumor suppressor IFIT2. BMC Cancer.
14:6332014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kennecke H, Yerushalmi R, Woods R, Cheang
MC, Voduc D, Speers CH, Nielsen TO and Gelmon K: Metastatic
behavior of breast cancer subtypes. J Clin Oncol. 28:3271–3277.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lassmann S, Shen Y, Jütting U, Wiehle P,
Walch A, Gitsch G, Hasenburg A and Werner M: Predictive value of
Aurora-A/STK15 expression for late stage epithelial ovarian cancer
patients treated by adjuvant chemotherapy. Clin Cancer Res.
13:4083–4091. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lakshmaiah KC, Jacob LA, Aparna S,
Lokanatha D and Saldanha SC: Epigenetic therapy of cancer with
histone deacetylase inhibitors. J Cancer Res Ther. 10:469–478.
2014.PubMed/NCBI
|
|
36
|
Wilmott JS, Colebatch AJ, Kakavand H,
Shang P, Carlino MS, Thompson JF, Long GV, Scolyer RA and Hersey P:
Expression of the class 1 histone deacetylases HDAC8 and 3 are
associated with improved survival of patients with metastatic
melanoma. Mod Pathol. 28:884–894. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tian Y, To KF, Lai P, Cheung YS, Wong VWS,
Chan HLY and Cheng ASL: Histone deacetylase 8 is a novel chromatin
modulator in NAFLD-associated hepatocarcinogenesis. Clin
Gastroenterol Hepatol. 13:2192015. View Article : Google Scholar
|
|
38
|
Balasubramanian S, Ramos J, Luo W,
Sirisawad M, Verner E and Buggy JJ: A novel histone deacetylase 8
(HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell
lymphomas. Leukemia. 22:1026–1034. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Matthews GM, Newbold A and Johnstone RW:
Intrinsic and extrinsic apoptotic pathway signaling as determinants
of histone deacetylase inhibitor antitumor activity. Adv Cancer
Res. 116:165–197. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Labi V, Grespi F, Baumgartner F and
Villunger A: Targeting the Bcl-2-regulated apoptosis pathway by BH3
mimetics: A breakthrough in anticancer therapy? Cell Death Differ.
15:977–987. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ginés A, Bystrup S, Ruiz de Porras V,
Guardia C, Musulén E, Martínez-Cardús A, Manzano JL, Layos L, Abad
A and Martínez-Balibrea E: PKM2 subcellular localization is
involved in oxaliplatin resistance acquisition in HT29 human
colorectal cancer cell lines. PLoS One. 10:e01238302015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Graab U, Hahn H and Fulda S:
Identification of a novel synthetic lethality of combined
inhibition of hedgehog and PI3K signaling in rhabdomyosarcoma.
Oncotarget. 6:8722–8735. 2015. View Article : Google Scholar : PubMed/NCBI
|